Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation from Whole Frozen Bat Specimens
2.2. Anal Swab and Fecal Sample Collection
2.3. RNA Extraction and NGS Library Preparation of Museum Samples and Anal Swabs
2.4. RNA Extraction and NGS Library Preparation of Fecal Samples
2.5. Metagenomic Data Analysis
2.6. Recombination Analysis
2.7. Phylogenetic Analysis
2.8. DNA Extraction and 16S Library Preparation
2.9. 16S rRNA Reads Processing and Statistical Analysis
2.10. Geospatial Mapping
3. Results
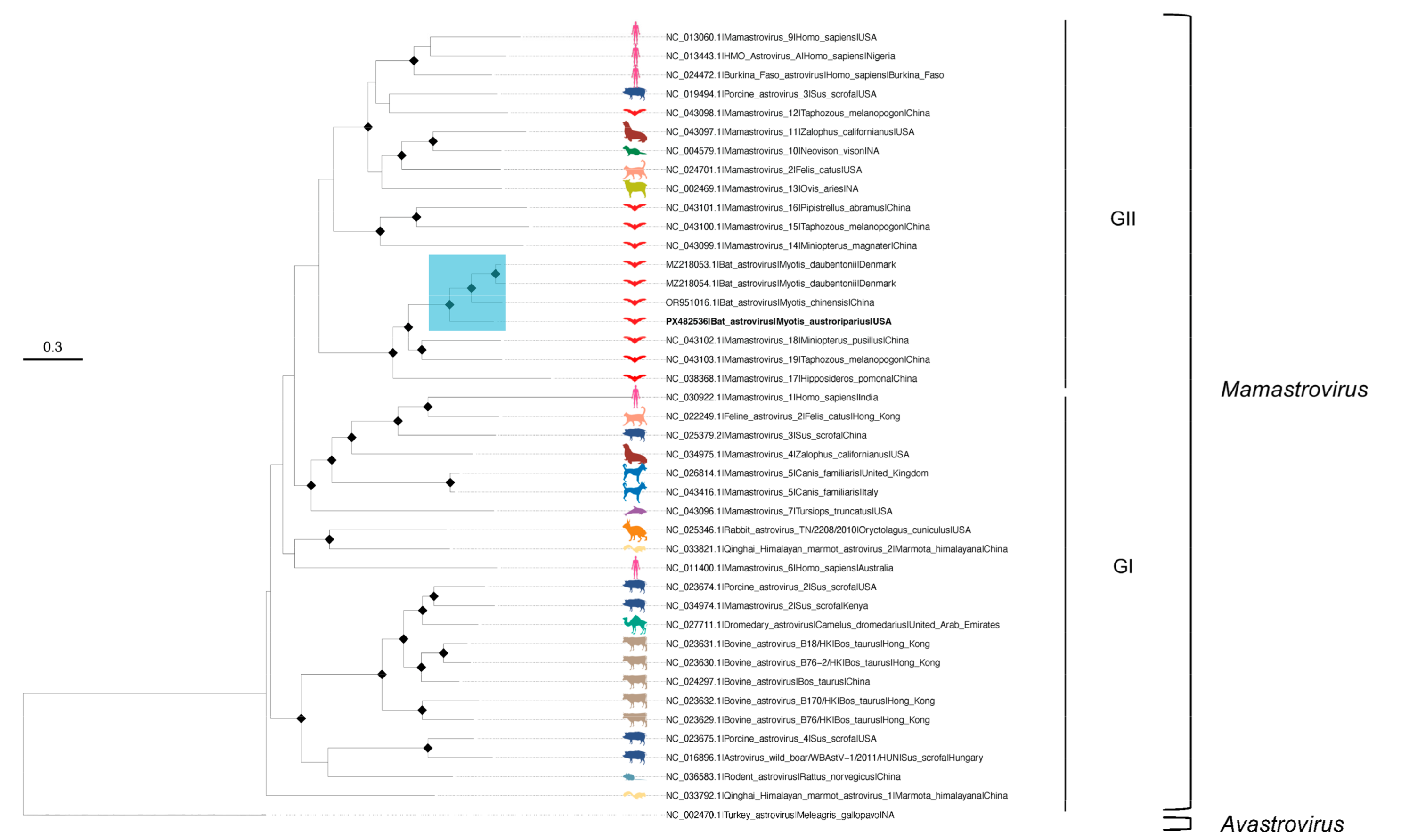
3.1. First Detection of Astrovirus Detection in Florida Bats
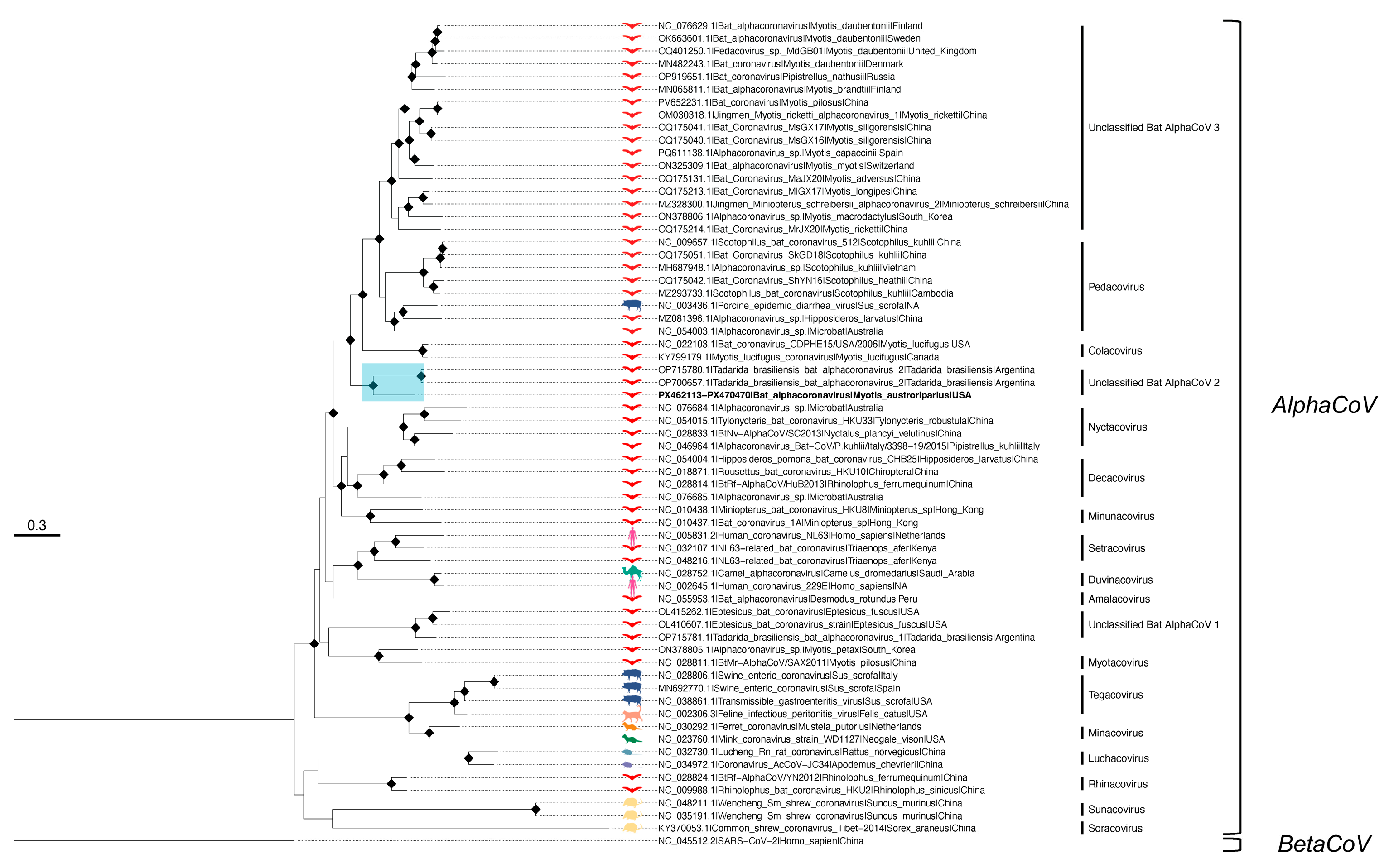
3.2. Co-Infection of an M. austroriparius with Astrovirus and a Bat Alphacoronavirus
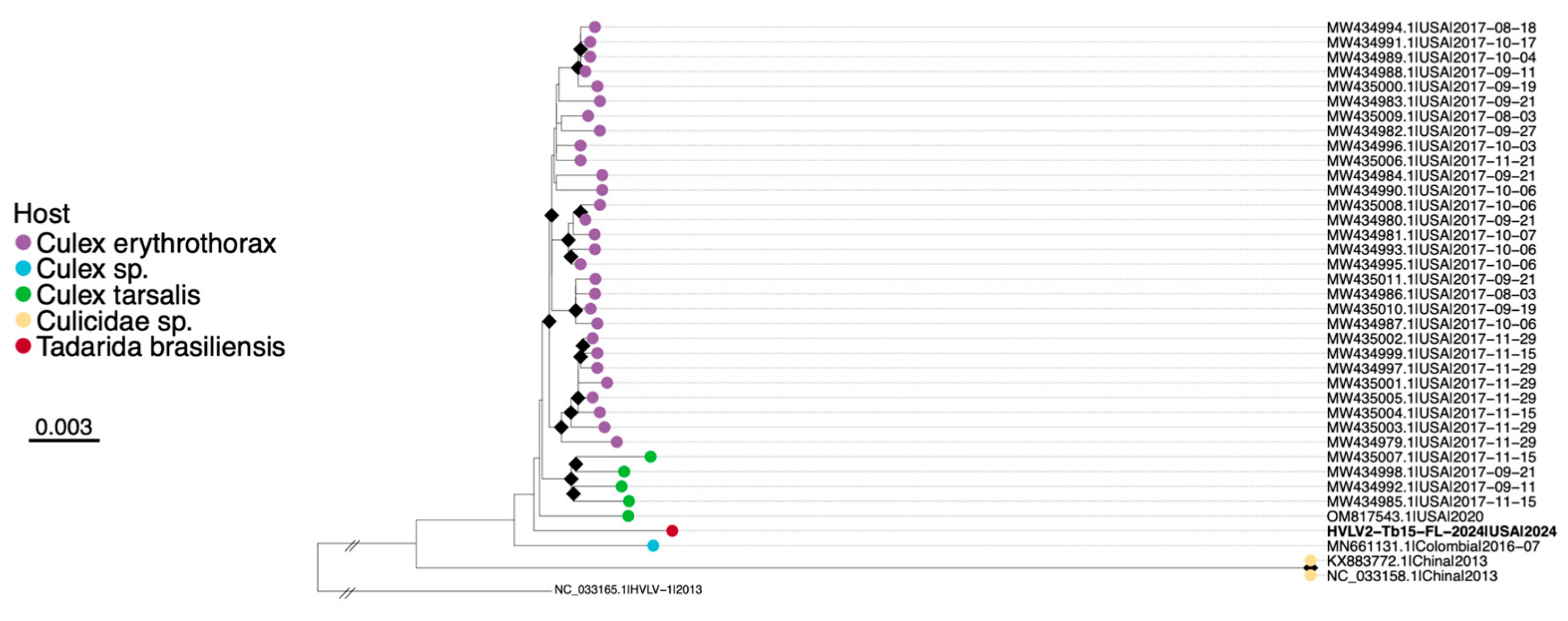
3.3. Detection of Hubei Virga-like Virus 2 from T. brasiliensis Feces
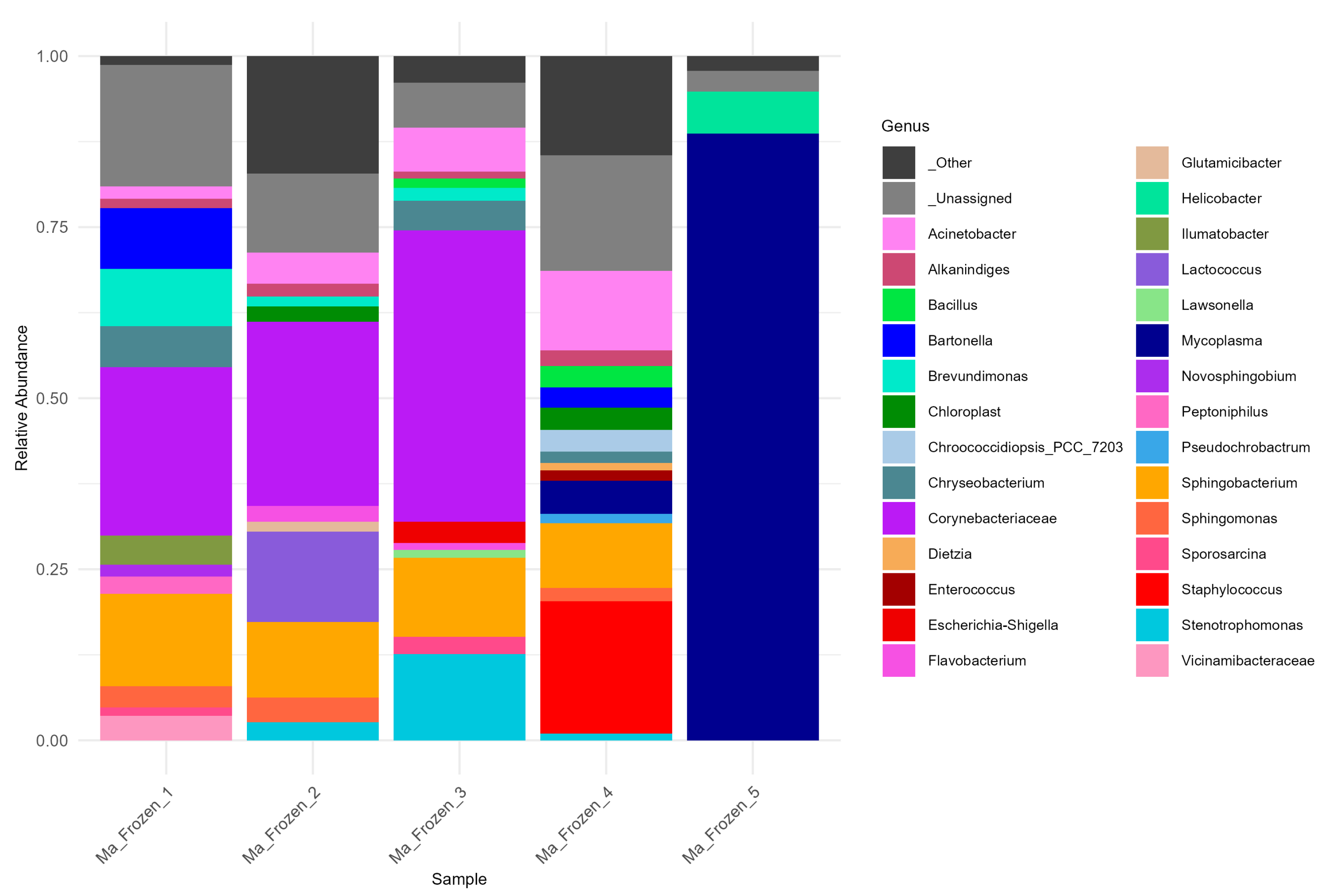
3.4. Microbiome Shifts in AlphaCoV and AstV Co-Infected Bat
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AlphaCoV | Alphacoronavirus |
| AstV | Astrovirus |
| HVLV2 | Hubei virga-like virus 2 |
| mNGS | Metagenomic next-generation sequencing |
| RdRp | RNA-dependent RNA polymerase |
| FLMNH | Florida Museum of Natural History |
| FWC | Florida Fish and Wildlife Conservation Commission |
| ICTV | International Committee on Taxonomy of Viruses |
| NGS | Next-generation sequencing |
| ACoV-2-Tb | Tadarida brasiliensis bat alphacoronavirus 2 |
References
- Brook, C.E.; Dobson, A.P. Bats as “special” Reservoirs for Emerging Zoonotic Pathogens. Trends Microbiol. 2015, 23, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Irving, A.T.; Ahn, M.; Goh, G.; Anderson, D.E.; Wang, L.-F. Lessons from the Host Defences of Bats, a Unique Viral Reservoir. Nature 2021, 589, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Imtiaz, M.A.; Islam, M.M.; Tanzin, A.Z.; Islam, A.; Hassan, M.M. Major Bat-Borne Zoonotic Viral Epidemics in Asia and Africa: A Systematic Review and Meta-Analysis. Vet. Med. Sci. 2022, 8, 1787–1801. [Google Scholar] [CrossRef]
- Sparrer, M.N.; Hodges, N.F.; Sherman, T.; VandeWoude, S.; Bosco-Lauth, A.M.; Mayo, C.E. Role of Spillover and Spillback in SARS-CoV-2 Transmission and the Importance of One Health in Understanding the Dynamics of the COVID-19 Pandemic. J. Clin. Microbiol. 2023, 61, e0161022. [Google Scholar] [CrossRef]
- Van Brussel, K.; Holmes, E.C. Zoonotic Disease and Virome Diversity in Bats. Curr. Opin. Virol. 2022, 52, 192–202. [Google Scholar] [CrossRef]
- Wang, J.; Pan, Y.-F.; Yang, L.-F.; Yang, W.-H.; Lv, K.; Luo, C.-M.; Wang, J.; Kuang, G.-P.; Wu, W.-C.; Gou, Q.-Y.; et al. Individual Bat Virome Analysis Reveals Co-Infection and Spillover among Bats and Virus Zoonotic Potential. Nat. Commun. 2023, 14, 4079. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, P.; Han, Y.; Zhao, W.; Zhao, L.; Li, R.; Zhang, J.; Zhang, S.; Lu, J.; Daszak, P.; et al. Unveiling Bat-Borne Viruses: A Comprehensive Classification and Analysis of Virome Evolution. Microbiome 2024, 12, 235. [Google Scholar] [CrossRef]
- Tan, C.W.; Yang, X.; Anderson, D.E.; Wang, L.-F. Bat Virome Research: The Past, the Present and the Future. Curr. Opin. Virol. 2021, 49, 68–80. [Google Scholar] [CrossRef]
- Bazzoni, E.; Cacciotto, C.; Zobba, R.; Pittau, M.; Martella, V.; Alberti, A. Bat Ecology and Microbiome of the Gut: A Narrative Review of Associated Potentials in Emerging and Zoonotic Diseases. Animals 2024, 14, 3043. [Google Scholar] [CrossRef]
- Dhivahar, J.; Parthasarathy, A.; Krishnan, K.; Kovi, B.S.; Pandian, G.N. Bat-Associated Microbes: Opportunities and Perils, an Overview. Heliyon 2023, 9, e22351. [Google Scholar] [CrossRef]
- Jones, D.N.; Ravelomanantsoa, N.A.F.; Yeoman, C.J.; Plowright, R.K.; Brook, C.E. Do Gastrointestinal Microbiomes Play a Role in Bats’ Unique Viral Hosting Capacity? Trends Microbiol. 2022, 30, 632–642. [Google Scholar] [CrossRef]
- Luo, J.; Liang, S.; Jin, F. Gut Microbiota in Antiviral Strategy from Bats to Humans: A Missing Link in COVID-19. Sci. China Life Sci. 2021, 64, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Popov, I.V.; Mazanko, M.S.; Kulaeva, E.D.; Golovin, S.N.; Malinovkin, A.V.; Aleshukina, I.S.; Aleshukina, A.V.; Prazdnova, E.V.; Tverdokhlebova, T.I.; Chikindas, M.L.; et al. Gut Microbiota of Bats: Pro-Mutagenic Properties and Possible Frontiers in Preventing Emerging Disease. Sci. Rep. 2021, 11, 21075. [Google Scholar] [CrossRef] [PubMed]
- Dimkić, I.; Fira, D.; Janakiev, T.; Kabić, J.; Stupar, M.; Nenadić, M.; Unković, N.; Grbić, M.L. The Microbiome of Bat Guano: For What Is This Knowledge Important? Appl. Microbiol. Biotechnol. 2021, 105, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Boyles, J.G.; Cryan, P.M.; McCracken, G.F.; Kunz, T.H. Conservation. Economic Importance of Bats in Agriculture. Science 2011, 332, 41–42. [Google Scholar] [CrossRef]
- Long, R.F.; Simpson, T.; Ding, T.-S.; Heydon, S.; Reil, W. Bats Feed on Crop Pests in Sacramento Valley. Calif. Agric. 1998, 52, 8–10. [Google Scholar] [CrossRef]
- Kunz, T.H.; Braun de Torrez, E.; Bauer, D.; Lobova, T.; Fleming, T.H. Ecosystem Services Provided by Bats. Ann. N. Y. Acad. Sci. 2011, 1223, 1–38. [Google Scholar] [CrossRef]
- Dai, W.; Leng, H.; Li, J.; Li, A.; Li, Z.; Zhu, Y.; Li, X.; Jin, L.; Sun, K.; Feng, J. The Role of Host Traits and Geography in Shaping the Gut Microbiome of Insectivorous Bats. mSphere 2024, 9, e0008724. [Google Scholar] [CrossRef]
- Mena Canata, D.A.; Benfato, M.S.; Pereira, F.D.; Ramos Pereira, M.J.; Hackenhaar, F.S.; Mann, M.B.; Frazzon, A.P.G.; Rampelotto, P.H. Comparative Analysis of the Gut Microbiota of Bat Species with Different Feeding Habits. Biology 2024, 13, 363. [Google Scholar] [CrossRef]
- Carrillo-Araujo, M.; Taş, N.; Alcántara-Hernández, R.J.; Gaona, O.; Schondube, J.E.; Medellín, R.A.; Jansson, J.K.; Falcón, L.I. Phyllostomid Bat Microbiome Composition Is Associated to Host Phylogeny and Feeding Strategies. Front. Microbiol. 2015, 6, 447. [Google Scholar] [CrossRef]
- Dietrich, M.; Kearney, T.; Seamark, E.C.J.; Paweska, J.T.; Markotter, W. Synchronized Shift of Oral, Faecal and Urinary Microbiotas in Bats and Natural Infection Dynamics during Seasonal Reproduction. R. Soc. Open Sci. 2018, 5, 180041. [Google Scholar] [CrossRef]
- Federici, L.; Masulli, M.; De Laurenzi, V.; Allocati, N. An Overview of Bats Microbiota and Its Implication in Transmissible Diseases. Front. Microbiol. 2022, 13, 1012189. [Google Scholar] [CrossRef]
- Lebeuf-Taylor, E.; Cosby, A.; Webber, Q.; Cottenie, K. Social Structuring of the Gut Microbiome in Communally Roosting Bats. PLoS ONE 2025, 20, e0325710. [Google Scholar] [CrossRef]
- Lutz, H.L.; Jackson, E.W.; Webala, P.W.; Babyesiza, W.S.; Kerbis Peterhans, J.C.; Demos, T.C.; Patterson, B.D.; Gilbert, J.A. Ecology and Host Identity Outweigh Evolutionary History in Shaping the Bat Microbiome. mSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Xiao, G.; Liu, S.; Xiao, Y.; Zhu, Y.; Zhao, H.; Li, A.; Li, Z.; Feng, J. Seasonal Changes in Gut Microbiota Diversity and Composition in the Greater Horseshoe Bat. Front. Microbiol. 2019, 10, 2247. [Google Scholar] [CrossRef] [PubMed]
- Bassi, C.; Guerriero, P.; Pierantoni, M.; Callegari, E.; Sabbioni, S. Novel Virus Identification through Metagenomics: A Systematic Review. Life 2022, 12, 2048. [Google Scholar] [CrossRef]
- Buddle, S.; Forrest, L.; Akinsuyi, N.; Martin Bernal, L.M.; Brooks, T.; Venturini, C.; Miller, C.; Brown, J.R.; Storey, N.; Atkinson, L.; et al. Evaluating Metagenomics and Targeted Approaches for Diagnosis and Surveillance of Viruses. Genome Med. 2024, 16, 111. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Li, Y.; Yang, X.; Zhang, H.; Zhou, P.; Zhang, Y.; Shi, Z. Metagenomic Analysis of Viruses from Bat Fecal Samples Reveals Many Novel Viruses in Insectivorous Bats in China. J. Virol. 2012, 86, 4620–4630. [Google Scholar] [CrossRef]
- Mishra, N.; Fagbo, S.F.; Alagaili, A.N.; Nitido, A.; Williams, S.H.; Ng, J.; Lee, B.; Durosinlorun, A.; Garcia, J.A.; Jain, K.; et al. A Viral Metagenomic Survey Identifies Known and Novel Mammalian Viruses in Bats from Saudi Arabia. PLoS ONE 2019, 14, e0214227. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, Z.; Liu, Y.; Ke, C.; Feng, J.; He, B.; Jiang, T. The Links between Dietary Diversity and RNA Virus Diversity Harbored by the Great Evening Bat (Ia Io). Microbiome 2024, 12, 246. [Google Scholar] [CrossRef]
- Li, L.; Victoria, J.G.; Wang, C.; Jones, M.; Fellers, G.M.; Kunz, T.H.; Delwart, E. Bat Guano Virome: Predominance of Dietary Viruses from Insects and Plants plus Novel Mammalian Viruses. J. Virol. 2010, 84, 6955–6965. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Aguilar, I.; Lorenzo, C.; Santos-Moreno, A.; Naranjo, E.J.; Navarrete-Gutiérrez, D. Coronaviruses in Bats: A Review for the Americas. Viruses 2021, 13, 1226. [Google Scholar] [CrossRef] [PubMed]
- Kotwa, J.D.; Bhuinya, A.; Yim, W.; Gallo, G.; Singh, P.; Liu, Q.; Kumar, A.; Chien, E.; Hou, A.; Chee, H.-Y.; et al. High Host Specificity of Alphacoronaviruses in Nearctic, Insectivorous Bats. NPJ Viruses 2025, 3, 38. [Google Scholar] [CrossRef]
- Warmuth, V.M.; Metzler, D.; Zamora-Gutierrez, V. Human Disturbance Increases Coronavirus Prevalence in Bats. Sci. Adv. 2023, 9, eadd0688. [Google Scholar] [CrossRef]
- Bonny, T.S.; Driver, J.P.; Paisie, T.; Salemi, M.; Morris, J.G.; Shender, L.A.; Smith, L.; Enloe, C.; Oxenrider, K.; Gore, J.A.; et al. Detection of Alphacoronavirus vRNA in the Feces of Brazilian Free-Tailed Bats (Tadarida brasiliensis) from a Colony in Florida, USA. Diseases 2017, 5, 7. [Google Scholar] [CrossRef]
- Leong, R.; Hoarau, A.O.G.; Carcauzon, V.; Köster, M.; Dietrich, M.; Tortosa, P.; Lebarbenchon, C. High Astrovirus Diversity in an Endemic Bat Species Suggests Multiple Spillovers from Synanthropic Rodents and Birds. J. Virol. 2025, 99, e0135724. [Google Scholar] [CrossRef]
- Li, Y.; Altan, E.; Reyes, G.; Halstead, B.; Deng, X.; Delwart, E. Virome of Bat Guano from Nine Northern California Roosts. J. Virol. 2021, 95. [Google Scholar] [CrossRef]
- Marks, C.S.; Marks, G.E. Bats of Florida; University Press of Florida: Gainesville, FL, USA, 2006; ISBN 978-0-8130-2985-6. [Google Scholar]
- Hughes, M.J.; Braun de Torrez, E.C.; Ober, H.K. Big Bats Binge Bad Bugs: Variation in Crop Pest Consumption by Common Bat Species. Agric. Ecosyst. Environ. 2021, 314, 107414. [Google Scholar] [CrossRef]
- Hughes, M.J.; Braun de Torrez, E.C.; Buckner, E.A.; Ober, H.K. Consumption of Endemic Arbovirus Mosquito Vectors by Bats in the Southeastern United States. J. Vector Ecol. 2022, 47, 153–165. [Google Scholar] [CrossRef]
- Webb, E.N.; Ober, H.K.; Braun de Torrez, E.C. Moth Munchers: Limited Intraspecific Variation in Summer Diet of the Endangered Florida Bonneted Bat (Eumops floridanus). J. Mammal. 2025, 106, 1139–1150. [Google Scholar] [CrossRef]
- Sikes, R. Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 97, 663–688. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Eden, J.-S. RAPIDprep: A Simple, Fast Protocol for RNA Metagenomic Sequencing of Clinical Samples, V2; Protocols.io: Berkeley, CA, USA, 2022. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-Scale Protein Function Classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Blum, M.; Andreeva, A.; Florentino, L.C.; Chuguransky, S.R.; Grego, T.; Hobbs, E.; Pinto, B.L.; Orr, A.; Paysan-Lafosse, T.; Ponamareva, I.; et al. InterPro: The Protein Sequence Classification Resource in 2025. Nucleic Acids Res. 2025, 53, D444–D456. [Google Scholar] [CrossRef]
- Brister, J.R.; Ako-Adjei, D.; Bao, Y.; Blinkova, O. NCBI Viral Genomes Resource. Nucleic Acids Res. 2015, 43, D571–D577. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A Computer Program for Analyzing Recombination in, and Removing Signals of Recombination from, Nucleotide Sequence Datasets. Virus Evol. 2021, 7, veaa087. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Yu, G. Using Ggtree to Visualize Data on Tree-Like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef]
- Strimmer, K.; von Haeseler, A. Likelihood-Mapping: A Simple Method to Visualize Phylogenetic Content of a Sequence Alignment. Proc. Natl. Acad. Sci. USA 1997, 94, 6815–6819. [Google Scholar] [CrossRef]
- Mavian, C.; Marini, S.; Prosperi, M.; Salemi, M. A Snapshot of SARS-CoV-2 Genome Availability up to April 2020 and Its Implications: Data Analysis. JMIR Public. Health Surveill. 2020, 6, e19170. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of General 16S Ribosomal RNA Gene PCR Primers for Classical and Next-Generation Sequencing-Based Diversity Studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Glöckner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 Years of Serving the Community with Ribosomal RNA Gene Reference Databases and Tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- An Ordination of the Upland Forest Communities of Southern Wisconsin—Bray—1957—Ecological Monographs—Wiley Online Library. Available online: https://esajournals.onlinelibrary.wiley.com/doi/10.2307/1942268 (accessed on 11 September 2025).
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative Beta Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- McKight, P.E.; Najab, J. Kruskal-Wallis Test. In The Corsini Encyclopedia of Psychology; Weiner, I.B., Craighead, W.E., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010; ISBN 978-0-470-47921-6. [Google Scholar]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley Statsref: Statistics Reference Online; Balakrishnan, N., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; John Wiley & Sons, Ltd: Chichester, UK, 2014; pp. 1–15. ISBN 978-1-118-44511-2. [Google Scholar]
- Simmonds, P.; Adriaenssens, E.M.; Lefkowitz, E.J.; Oksanen, H.M.; Siddell, S.G.; Zerbini, F.M.; Alfenas-Zerbini, P.; Aylward, F.O.; Dempsey, D.M.; Dutilh, B.E.; et al. Changes to Virus Taxonomy and the ICTV Statutes Ratified by the International Committee on Taxonomy of Viruses (2024). Arch. Virol. 2024, 169, 236. [Google Scholar] [CrossRef]
- Donato, C.; Vijaykrishna, D. The Broad Host Range and Genetic Diversity of Mammalian and Avian Astroviruses. Viruses 2017, 9, 102. [Google Scholar] [CrossRef]
- King, A.M.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses, 1st ed.; Elsevier: London, UK, 2011; ISBN 978-0-12-384684-6. [Google Scholar]
- Cerri, A.; Bolatti, E.M.; Zorec, T.M.; Montani, M.E.; Rimondi, A.; Hosnjak, L.; Casal, P.E.; Di Domenica, V.; Barquez, R.M.; Poljak, M.; et al. Identification and Characterization of Novel Alphacoronaviruses in Tadarida brasiliensis (Chiroptera, Molossidae) from Argentina: Insights into Recombination as a Mechanism Favoring Bat Coronavirus Cross-Species Transmission. Microbiol. Spectr. 2023, 11, e0204723. [Google Scholar] [CrossRef]
- Ober, H.K.; Mazzotti, F.J. Conservation of Bats in Florida. EDIS 2024, 2008. [Google Scholar] [CrossRef]
- Perry, M.; Rogers, L.; Wilder, K. Florida Fastest-Growing State for First Time Since 1957. Available online: https://www.census.gov/library/stories/2022/12/florida-fastest-growing-state.html (accessed on 7 July 2025).
- Eby, P.; Peel, A.J.; Hoegh, A.; Madden, W.; Giles, J.R.; Hudson, P.J.; Plowright, R.K. Pathogen Spillover Driven by Rapid Changes in Bat Ecology. Nature 2023, 613, 340–344. [Google Scholar] [CrossRef] [PubMed]
- Plowright, R.K.; Eby, P.; Hudson, P.J.; Smith, I.L.; Westcott, D.; Bryden, W.L.; Middleton, D.; Reid, P.A.; McFarlane, R.A.; Martin, G.; et al. Ecological Dynamics of Emerging Bat Virus Spillover. Proc. Biol. Sci. 2015, 282, 20142124. [Google Scholar] [CrossRef] [PubMed]
- Gore, J.A.; Studenroth, K.R. Status and Management of Bats Roosting in Bridges in Florida; Florida Department of Transportation: Tallahassee, FL, USA, 2005. [Google Scholar]
- Li, H.; Wilkins, K.T. Selection of Building Roosts by Mexican Free-Tailed Bats (Tadarida brasiliensis) in an Urban Area. Acta Chiropterologica 2015, 17, 321–330. [Google Scholar] [CrossRef]
- Rice, D.W. Life History and Ecology of Myotis Austroriparius in Florida. J. Mammal. 1957, 38, 15. [Google Scholar] [CrossRef]
- Smith, L.M.; Doonan, T.J.; Gore, J.A. Bats Roost in Culverts during Hibernation and Maternity Season in North Florida. J. N. Am. Bat Res. 2024, 2, 1–18. [Google Scholar]
- Seltmann, A.; Corman, V.M.; Rasche, A.; Drosten, C.; Czirják, G.Á.; Bernard, H.; Struebig, M.J.; Voigt, C.C. Seasonal Fluctuations of Astrovirus, but Not Coronavirus Shedding in Bats Inhabiting Human-Modified Tropical Forests. Ecohealth 2017, 14, 272–284. [Google Scholar] [CrossRef]
- Lazov, C.M.; Belsham, G.J.; Bøtner, A.; Rasmussen, T.B. Full-Genome Sequences of Alphacoronaviruses and Astroviruses from Myotis and Pipistrelle Bats in Denmark. Viruses 2021, 13, 1073. [Google Scholar] [CrossRef]
- Niu, Y.; McKee, C.D. Bat Viral Shedding: A Review of Seasonal Patterns and Risk Factors. Vector Borne Zoonotic Dis. 2025, 25, 229–239. [Google Scholar] [CrossRef]
- Figueiroa, T.; Galvão Bueno, M.; Bento Moura, P.E.; de Oliveira, M.B.; Passos Cordeiro, J.L.; Santos-Cavalcante, N.; Camacho Antevere Mazzarotto, G.A.; Wallau, G.L.; Corrêa da Silva Junior, L.; Resende, P.C.; et al. Alpha and Betacoronavirus Detection in Neotropical Bats from Northeast Brazil Suggests Wide Geographical Distribution and Persistence in Natural Populations. Animals 2025, 15, 332. [Google Scholar] [CrossRef]
- Moreira-Soto, A.; Taylor-Castillo, L.; Vargas-Vargas, N.; Rodríguez-Herrera, B.; Jiménez, C.; Corrales-Aguilar, E. Neotropical Bats from Costa Rica Harbour Diverse Coronaviruses. Zoonoses Public Health 2015, 62, 501–505. [Google Scholar] [CrossRef] [PubMed]
- Bittar, C.; Machado, R.R.G.; Comelis, M.T.; Bueno, L.M.; Beguelini, M.R.; Morielle-Versute, E.; Nogueira, M.L.; Rahal, P. Alphacoronavirus Detection in Lungs, Liver, and Intestines of Bats from Brazil. Microb. Ecol. 2020, 79, 203–212. [Google Scholar] [CrossRef] [PubMed]
- De Benedictis, P.; Schultz-Cherry, S.; Burnham, A.; Cattoli, G. Astrovirus Infections in Humans and Animals—Molecular Biology, Genetic Diversity, and Interspecies Transmissions. Infect. Genet. Evol. 2011, 11, 1529–1544. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.S.; Jeggo, M. Reservoirs and Vectors of Emerging Viruses. Curr. Opin. Virol. 2013, 3, 170–179. [Google Scholar] [CrossRef]
- Guito, J.C.; Prescott, J.B.; Arnold, C.E.; Amman, B.R.; Schuh, A.J.; Spengler, J.R.; Sealy, T.K.; Harmon, J.R.; Coleman-McCray, J.D.; Kulcsar, K.A.; et al. Asymptomatic Infection of Marburg Virus Reservoir Bats Is Explained by a Strategy of Immunoprotective Disease Tolerance. Curr. Biol. 2021, 31, 257–270.e5. [Google Scholar] [CrossRef]
- Banerjee, A.; Kulcsar, K.; Misra, V.; Frieman, M.; Mossman, K. Bats and Coronaviruses. Viruses 2019, 11, 41. [Google Scholar] [CrossRef]
- Seltmann, A.; Czirják, G.Á.; Courtiol, A.; Bernard, H.; Struebig, M.J.; Voigt, C.C. Habitat Disturbance Results in Chronic Stress and Impaired Health Status in Forest-Dwelling Paleotropical Bats. Conserv. Physiol. 2017, 5, cox020. [Google Scholar] [CrossRef]
- Fleischer, R.; Jones, C.; Ledezma-Campos, P.; Czirják, G.Á.; Sommer, S.; Gillespie, T.R.; Vicente-Santos, A. Gut Microbial Shifts in Vampire Bats Linked to Immunity Due to Changed Diet in Human Disturbed Landscapes. Sci. Total Environ. 2024, 907, 167815. [Google Scholar] [CrossRef]
- Cravens, Z.M.; Boyles, J.G. Illuminating the Physiological Implications of Artificial Light on an Insectivorous Bat Community. Oecologia 2019, 189, 69–77. [Google Scholar] [CrossRef]
- Ramadan, Y.N.; Alqifari, S.F.; Alshehri, K.; Alhowiti, A.; Mirghani, H.; Alrasheed, T.; Aljohani, F.; Alghamdi, A.; Hetta, H.F. Microbiome Gut-Brain-Axis: Impact on Brain Development and Mental Health. Mol. Neurobiol. 2025, 62, 10813–10833. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, Y.; Gu, T.; Zhou, J.; Chen, F.; Li, S. Investigating the Gut Bacteria Structure and Function of Hibernating Bats through 16S rRNA High-Throughput Sequencing and Culturomics. mSystems 2025, 10, e0146324. [Google Scholar] [CrossRef] [PubMed]
- Corduneanu, A.; Wu-Chuang, A.; Maitre, A.; Obregon, D.; Sándor, A.D.; Cabezas-Cruz, A. Structural Differences in the Gut Microbiome of Bats Using Terrestrial vs. Aquatic Feeding Resources. BMC Microbiol. 2023, 23, 93. [Google Scholar] [CrossRef] [PubMed]
- De Leon, M.P.; Montecillo, A.D.; Pinili, D.S.; Siringan, M.A.T.; Park, D.-S. Bacterial Diversity of Bat Guano from Cabalyorisa Cave, Mabini, Pangasinan, Philippines: A First Report on the Metagenome of Philippine Bat Guano. PLoS ONE 2018, 13, e0200095. [Google Scholar] [CrossRef] [PubMed]
- Heard, D.J.; De Young, J.L.; Goodyear, B.; Ellis, G.A. Comparative Rectal Bacterial Flora of Four Species of Flying Fox (Pteropus sp.). J. Zoo. Wildl. Med. 1997, 28, 471–475. [Google Scholar]
- Selvin, J.; Lanong, S.; Syiem, D.; De Mandal, S.; Kayang, H.; Kumar, N.S.; Kiran, G.S. Culture-Dependent and Metagenomic Analysis of Lesser Horseshoe Bats’ Gut Microbiome Revealing Unique Bacterial Diversity and Signatures of Potential Human Pathogens. Microb. Pathog. 2019, 137, 103675. [Google Scholar] [CrossRef]
- Li, A.; Li, Z.; Dai, W.; Parise, K.L.; Leng, H.; Jin, L.; Liu, S.; Sun, K.; Hoyt, J.R.; Feng, J. Bacterial Community Dynamics on Bats and the Implications for Pathogen Resistance. Environ. Microbiol. 2022, 24, 1484–1498. [Google Scholar] [CrossRef]
- Li, Z.; Li, A.; Dai, W.; Leng, H.; Liu, S.; Jin, L.; Sun, K.; Feng, J. Skin Microbiota Variation among Bat Species in China and Their Potential Defense against Pathogens. Front. Microbiol. 2022, 13, 808788. [Google Scholar] [CrossRef]
- Banskar, S.; Mourya, D.T.; Shouche, Y.S. Bacterial Diversity Indicates Dietary Overlap among Bats of Different Feeding Habits. Microbiol. Res. 2016, 182, 99–108. [Google Scholar] [CrossRef]
- Darby, E.M.; Moran, R.A.; Holden, E.; Morris, T.; Harrison, F.; Clough, B.; McInnes, R.S.; Schneider, L.; Frickel, E.M.; Webber, M.A.; et al. Differential Development of Antibiotic Resistance and Virulence between Acinetobacter Species. mSphere 2024, 9, e0010924. [Google Scholar] [CrossRef]
- Regalado, N.G.; Martin, G.; Antony, S.J. Acinetobacter lwoffii: Bacteremia Associated with Acute Gastroenteritis. Travel. Med. Infect. Dis. 2009, 7, 316–317. [Google Scholar] [CrossRef]
- Rosa, R.; Mills, J.; Munoz-Price, L.S. Clinical and Microbiological Characteristics of Acinetobacter lwoffii Bacteremia Compared with Acinetobacter baumannii. Open Forum Infect. Dis. 2015, 2, 1780. [Google Scholar] [CrossRef]
- Brila, I.; Lavrinienko, A.; Tukalenko, E.; Kallio, E.R.; Mappes, T.; Watts, P.C. Idiosyncratic Effects of Coinfection on the Association between Systemic Pathogens and the Gut Microbiota of a Wild Rodent, the Bank Vole Myodes Glareolus. J. Anim. Ecol. 2023, 92, 826–837. [Google Scholar] [CrossRef] [PubMed]
- Cortez, V.; Margolis, E.; Schultz-Cherry, S. Astrovirus and the Microbiome. Curr. Opin. Virol. 2019, 37, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, A.O.G.; Mavingui, P.; Lebarbenchon, C. Coinfections in Wildlife: Focus on a Neglected Aspect of Infectious Disease Epidemiology. PLoS Pathog. 2020, 16, e1008790. [Google Scholar] [CrossRef] [PubMed]
- Schmid, D.W.; Risely, A. Co-Infections Mask Pathogen-Specific Associations with the Gut Microbiota in Wild Voles. J. Anim. Ecol. 2023, 92, 790–793. [Google Scholar] [CrossRef]
- Hetta, H.F.; Ramadan, Y.N.; Alharbi, A.A.; Alsharef, S.; Alkindy, T.T.; Alkhamali, A.; Albalawi, A.S.; El Amin, H. Gut Microbiome as a Target of Intervention in Inflammatory Bowel Disease Pathogenesis and Therapy. Immuno 2024, 4, 400–425. [Google Scholar] [CrossRef]
- Johnson, C.; Hargest, V.; Cortez, V.; Meliopoulos, V.A.; Schultz-Cherry, S. Astrovirus Pathogenesis. Viruses 2017, 9, 22. [Google Scholar] [CrossRef]
- Wasimuddin; Brändel, S.D.; Tschapka, M.; Page, R.; Rasche, A.; Corman, V.M.; Drosten, C.; Sommer, S. Astrovirus Infections Induce Age-Dependent Dysbiosis in Gut Microbiomes of Bats. ISME J. 2018, 12, 2883–2893. [Google Scholar] [CrossRef]
- Wray, A.K.; Jusino, M.A.; Banik, M.T.; Palmer, J.M.; Kaarakka, H.; White, J.P.; Lindner, D.L.; Gratton, C.; Peery, M.Z. Incidence and Taxonomic Richness of Mosquitoes in the Diets of Little Brown and Big Brown Bats. J. Mammal. 2018, 99, 668–674. [Google Scholar] [CrossRef]
- Kondo, H.; Chiba, S.; Maruyama, K.; Andika, I.B.; Suzuki, N. A Novel Insect-Infecting Virga/Nege-like Virus Group and Its Pervasive Endogenization into Insect Genomes. Virus Res. 2019, 262, 37–47. [Google Scholar] [CrossRef]
- Konstantinidis, K.; Dovrolis, N.; Kouvela, A.; Kassela, K.; Rosa Freitas, M.G.; Nearchou, A.; de Courcy Williams, M.; Veletza, S.; Karakasiliotis, I. Defining Virus-Carrier Networks That Shape the Composition of the Mosquito Core Virome of a Local Ecosystem. Virus Evol. 2022, 8, veac036. [Google Scholar] [CrossRef]
- Liu, Q.; Cui, F.; Liu, X.; Fu, Y.; Fang, W.; Kang, X.; Lu, H.; Li, S.; Liu, B.; Guo, W.; et al. Association of Virome Dynamics with Mosquito Species and Environmental Factors. Microbiome 2023, 11, 101. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Shi, M.; Holmes, E.C. Using Metagenomics to Characterize an Expanding Virosphere. Cell 2018, 172, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Esterly, A.T.; Alemayehu, D.; Rusmisel, B.; Busam, J.; Shelton, T.L.; Sebay, T.; Zahiri, N.; Huston, J.W.; Clausnitzer, R.J.; Haas-Stapleton, E.J. Culex Erythrothorax (Diptera: Culicidae): Activity Periods, Insecticide Susceptibility and Control in California (USA). PLoS ONE 2020, 15, e0228835. [Google Scholar] [CrossRef] [PubMed]
- Gorris, M.E.; Bartlow, A.W.; Temple, S.D.; Romero-Alvarez, D.; Shutt, D.P.; Fair, J.M.; Kaufeld, K.A.; Del Valle, S.Y.; Manore, C.A. Updated Distribution Maps of Predominant Culex Mosquitoes across the Americas. Parasit. Vectors 2021, 14, 547. [Google Scholar] [CrossRef] [PubMed]
- Madhav, M.; Blasdell, K.R.; Trewin, B.; Paradkar, P.N.; López-Denman, A.J. Culex-Transmitted Diseases: Mechanisms, Impact, and Future Control Strategies Using Wolbachia. Viruses 2024, 16, 1134. [Google Scholar] [CrossRef]
- Puig-Montserrat, X.; Flaquer, C.; Gómez-Aguilera, N.; Burgas, A.; Mas, M.; Tuneu, C.; Marquès, E.; López-Baucells, A. Bats Actively Prey on Mosquitoes and Other Deleterious Insects in Rice Paddies: Potential Impact on Human Health and Agriculture. Pest. Manag. Sci. 2020, 76, 3759–3769. [Google Scholar] [CrossRef]
- Moser, S.K.; Barnard, M.; Frantz, R.M.; Spencer, J.A.; Rodarte, K.A.; Crooker, I.K.; Bartlow, A.W.; Romero-Severson, E.; Manore, C.A. Scoping Review of Culex Mosquito Life History Trait Heterogeneity in Response to Temperature. Parasit. Vectors 2023, 16, 200. [Google Scholar] [CrossRef]
- Day, J.F. The Florida SLE Mosquito, Culex (Culex) Nigripalpus Theobald (Insecta: Diptera: Culicidae). EDIS 1997, 1997. [Google Scholar] [CrossRef]
- Tavares, Y.; Bauer, A.; Reeves, L.; Campbell, L. Fact Sheet: West Nile Virus. EDIS 2022, 2022. [Google Scholar] [CrossRef]
- Falvo, C.A.; Crowley, D.E.; Benson, E.; Hall, M.N.; Schwarz, B.; Bohrnsen, E.; Ruiz-Aravena, M.; Hebner, M.; Ma, W.; Schountz, T.; et al. Diet-Induced Changes in Metabolism Influence Immune Response and Viral Shedding in Jamaican Fruit Bats. Proc. Biol. Sci. 2025, 292, 20242482. [Google Scholar] [CrossRef]
- Wetzler, G.; Boyles, J. The Energetics of Mosquito Feeding by Insectivorous Bats. Can. J. Zool. 2017, 96, 373–377. [Google Scholar] [CrossRef]
- Perea, S.; Meinecke, C.D.; Larsen-Gray, A.L.; Greene, D.U.; Villari, C.; Gandhi, K.J.K.; Castleberry, S.B. Winter Diet of Bats in Working Forests of the Southeastern U.S. Coastal Plain. Sci. Rep. 2024, 14, 12778, Erratum in Sci. Rep. 2024, 14, 14313. [Google Scholar] [CrossRef] [PubMed]
- Bourgarel, M.; Noël, V.; Pfukenyi, D.; Michaux, J.; André, A.; Becquart, P.; Cerqueira, F.; Barrachina, C.; Boué, V.; Talignani, L.; et al. Next-Generation Sequencing on Insectivorous Bat Guano: An Accurate Tool to Identify Arthropod Viruses of Potential Agricultural Concern. Viruses 2019, 11, 1102. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.W.; Phelps, K.L.; Allard, M.W.; Cook, J.A.; Dunnum, J.L.; Ferguson, A.W.; Gelang, M.; Khan, F.A.A.; Paul, D.L.; Reeder, D.M.; et al. Preserve a Voucher Specimen! The Critical Need for Integrating Natural History Collections in Infectious Disease Studies. MBio 2021, 12. [Google Scholar] [CrossRef] [PubMed]
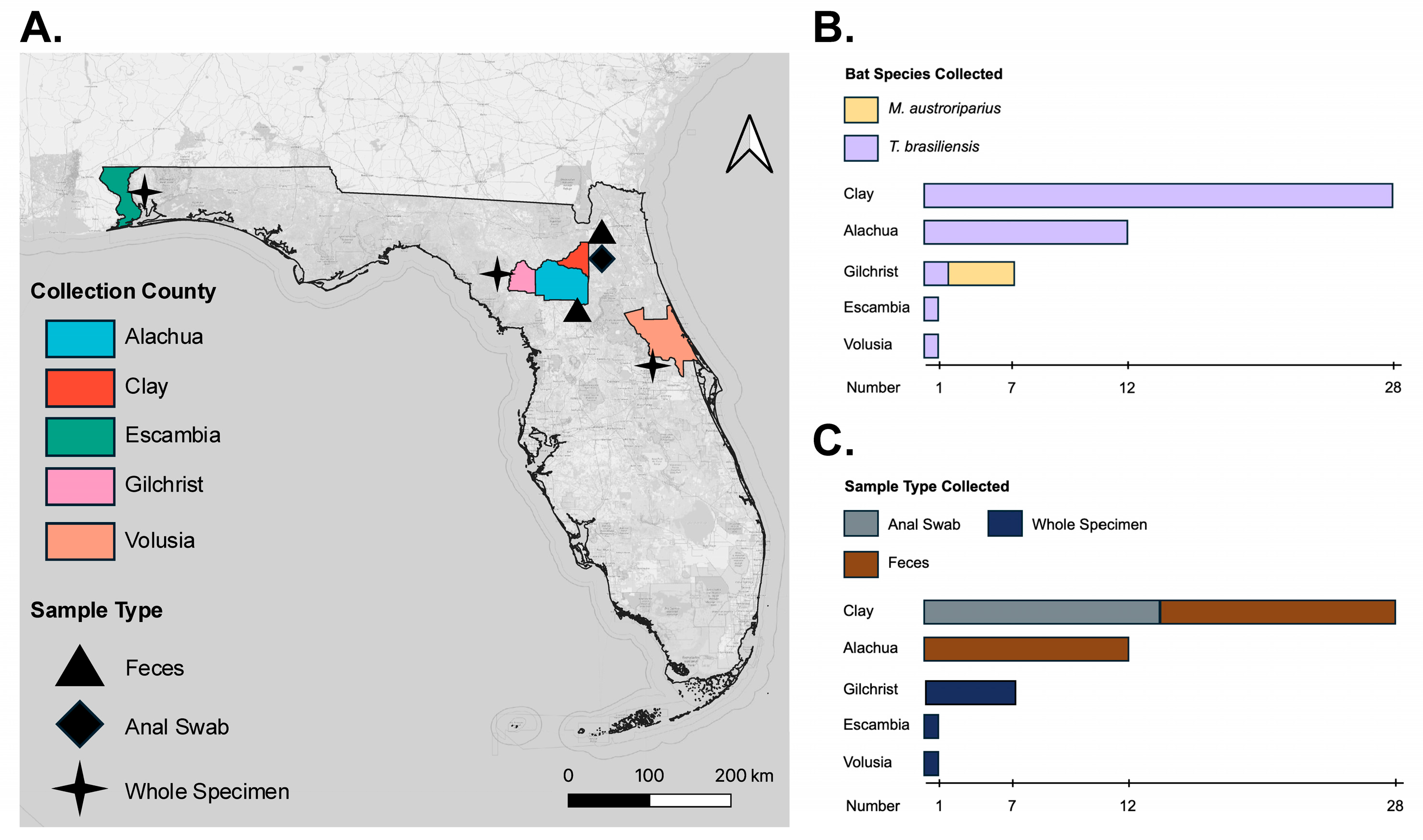
| Sample Type | Bat Species (N) | Collection Year | Collection County |
|---|---|---|---|
| Feces | T. brasiliensis (12) | 2024 | Alachua |
| T. brasiliensis (14) | 2022 | Clay | |
| Anal Swab | T. brasiliensis (14) | 2022 | Clay |
| Museum Specimen | M. austroriparius (5) | 2021 | Gilchrist |
| T. brasiliensis (5) | 2021 | Escambia, Gilchrist, Volusia |
| Virus | Bat Species (Bat ID) | Contig (# bp) | Mean Coverage Depth | BLAST Best Match (Accession) | Percent Identity | Genome Region |
|---|---|---|---|---|---|---|
| AlphaCoV | M. austroriparius (Ma_Frozen_3) | k119_6311 (925) | 21 | ACoV-2-Tb (OP700657.1) | 81 | 5′, ORF1a |
| k119_670 (5309) | 178 | ACoV-2-Tb (OP700657.1) | 81 | S, ORF3, E, M, N, ORF7, ORF8, 3′ | ||
| M. austroriparius (Ma_Frozen_4) | k119_1119 (1327) | 12 | ACoV-2-Tb (OP700657.1) | 84 | N, ORF7, ORF8, 3′ | |
| AstV | M. austroriparius (Ma_Frozen_4) | k119_11258 (3292) | 209 | Bat AstV (MZ218054.1) | 80 | ORF1b, ORF2 |
| HVLV2 | T. brasiliensis (Tb_Guano_15) | k141_4344 (9829) | 194 | HVLV2 (MW434995.1) | 99 | Polyprotein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paoli, J.E.; Thongthum, T.; Bassett, M.; Beardsley, J.; Tagliamonte, M.S.; Cash, M.N.; Spertus Newman, J.; Smith, L.M.; Anderson, B.D.; Salemi, M.; et al. Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts. Microorganisms 2025, 13, 2625. https://doi.org/10.3390/microorganisms13112625
Paoli JE, Thongthum T, Bassett M, Beardsley J, Tagliamonte MS, Cash MN, Spertus Newman J, Smith LM, Anderson BD, Salemi M, et al. Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts. Microorganisms. 2025; 13(11):2625. https://doi.org/10.3390/microorganisms13112625
Chicago/Turabian StylePaoli, Julia E., Thanaporn Thongthum, Maclean Bassett, Jakob Beardsley, Massimiliano S. Tagliamonte, Melanie N. Cash, Jason Spertus Newman, Lisa M. Smith, Benjamin D. Anderson, Marco Salemi, and et al. 2025. "Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts" Microorganisms 13, no. 11: 2625. https://doi.org/10.3390/microorganisms13112625
APA StylePaoli, J. E., Thongthum, T., Bassett, M., Beardsley, J., Tagliamonte, M. S., Cash, M. N., Spertus Newman, J., Smith, L. M., Anderson, B. D., Salemi, M., Subramaniam, K., von Fricken, M. E., Braun de Torrez, E., Mathis, V., & Mavian, C. N. (2025). Virome and Microbiome of Florida Bats Illuminate Viral Co-Infections, Dietary Viral Signals, and Gut Microbiome Shifts. Microorganisms, 13(11), 2625. https://doi.org/10.3390/microorganisms13112625







