Abstract
Bacillus safensis strains have emerged as versatile microbial platforms for bioproduction, combining the benefits of probiotic utility and biocontrol. In this study, we describe the isolation and in-depth characterization of a previously unreported B. safensis strain, LG01. The genome of this strain comprises a circular chromosome encoding 13 secondary metabolite biosynthetic gene clusters, 144 carbohydrate-active enzymes, 2 antibiotic resistance loci, and 1 prophage region, indicative of strong antimicrobial and metabolic capacity. Its protein secretion systems support nutrient acquisition, colonization, quorum sensing, and antibiotic synthesis. Our phenotypic assays confirmed the antifungal and antibacterial activity, proteolytic and cellulolytic functions, and robust biofilm formation of the strain. By performing a comparative genomic analysis, we identified 78 strain-specific genes enriched in the bacteriocin immunity and sporulation pathways. Signals of positive selection in the membrane and transcriptional regulator genes further reflect the adaptive evolution underlying the strain’s ecological fitness. Together, these findings advance our understanding of the genomic features of B. safensis LG01 and highlight its promise as a candidate for biocontrol and probiotic applications.
1. Background
Bacillus species are genetically diverse and can thrive in various environments [1]. Bacillus safensis is a Gram-positive bacterium first isolated and reported in 2006. The DNA sequence similarity of its gyrB gene was only 54–66% and 91.2% when compared to previously known species, leading to its ultimate classification as B. safensis [2]. This strain has many desirable properties and wide application prospects; for example, genomic characterization of B. safensis RP10, which was isolated from Atacama Desert soil, revealed its adaptations to high heavy metal concentrations, saline conditions, and limited carbon/energy sources. These findings suggest its potential for bioremediation of metal-contaminated environments and enhancing plant stress resilience [3]. These findings suggest its potential for bioremediation of metal-contaminated environments and enhancing plant stress resilience. When sent to the International Space Station, contrary to other strains, B. safensis JPL-MERTA-8-2 showed a 60% improvement in growth compared to on Earth [4], reflecting its strong stress resistance. B. safensis DVL-43 produces a novel and organic solvent-stable lipase [5], while B. safensis CFA-06 can produce catalases and oxidoreductases to degrade aromatic compounds and petroleum aromatic fractions [6], reflecting its strong degradation effect on oil. A mutant B. safensis LAU13 isolated with ultraviolet radiation was found to secrete highly active keratinase to degrade chicken feathers without damaging the skin [7]. B. safensis, as a rhizobacteria, can reduce abiotic stresses and thus promote plant growth [8], which would be extremely useful tools in sustainable agriculture. An RNase produced by B. safensis RB-5 showed antiproliferative activities towards many human transformed cells, and widely retain normal cells [9], surfactin produced by B. safensis F4 showed an antitumor activity against T47D breast cancer cells and B16F10 mouse melanoma cells [10], reflecting its potential to be developed into new antitumoral drugs. B. safensis S7, and B. safensis N4 have a strong metabolic effect on heavy metal ions in soil and thus provided a basic reference for remediation of heavy metal contaminated soil by bacteria. Furthermore, B. safensis also possess strong antimicrobial ability. B. safensis strains demonstrate significant potential as biocontrol agents and sources of next-generation biopesticides. Strain STJP (NAIMCC-B-02323) exhibits strong antagonism against the phytopathogen Alternaria alternata [11], while strain B21 produces the antifungal compounds iturin A2 and iturin A6, which impair hyphal growth in Magnaporthe oryzae by altering membrane permeability [12]. Additionally, strain C3 shows antibacterial activity against Escherichia coli and Pseudomonas syringae [13], and strain F4 displays broad-spectrum efficacy against several pathogenic strains [10]. B. safensis therefore represents a promising biological alternative to synthetic pesticides for sustainable plant disease management.
In this study, we analyzed a novel B. safensis strain, LG01, for its potential probiotic and biocontrol properties. Our genomic analysis identified the biosynthetic gene clusters for antimicrobial compounds, with experimental validation confirming both functional traits. Additionally, comparative genomic studies and whole-genome sequencing were conducted to determine which of the four subclusters the orthologous and specific genes of B. safensis belonged to. This work sheds light on the evolutionary processes of B. safensis isolates and provides a solid foundation for utilizing LG01 as a new biocontrol and probiotic agent, contributing to the current limited literature on this strain.
2. Materials and Methods
2.1. Bacterial Strain and Cultivation Conditions
Strains were identified through polyphasic characterization, combining cultural, morphological, and molecular analyses. The complete genome sequence has been deposited in GenBank under accession number CP109651.1. For routine cultivation, strains were grown in LB medium at 37 °C for 20 h under microaerophilic conditions. Long-term storage was maintained at −80 °C.
2.2. DNA Extraction, Sequencing, and Genome Assembly
Genomic DNA extraction, sequencing and assembly were performed according to established protocols [14]. Genomic DNA was extracted following the manufacturer’s standard protocol for Oxford Nanopore Technologies sequencing platforms (Oxford Nanopore Technologies, Oxford, UK) [15]. DNA quality was assessed using a Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and NanoDrop system (ThermoFisher Scientific, Wilmington, DE, USA) following manufacturer-recommended protocols. Next, we utilized the standard protocols for the Illumina second-generation high-throughput sequencing platform (Illumina, San Diego, CA, USA) to sequence the total DNA of the samples and used Mecat2 to optimize the original data, obtaining the more accurate third-generation sequencing data. To assemble the 395,699 filtered subsequences with an average length of 2105.4 bp, the SMRT Link v5.1.0 software with default parameters was used for pure third-generation sequencing data, while the Unicycle v0.9.4 software was used for data consisting of data from both second- and third-generation sequencing [16]. Lastly, in order to attain a more accurate genome, we compared the original data and the assembled genome for further correction of errors using Arrow v19.0.0 software [17]. Plasmid sequences were identified through a two-step computational process: unmapped reads were first extracted from whole-genome alignment data using SAMTOOLS v1.19.2, followed by de novo assembly of these reads with SPAdes v4.1.0 software [18].
2.3. Functional Annotation
To delineate the functional repertoire of the genome, a systematic functional annotation was conducted [14]. Gene prediction was performed using Glimmer3 v3.02 or Prodigal v2.6.3 for genomes with GC content exceeding 70%, while tRNA identification was carried out with tRNAscan-SE v1.3.1 [19]; and using the rRNAmmer v1.2 was used for identifying the rRNAs [20]. sRNA predictions were first aligned against the Rfam database and then filtered using the cmsearch program. Genomic islands were predicted using IslandPath-DIMOB [21], CRISPR elements with CRISPRdigger [22] and prophages with PHAST [23]. Gene annotation of LG01 was performed through sequence homology analysis using BLAST v2.17.0 and DIAMOND v2.1.9 against multiple databases: NCBI Nr, Swiss-Prot, COG, KEGG, GO, Pfam, CAZy, PHI, VFDB, and CARD. Gene Ontology functional classification was performed using Blast2GO v2.5 [24]. Protein family (Pfam) database annotation based on the Pfam was applicable with a hmmer [25]. In addition, the superior annotation was annotated on the basis of the following databases: Carbohydrate-Active enZYmes Database (CAZyme Database), Pathogen Host Interactions Database (PHI Database), Virulence Factors of Pathogenic Bacteria Database (VFDB Database), Comprehensive Antibiotic Resistance Database (CARD Database). To identify secreted proteins containing signal peptides at the N-terminus rather than transmembrane helices, SignalP, LipoP, TMHMM PSORTb and other software were used [26]. The Circos v0.66 software was used for visual display to obtain the whole genome cycle map [27]. AntiSMASH v5.0.0 was used to predict the gene clusters of secondary metabolites [28]. Phylogenetic analysis was conducted with MEGA X and iTOL using the complete genome sequences of B. safensis strains, with all analyses performed under default parameters [29,30].
2.4. Antimicrobial Activity and Probiotic Properties
Antibacterial activity was determined using an agar well diffusion method according to established protocols, while probiotic properties were evaluated [31]. A modified agar well diffusion assay was employed to evaluate protease and cellulase production. Proteolysis was assessed on skim milk agar, while cellulose degradation was determined on CMC-Na plates followed by Congo red staining. Biofilm formation was quantified via crystal violet staining. We inoculated 200 µL of an overnight culture into a 96-well plate and removed planktonic cells with PBS washing. The adherent biofilms were then fixed with methanol, stained with crystal violet, and solubilized in glacial acetic acid for absorbance measurement at 595 nm, with a blank medium serving as the negative control. The antibiotic susceptibility of B. safensis was assessed against ten different antibiotics using a standard disk diffusion assay. Briefly, bacterial suspensions were spread on antibiotic-free LB agar plates (100 μL per plate). Three antibiotic-impregnated disks and three sterile filter disks (as controls) were evenly spaced on each plate. After 24 h of incubation at 37 °C, the diameters of the inhibition zones were measured. PathogenFinder v1.1 was used to predict the human pathogenicity of B. safensis LG01 from its whole-genome sequence.
2.5. Comparative Genomic Analysis
Comparative genomics was applied to delineate the pan-genome structure and identify lineage-specific genes [14]. DNA alignments and phylogenetic trees were created using the gyrB genes from 13 B. safensis strains that underwent complete genome sequencing [32]. The phylogenetic tree was reconstructed using the Maximum Likelihood method, with the strains divided into four classes for comparative analysis. The pan-genome analysis in this study was based on strains selected from the NCBI database. Sequence comparisons were conducted using a 95% megablast identity threshold with standard parameters. For synteny visualization, TBtools v1.09854 was employed with its default settings [33]. Orthologous clusters were delineated across the four strain classes through OrthoVenn2 analysis, applying classification thresholds of 1 × 10−2 for E-value and 1.5 for inflation value [34]. All-against-all protein alignments were performed with DIAMOND v0.9.24 using a BLASTX E-value cutoff of 10−3 to refine orthologous relationships. Functional characterization of the identified orthologous clusters was completed through Gene Ontology annotation using the AmiGO2 database (http://geneontology.org, accessed on 11 June 2025).
3. Result and Discussion
3.1. Genome Sequencing and Assembly
B. safensis LG01 was sequenced and assembled to obtain integrated genomic information, followed by nanopore sequencing to acquire raw data. Following quality control with Mecat2, 0.83 Gb of high-quality data was retained for assembly, yielding a complete circular chromosome of 3,664,251 bp (Figure 1). The genome has a GC content of 41.73% and encodes 3836 predicted genes, along with 81 tRNAs, 24 rRNAs, 6 sRNAs, one prophage region, and 5 genomic islands (Table 1). No CRISPR arrays or plasmids were detected.
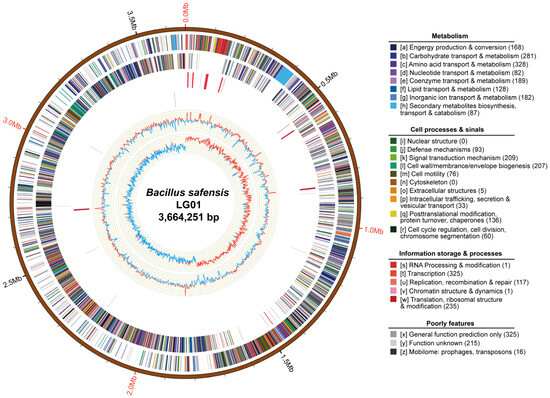
Figure 1.
Circular genome map of Bacillus safensis LG01. From outer to inner: genomic scale (5 kb intervals); coding sequences (CDSs) on forward (outer ring) and reverse (inner ring) strands, CDSs are colored based on major COG functional categories; tRNA (blue) and rRNA (purple) genes; in the GC content track, light-yellow shading highlights regions where GC content exceeds the genomic average, with peak height reflecting the degree of deviation, blue shading indicates regions below the average; GC skew (dark grey, G > C; red, C > G).

Table 1.
Genome overview of the B. safensis LG01 genome.
3.2. Functional Annotation
Chromosomal COG analysis identified 215 genes of unknown function, while the annotated gene set was dominated by ten functional categories including amino acid transport and metabolism, transcription, and carbohydrate transport and metabolism (Figure 1). To functionally characterize the predicted genes of B. safensis LG01, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. The GO analysis revealed significant enrichment in three principal categories: biological processes (e.g., metabolic, cellular, and single-organism processes), cellular components (e.g., membrane and cell parts), and molecular functions (e.g., catalytic activity, binding, and transporter activity) (Figure 2A). The KEGG pathway analysis further indicated strong enrichment in ABC transporters and amino acid biosynthesis, with additional representation in two-component systems and carbon metabolism (Figure S3).
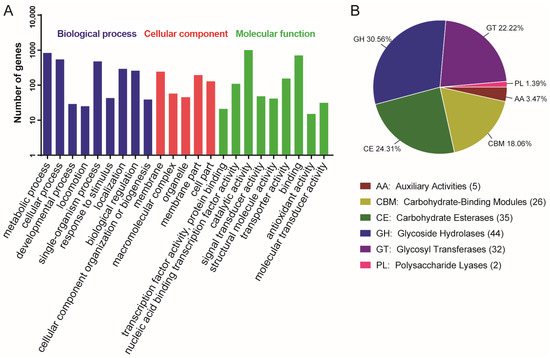
Figure 2.
Genome functional annotation of B. safensis LG01. (A) GO pathway functional Categorization. (B) The result of carbohydrate enzyme analysis of B. safensis LG01.
3.3. CAZyme Repertoire and Functional Analysis
Carbohydrate-active enzymes (CAZymes) facilitate the synthesis, modification, and degradation of complex carbohydrates [35]. They are categorized into six major classes according to the sequences and structural features of their functional domains. We located a total of 144 genes from the CAZymes family in the LG01 genome sequence. Glycoside hydrolases (30.56%) and carbohydrate esterases (24.31%) were the most dominant, followed by glycosyl transferases (22.22%), carbohydrate-binding modules (18.06%), auxiliary activities (3.47%), and polysaccharide lyases (1.39%) (Figure 2B). CAZymes serve as key genomic markers of an organism’s adaptation to specific ecological niches [36]; of these, glycoside hydrolases catalyze the cleavage or rearrangement of glycosidic bonds in diverse glycoconjugates and polysaccharides, while carbohydrate esterases remove ester-linked decorations from carbohydrates through de-O- or de-N-acylation [37]. By stripping acyl groups from polysaccharides, these esterases not only facilitate subsequent hydrolysis by glycoside hydrolases but also enhance overall biomass saccharification efficiency [38]. The range of CAZymes identified in the present study likely underlies the environmental resilience of the strains examined.
3.4. Secondary Metabolism Gene Cluster Analysis and Biocontrol Functions
Studies have shown that B. safensis possesses inhibitory capabilities and should be considered as a biocontrol agent for fungi [12,39]; it has also been investigated for its strong inhibitory effect on a variety of bacteria [40]. Recently, B. safensis has been shown to harbor a broad spectrum of antimicrobial substance-coding genes in its genomes [41]. In this study, 13 gene clusters associated with the biosynthesis of secondary metabolites, which are significant for the biocontrol capacity against pathogens, were discovered through genome mining of B. safensis LG01 (Table 2). Genome annotation revealed that B. safensis LG01 encodes diverse biosynthetic pathways for specialized metabolites, including terpenes, β-lactones, RRE-element-containing and linear azol(in)e-containing peptides, sactipeptide, ranthipeptide, and siderophore. In LG01, gene clusters for Bacillus subtilis subsp. subtilis str. 168 [42], Bacillus licheniformis DSM 13 [43], Bacillus velezensis FZB42 [44], Halobacillus halophilus DSM 2266, Bacillus pumilus ATCC 7061, and Bacillus subtilis subsp. spizizenii ATCC 6633 demonstrated high homology, suggesting the possibility that secondary metabolism gene clusters underwent horizontal transfer between LG01 and these strains [45]. Furthermore, six additional gene clusters encoded enzymes whose functions were not yet known, suggesting that these gene clusters of LG01 are specific to the strain.

Table 2.
Gene clusters involved in synthesis of secondary metabolites in B. safensis LG01.
B. safensis LG01 displayed broad-spectrum antimicrobial activity, inhibiting representative fungal and Gram-positive bacterial pathogens. Notably, it also showed significant inhibition against the Gram-negative pathogen Legionella pneumophila, although it was inactive against other Gram-negative strains such as Escherichia coli and Pseudomonas aeruginosa (Table 3). L. pneumophila, when inhaled via contaminated aerosols, evades host immunity by replicating within alveolar macrophages and ultimately causes cell death, a key mechanism in legionellosis pathogenesis [46]. Elucidating the inhibitory mechanism of B. safensis LG01 against L. pneumophila may offer a novel strategy for combating infections. The selective activity against L. pneumophila could be attributed to the action of specific antimicrobial compounds, such as lipopeptides (surfactin), which are known to disrupt membrane integrity. However, the lack of activity against E. coli and P. aeruginosa may be due to the formidable outer membrane barrier of these bacteria, which acts as a primary defense against macromolecules, coupled with the potential role of efficient efflux pump systems (particularly in P. aeruginosa) that can expel toxic compounds. We emphasize that the potent inhibition of L. pneumophila a significant human pathogen positions LG01 as a highly promising candidate for the development of narrow-spectrum anti-Legionella agents, thereby potentially mitigating the resistance issues associated with broad-spectrum antibiotic use. Elucidating the inhibitory mechanism of B. safensis LG01 against L. pneumophila may offer a novel strategy for combating infections.

Table 3.
Antimicrobial activity of the B. safensis LG01.
3.5. Antibiotic Resistance
The ubiquity of Bacillus species in the environment means that antibiotic resistance in this genus poses a potential health risk [47]. In bacteria, antibiotic resistance evolves through two principal genetic strategies: target gene mutation and horizontal acquisition of resistance determinants [48]. The intrinsic resistance genes identified in LG01 are chromosomally encoded. The presence of these chromosomally encoded resistance genes likely enhances the strain’s ecological fitness, improving its ability to successfully colonize specific niches where these antibiotics are present.
Horizontal gene transfer, the non-hereditary passage of genetic material between organisms, significantly contributes to bacterial genome plasticity and adaptive evolution [49]. Obtaining foreign genes can effectively alter the genotype of a bacterium, potentially resulting in the development of new traits or even new species [50]. Genes acquired through horizontal gene transfer (HGT) are frequently localized within integrons and mobile genetic elements (MGEs), including transposons, insertion sequences, genomic islands, phages, plasmids, and integrative and conjugative elements (ICEs) [51]. As an important vector for the transfer of resistance genes between bacteria, plasmids play an important role in the global spread of antibiotic resistance [52]. However, there is no plasmid in B. safensis LG01 (Figure 1). However, no plasmids were identified in B. safensis LG01 (Figure 1); this reduces the risk of using this strain in humans, as these genes were present on genomic DNA rather than plasmid DNA [53].
Previous authors identified the B. subtilis subsp. subtilis strain RK with a novel class of aminoglycoside transferase (AHP5), which is responsible for resistance against aminoglycoside-based antibiotics [47]. In Staphylococcus aureus, MFS transporters actively export structurally different antimicrobial agents from the cells [54]. Our genomic analysis identified two key antibiotic resistance determinants: a major facilitator superfamily (MFS) transporter gene (mdr) that mediates multidrug efflux, and an undecaprenyl pyrophosphate phosphatase gene (baca) involved in bacitracin resistance through cell wall biosynthesis modulation (Table S3). These genes may allow LG01 to adapt to different survival conditions, hosts, and environments.
Antibiotic susceptibility testing revealed that B. safensis was susceptible to nine of the ten antibiotics tested, including penicillin, ampicillin, ceftriaxone, tetracycline, chloramphenicol, gentamicin, erythromycin, ciprofloxacin, and trimethoprim-sulfamethoxazole. No zone of inhibition was observed around the lincomycin (2 μg) disk. Quantitative measurements of the inhibition zones confirmed diameters of 20–40 mm for the nine effective antibiotics (Table S4). These results demonstrate that the strain exhibits high susceptibility to antibiotics and lacks broad-spectrum resistance, indicating that it can be effectively controlled in practical applications and is unlikely to contribute to the spread of antibiotic resistance.
Analysis of the B. safensis LG01 genome using the tool PathogenFinder v1.1 predicted that 33 out of its 3659 coding sequences belong to non-pathogenic families, with no sequences identified as part of pathogenic lineages. These results indicate that B. safensis LG01 is non-pathogenic to humans.
3.6. Genomic Islands
Genomic islands (GIs) horizontally acquired chromosomal segments typically spanning 10–500 kbp [49], serve as valuable markers for comparative genomics and the identification of strain-specific adaptive genes [1]. Within the genomic architecture of B. safensis LG01, we identified five such GIs (Figure S1). GI region 1 comprised 21 coding sequences (CDSs), including an ABC-2 type transport system-associated protein, aminotransferase, mutase, endoribonuclease, permease, and transcriptional regulator. GI region 2 contained 11 CDSs encoding the SMI1/KNR4 family protein, transcriptional regulators, reductase, acetyltransferase, phosphatase, transposase or phage integrase, and phosphotransferase. GI region 3 contained 18 CDSs encoding integrase/recombinase, methyltransferase, flagellar-associated protein, transcriptional pleiotropic repressor, and proteases. GI region 4 contained 27 CDSs, including those encoding synthase, endoribonucleases, oxidoreductases, transcriptional regulators, dehydrogenases, integrase/recombinase, acetyltransferase, DNA helicase, azoreductases, NarL family-associated protein, and glyoxalase/bleomycin resistance protein/dioxygenase. GI region 5 was located in the area from 2,725,696 to 2,738,546 bp, with a length of 12,850 bp; it contained one Alanine tRNA coding gene and 22 CDSs consisting of lipoproteins, transcriptional regulators, amidases, acetyltransferase, and bhlA. BhlA has shown efficacy against multiple drug-resistant bacteria and possesses a broad spectrum of antibacterial activity [55]. GIs confer adaptive advantages to host bacteria through acquisition of novel metabolic pathways, antibiotic resistance determinants, and virulence factors, thereby enhancing fitness and resilience under diverse abiotic stresses [49].
3.7. Prophage Elements
Bacteriophages, which multiply passively with their bacterial hosts, are carried by a large number of bacteria and integrate into their genomes as prophages [56]. Lysogeny is ubiquitous and widely distributed throughout the murine gut microbiota [57]. Phages reside in 40–50% of bacterial genomes as prophage elements, which are the primary source of bacterial diversity within species and the main drivers of horizontal gene transfer [58]. Prophage-mediated DNA rearrangement is a common phenomenon in spore-forming bacteria [59], and accurate prophage annotation is therefore essential for complete functional and genomic characterization of bacterial strains [60]. A single prophage region was identified in the LG01 genome (Figure S2), located from 2,723,924 to 2,759,923 bp, with a length of 35,999 bp, and featuring one Alanine tRNA coding gene and 53 CDSs consisting of transcriptional regulators, lipoproteins, integrases, terminases, amidases, phosphatases, bhlA, acetyltransferases, phage portal proteins, putative bacteriophage proteins, transglycosylase, RNA polymerase, and resolvase. These prophage sequences may confer antibiotic resistance, enhance adhesion, and promote environmental adaptation, and they likely play an active role in cell physiology. They also contribute to LG01’s ability to adjust to changes in ecological environments. The functional influence of prophage elements on the diversification of beneficial traits remains uncharacterized.
3.8. Host Microbe Secreted Effectors
Surface and secreted proteins are key mediators of host adaptation in bacteria, enabling nutrient acquisition, host cell interactions, and tissue specific colonization [61]. B. safensis LG01 encodes an array of surface and secreted proteins that facilitate host bacterium interactions (Table S5). Key genes implicated in these functions include: BsLG_04104, encoding a beta-N-acetylhexosaminidase involved in chitin degradation and glycoconjugate metabolism; BsLG_01459 [62], encoding a gamma-glutamyltranspeptidase that modifies glutathione derivatives; and BsLG_00941, encoding a WxL domain-containing surface protein potentially involved in virulence and host protein interaction [63]. Multiple hydrolytic enzymes including peptidases, subtilisin, pectate lyase, and xylan hydrolase (encoded by genes BsLG_00300, BsLG_02651, BsLG_01213, BsLG_00711, BsLG_01808, BsLG_04032, BsLG_04364, BsLG_04446 and BsLG_02332) may support nutrient digestion from complex dietary substrates [64,65]. Furthermore, LG01 harbors global regulators linked to biofilm formation and colonization, such as Spo0A (BsLG_02744), wall teichoic acid biosynthesis proteins GtaB (BsLG_04097, BsLG_04094), and the LuxS/AI-2 quorum-sensing system (BsLG_03440) [66,67,68]. Notably, several Rap family phosphorelay-integrating regulators (BsLG_04580, BsLG_02742, BsLG_04169, BsLG_01426, BsLG_02061, BsLG_04411, BsLG_02106) were identified, suggesting that in LG01, Rap protein and Phr signaling play prominent roles in modulating quorum sensing and sporulation [69].
3.9. Probiotic Properties
The rise of antibiotic-resistant pathogens underscores the need for alternative antimicrobial strategies. B. safensis LG01 emerges as a promising probiotic candidate, harboring numerous biosynthetic gene clusters for antimicrobial secondary metabolites (Table 2) and exhibiting potent antibacterial and antifungal activity (Table 3). B. safensis VQV8 exhibits potent anti-Vibrio activity and demonstrates safety profiles compatible with probiotic applications [70]. The LG01 strain produces proteases and cellulases which function to degrade dietary proteins and cellulose, consistent with the growth-promoting effects observed in the enzyme-producing strain NPUST1 in tilapia models [71]. LG01 also forms structured biofilms that promote host colonization and ecological persistence (Table 4), while also potentially excluding pathogens as demonstrated in other B. safensis strains [72]. Biofilms provide microbes with adaptive advantages by facilitating nutrient acquisition and enhancing stress resilience [13]. In related strains, such biofilms contribute to the exclusion of foodborne pathogens like Listeria monocytogenes on the surfaces of produce, though the molecular mechanisms underlying this competitive exclusion require further characterization [73].

Table 4.
Probiotic properties of the B. safensis LG01.
Our safety evaluation identified two intrinsic antibiotic resistance genes in LG01 (Table S3). Consistent with this genus profile [70], the VQV8 strain shows susceptibility to common antibiotics [53]. Collectively, these functional and genomic attributes support the potential of B. safensis LG01 as a versatile probiotic.
3.10. Comparative Genomic Analysis
3.10.1. Phylogenetic Analysis and Genomic Collinearity
The field of molecular phylogenetics focuses on inferring organismal evolutionary history through comparative analyses of molecular data [74], providing answers to various biological questions [75]. 16S rRNA gene sequencing serves as a powerful diagnostic tool, enabling identification of novel pathogens, uncultured bacteria, phenotypically ambiguous strains, and rarely isolated species [76]. This method is not without its drawbacks, especially when it comes to differentiating closely related species. Molecular phylogenetic studies are increasingly utilizing protein-coding genes such as gyrA and gyrB as molecular chronometers in neighbor-joining trees, complementing the use of 16S rRNA. While 16S rRNA remains effective for delineating deeper-branching taxonomic relationships [32], gyrA and gyrB sequences exhibit superior discriminatory power for resolving closely related lineages at intra- and intergeneric levels [14]. Notably, gyrB sequence polymorphisms have proven particularly effective in distinguishing phylogenetic groups within the B. safensis complex [77,78].
To evaluate the phylogenetic position of LG01, we reconstructed a gyrB-based Maximum Likelihood tree with MEGA X, comparing it against other available B. safensis genomes [29]. Thirteen B. safensis strains were used in this analysis, and the genome of the LG01 strain was compared to every fully sequenced B. safensis strain available in the GenBank database (Figure 3). Additionally, we have provided a comparative table (Table S6) that systematically outlines the unique genomic features and functional attributes of LG01 compared to other closely related Bacillus strains, including type strains and characterized isolates. Phylogenetically segregated into four distinct classes (I–IV), these strains were subsequently subjected to whole-genome comparative analysis using TBtools, revealing conserved syntenic blocks across the taxonomic divisions (Figure S4).
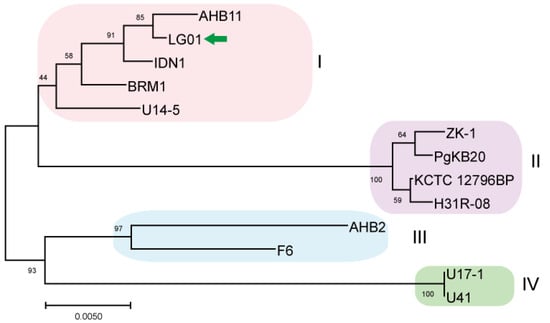
Figure 3.
Maximum-Likelihood phylogenetic tree of B. safensis LG01 reconstructed from gyrB sequences. The dataset for phylogenetic reconstruction comprised 13 nucleotide sequences derived from the complete genomes of available B. safensis strains and was conducted using the Maximum Likelihood method. Phylogenetic analysis revealed four subclusters among the 13 strains. The target strain is marked with a green arrow for clarity.
B. safensis Class I comprises five genomically distinct strains AHB11, LG01, IDN1, BRM1, and the psychrotrophic Antarctic isolate U14-5 from Lake Untersee. Our pairwise comparison identified AHB11 as sharing 1687 collinear blocks with BRM1, 1530 with IDN1, 1718 with LG01, and 2019 with U14-5. BRM1 displayed 1373 collinear blocks with IDN1, 1439 with LG01, and 1748 with U14-5. The IDN1-LG01 comparison revealed 1307 collinear blocks, while IDN1 shared 1606 blocks with U14-5. Notably, the LG01-U14-5 comparison showed 1823 collinear blocks, representing one of the highest syntenic relationships within this class. Comparative analysis established U14-5 as the genomic cornerstone of Class I, demonstrating the maximum number of collinear blocks with all other members: 2019 with AHB11, 1748 with BRM1, 1606 with IDN1, and 1823 with LG01. This consistent syntenic predominance suggests that U14-5 may represent an evolutionarily conserved lineage within the B. safensis complex (Table S1) and has the highest syntenic relationship with other strains and was also compared based on Average Nucleotide Identity (ANI) (Table S2). B. safensis ZK-1, PgKB20, KCTC 12796BP, and H31R 08 make up Class II. Whole-genome alignment revealed that H31R 08 shares 2561 collinear blocks with KCTC 12796BP, 2760 with PgKB20, and 2672 with ZK-1. KCTC 12796BP exhibited 2682 collinear blocks with PgKB20 and 2602 with ZK-1. The comparison of PgKB20 and ZK-1 showed 2765 collinear blocks, the highest value observed in Class II. Consistently, PgKB20 demonstrated peak syntenic conservation with all other class members: 2760 blocks with H31R 08, 2682 with KCTC 12796BP, and 2765 with ZK-1, establishing it as the genomic reference for this phylogenetic group. Class III, represented by strains F6 and AHB2, showed substantial genomic conservation, with 2665 collinear blocks between them. Class IV, consisting of U41 and U17-1, exhibited 1520 collinear blocks, indicating more divergent genome architecture compared to other classes. These results demonstrate distinct patterns of syntenic conservation across the phylogenetic spectrum, with PgKB20 serving as the genomic anchor for Class II, while Classes III and IV show characteristic levels of genome structural conservation reflective of their phylogenetic distances.
3.10.2. Comparative Genomic Analysis of Four Sub Clusters
Comparative genomic analysis across the four B. safensis phylogenetic classes identified 2975 core orthologous clusters, representing the essential genetic repertoire of this species complex. This conserved genomic foundation exhibits clear phylogenetic stratification, with Class I and II strains showing greater orthology conservation than the more divergent Classes III and IV (Figure 4A). Compared to the other three sub clusters, a total of 3047 genes were identified in Class I, with no specific gene clusters and 16 specific singletons, 11 of which were annotated in the GO terms and five of which were not annotated; in Class II, a total of 3425 genes were identified, with one specific gene cluster (including four genes) and 33 specific singletons, 17 of which were annotated in the GO terms and 17 of which were not annotated; in Class III, a total of 3441 genes were identified, with two specific gene clusters (including eight genes) and 59 specific singletons, 38 of which were annotated in the GO terms and 23 of which were not; in Class IV, a total of 3446 genes were identified, with eight specific gene clusters (including 17 genes) and 295 specific singletons, 92 of which were annotated in the GO terms and 211 of which were not (Figure 4B,C).
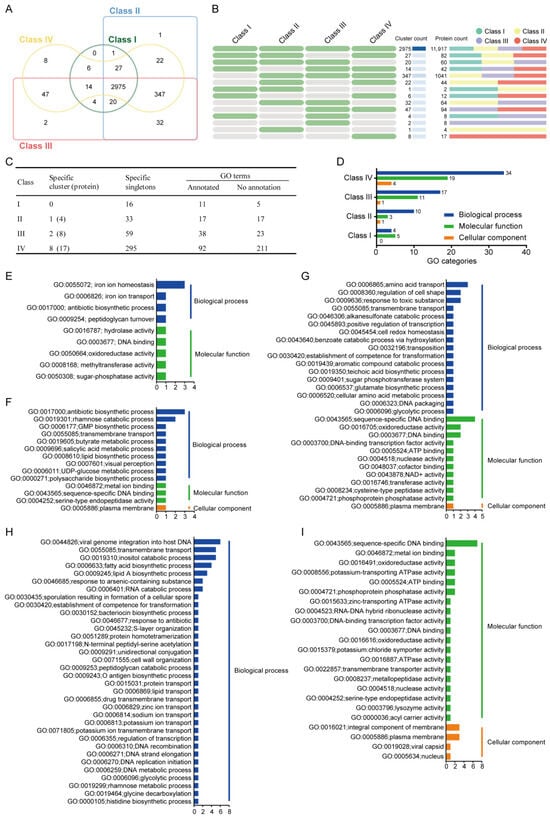
Figure 4.
Comparative analysis of orthologous gene clusters across four B. safensis subclusters. (A,B) Venn diagrams illustrate shared and unique orthologous gene clusters. Green cells denote present; gray absent across classes. (C,D) GO functional annotation of orthologous genes across classes. (E–I) Class-specific functional profiles for Class I (E), Class II (F), Class III (G), and Class IV (H,I).
Gene Ontology enrichment analysis of subclass-specific gene clusters and singletons revealed distinct functional profiles across the phylogenetic classes. In Class I strains, significant enrichment was observed in two primary GO categories: biological processes and molecular functions, indicating specific evolutionary adaptations in these functional domains (Figure 4D). The enriched biological process terms included iron ion homeostasis (GO:0055072), iron ion transport (GO:0006826), antibiotic biosynthetic process (GO:0017000), and peptidoglycan turnover (GO:0009254). The enriched molecular function terms included hydrolase activity (GO:0016787), DNA binding (GO:0003677), oxidoreductase activity (GO:0050664), methyltransferase activity (GO:0008168), and sugar-phosphatase activity (GO:0050308) (Figure 4E), showing that Class I strains have a better capacity for iron ion homeostasis and transport, which likely contributes to their improved resistance to heavy metal detoxification. The maximum metal tolerance of B. safensis ST7 to Fe (III) was 250 mg/L, and it was able to grow in the presence of high concentrations of this element [79]. Therefore, Class I is a potential candidate for restoring soil polluted by multiple heavy metals.
In Class II, GO terms were enriched in the following three categories: biological processes, molecular functions, and cellular components (Figure 4D). Gene Ontology analysis revealed functional specialization in Class II strains, with significant enrichment in antibiotic biosynthesis (GO:0017000) and rhamnose catabolism (GO:0019301) for biological processes. Molecular functions were characterized by metal ion binding (GO:0046872), sequence-specific DNA binding (GO:0043565), and serine-type endopeptidase activity (GO:0004252), while cellular components showed predominant localization to the plasma membrane (GO:0005886) (Figure 4F). The significantly enriched antibiotic biosynthetic process likely indicates the production of metabolites with antimicrobial activity in Class II. Many microorganisms are capable of utilizing rhamnose as a carbon source [80]. Thus, these clusters might enhance the environmental adaptation and food acquisition capabilities of the Class II strains, thereby improving their potential as new biological control agents.
In Class III, significantly enriched biological process GO terms included amino acid transport (GO:0006865), regulation of cell shape (GO:0008360), and response to toxic substances (GO:0009636). The significantly enriched molecular function terms included sequence-specific DNA binding (GO:0043565), oxidoreductase activity (GO:0016705), and DNA binding (GO:0003677). Plasma membranes were the only enriched cellular component term (GO:0005886) (Figure 4G). The Class III strains F6 and AHB2 were isolated from chicken feces and beehives, respectively (Table S1).
In Class IV, GO terms included viral genome integration into host DNA (GO:0044826), transmembrane transport (GO:0055085), inositol catabolic process (GO:0019310), fatty acid biosynthetic process (GO:0006633), lipid A biosynthetic process (GO:0009245), response to arsenic-containing substance (GO:0046685), and RNA catabolic process (GO:0006401). The significantly enriched molecular function terms included sequence-specific DNA binding (GO:0043565), metal ion binding (GO:0046872), oxidoreductase activity (GO:0016491), potassium-transporting ATPase activity (GO:0008556), ATP binding (GO:0005524), and phosphoprotein phosphatase activity (GO:0004721). The significantly enriched cellular component terms included integral component of membrane (GO:0016021) and plasma membrane (GO:0005886) (Figure 4H). The Class IV strains U41 and U17-1 are psychrotrophic bacteria isolated from Lake Untersee in Antarctica (Table S1). Thus, these clusters might enhance the adaptability of Class IV strains to cold environments.
These quasi-specific genes represent the fundamental genetic determinants that facilitate their adaptation to distinct lifestyles, minimize intraspecific competition, and promote coexistence and proliferation.
4. Conclusions
Our study establishes B. safensis LG01 as a genomically versatile and functionally robust strain with dual potential for sustainable agriculture and probiotic development. The expanded repertoire of secondary metabolite clusters, coupled with its experimentally validated antimicrobial and enzymatic activities, underscores LG01’s capacity to thrive in competitive environments while exerting beneficial effects. Evolutionary genomic analyses further reveal adaptive mutations that may underpin its ecological success. These insights not only illuminate the genetic foundations of B. safensis as an emerging microbial platform but also position LG01 as a promising candidate for future development into targeted biocontrol agents or next-generation probiotics. The limitation of this study is the laboratory data. Further investigation into its functional efficacy in vivo and under real-world conditions will be essential to translate these genomic advantages into practical applications.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms13112605/s1, Figure S1: Genomic islands identified in B. safensis LG01. Figure S2: Prophage regions identified in B. safensis LG01. Figure S3: The result of classification statistics of KEGG pathway annotation of B. safensis LG01. Figure S4: Synteny map of four sub clusters of B. safensis strains. Table S1: Results of the whole genome sequence comparison. Table S2: Comparison of Average Nucleotide Identity (ANI) among LG01 and Other Strains. Table S3: Antibiotic resistance genes identified in B. safensis LG01. Table S4: Antimicrobial Susceptibility of B. safensi LG01. Table S5: Representative genes of B. safensis LG01 probably involved in host-bacteria interactions. Table S6: List of various bioactive metabolites synthesized by B. safensis LG01 and the closely related species. Ref [81] is included in Supplementary Materials.
Author Contributions
L.Y., Y.Y., J.R., Y.S. and Z.G. planned the experiments. L.Y., Y.Y., J.R., Y.S. and Z.J. performed the experiments and analysis. Z.G. and Y.L. wrote the draft of the manuscript. All authors contributed to the edits of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Guang Dong Cheung Kong Philanthropy Foundation (E2018096 to Y.J.L.), Key laboratory start-up project (Sixth Affiliated Hospital of Sun Yat-sen University) (2023WST02 to Y.J.L.), Guangdong Basic and Applied Basic Research Foundation (2023A1515010247, 2024A1515012253 to Z.H.G.), Guangzhou Basic and Applied Basic Research Foundation (2024A04J3491 to Z.H.G.), Fundamental Research Funds for the Central Universities, Sun Yat-sen University (24qnpy069 to Z.H.G.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in [GenBank] at [https://www.ncbi.nlm.nih.gov/], reference number [CP109651.1].
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abdelhafiz, Y.A.; Manaharan, T.; Bin Mohamad, S.; Merican, A.F. Whole genome sequencing and functional features of UMX-103: A new Bacillus strain with biosurfactant producing capability. Genes. Genom. 2017, 39, 877–886. [Google Scholar] [CrossRef]
- Satomi, M.; La Duc, M.T.; Venkateswaran, K. Bacillus safensis sp nov., isolated from spacecraft and assembly-facility surfaces. Int. J. Syst. Evol. Microbiol. 2006, 56, 1735–1740. [Google Scholar] [CrossRef]
- Strahsburger, E.; Zapata, F.; Pedroso, I.; Fuentes, D.; Tapia, P.; Ponce, R.; McClelland, M.; Valdes, J. Draft Genome Sequence of Bacillus safensis RP10, Isolated from Soil in the Atacama Desert, Chile. Microbiol. Resour. Announc. 2019, 8, e01150-19. [Google Scholar] [CrossRef]
- Coil, D.A.; Neches, R.Y.; Lang, J.M.; Brown, W.E.; Severance, M.; Cavalier, D.; Eisen, J.A. Growth of 48 built environment bacterial isolates on board the International Space Station (ISS). PeerJ 2016, 4, e1842. [Google Scholar] [CrossRef]
- Kumar, D.; Parshad, R.; Gupta, V.K. Application of a statistically enhanced, novel, organic solvent stable lipase from Bacillus safensis DVL-43. Int. J. Biol. Macromol. 2014, 66, 97–107. [Google Scholar] [CrossRef]
- da Fonseca, F.S.A.; Angolini, C.F.F.; Arruda, M.A.Z.; Junior, C.A.L.; Santos, C.A.; Saraiva, A.M.; Pilau, E.; Souza, A.P.; Laborda, P.R.; de Oliveira, P.F.L.; et al. Identification of oxidoreductases from the petroleum Bacillus safensis strain. Biotechnol. Rep. 2015, 8, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Lateef, A.; Adelere, I.A.; Gueguim-Kana, E.B. Bacillus safensis LAU 13: A new source of keratinase and its multi-functional biocatalytic applications. Biotechnol. Biotechnol. Equip. 2015, 29, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, U.; Chakraborty, B.N.; Chakraborty, A.P.; Dey, P.L. Water stress amelioration and plant growth promotion in wheat plants by osmotic stress tolerant bacteria. World J. Microbiol. Biotechnol. 2013, 29, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Katwal, S.; Sharma, B.; Sharma, A.; Puri, S.; Kamboj, N.; Kanwar, S.S. Purification, characterization and cytotoxic properties of a bacterial RNase. Int. J. Biol. Macromol. 2021, 166, 665–676. [Google Scholar] [CrossRef]
- Abdelli, F.; Jardak, M.; Elloumi, J.; Stien, D.; Cherif, S.; Mnif, S.; Aifa, S. Antibacterial, anti-adherent and cytotoxic activities of surfactin(s) from a lipolytic strain Bacillus safensis F4. Biodegradation 2019, 30, 287–300. [Google Scholar] [CrossRef]
- Prakash, J.; Arora, N.K. Novel metabolites from Bacillus safensis and their antifungal property against Alternaria alternata. Antonie van Leeuwenhoek 2021, 114, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Xu, H.; Li, L.; Chen, R.; Gao, X.; Xu, Z. Antifungal activity of endophytic Bacillus safensis B21 and its potential application as a biopesticide to control rice blast. Pestic. Biochem. Physiol. 2020, 162, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Romero-Severson, J.; Moran, T.E.; Shrader, D.G.; Fields, F.R.; Pandey-Joshi, S.; Thomas, C.L.; Palmer, E.C.; Shrout, J.D.; Pfrender, M.E.; Lee, S.W. A Seed-Endophytic Bacillus safensis Strain with Antimicrobial Activity Has Genes for Novel Bacteriocin-Like Antimicrobial Peptides. Front. Microbiol. 2021, 12, 734216. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.H.; Kuang, Z.Q.; Chen, J.H.; Chen, J.Y.; Liu, T.H.; She, Z.G.; Lu, Y.J. Comparative genomics analysis of Bacillus velezensis LOH112 isolated from a nonagenarian provides insights into its biocontrol and probiotic traits. Gene. 2022, 835, 146644. [Google Scholar] [CrossRef]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Burge, S.W.; Bateman, A.; Daub, J.; Eberhardt, R.Y.; Eddy, S.R.; Floden, E.W.; Gardner, P.P.; Jones, T.A.; Tate, J.; et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015, 43, D130–D137. [Google Scholar] [CrossRef]
- Langille, M.G.; Hsiao, W.W.; Brinkman, F.S. Evaluation of genomic island predictors using a comparative genomics approach. BMC Bioinform. 2008, 9, 329. [Google Scholar] [CrossRef]
- Bland, C.; Ramsey, T.L.; Sabree, F.; Lowe, M.; Brown, K.; Kyrpides, N.C.; Hugenholtz, P. CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007, 8, 209. [Google Scholar] [CrossRef] [PubMed]
- Akhter, S.; Aziz, R.K.; Edwards, R.A. PhiSpy: A novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 2012, 40, e126. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Profile hidden Markov models. Bioinformatics 1998, 14, 755–763. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H. Weber T: antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Mega, X. Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Liu, G.; Kong, Y.; Fan, Y.; Geng, C.; Peng, D.; Sun, M. Whole-genome sequencing of Bacillus velezensis LS69, a strain with a broad inhibitory spectrum against pathogenic bacteria. J. Biotechnol. 2017, 249, 20–24. [Google Scholar] [CrossRef]
- Owen, R.J. Bacterial taxonomics: Finding the wood through the phylogenetic trees. Methods Mol. Biol. 2004, 266, 353–383. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2013, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Bradley, E.L.; Ökmen, B.; Doehlemann, G.; Henrissat, B.; Bradshaw, R.E.; Mesarich, C.H. Secreted Glycoside Hydrolase Proteins as Effectors and Invasion Patterns of Plant-Associated Fungi and Oomycetes. Front. Plant Sci. 2022, 13, 853106. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.M.; Nascimento, A.S.; Polikarpov, I. Structural diversity of carbohydrate esterases. Biotechnol. Res. Innov. 2017, 1, 35–51. [Google Scholar] [CrossRef]
- Manzo, N.; D’Apuzzo, E.; Coutinho, P.M.; Cutting, S.M.; Henrissat, B.; Ricca, E. Carbohydrate-active enzymes from pigmented Bacilli: A genomic approach to assess carbohydrate utilization and degradation. BMC Microbiol. 2011, 11, 198. [Google Scholar] [CrossRef] [PubMed]
- Einloft, T.C.; de Oliveira, P.B.; Radünz, L.L.; Dionello, R.G. Biocontrol capabilities of three Bacillus isolates towards aflatoxin B1 producer A. flavus in vitro and on maize grains. Food Control 2021, 125, 107978. [Google Scholar] [CrossRef]
- Ren, P.; Peng, J.; Liu, S.; Yao, Z.; Zhu, G.; Lu, G.; Li, R. Isolation and Identification of a Bacillus safensis Strain GX-H6 and Its Biocontrol Effect on Bacterial Leaf Streak of Rice. Biotechnol. Bull. 2023, 39, 243–253. [Google Scholar]
- Mateus, J.R.; Dal’Rio, I.; Jurelevicius, D.; da Mota, F.F.; Marques, J.M.; Ramos, R.T.J.; da Silva, A.L.D.; Gagliardi, P.R.; Seldin, L. Bacillus velezensis T149-19 and Bacillus safensis T052-76 as Potential Biocontrol Agents against Foot Rot Disease in Sweet Potato. Agriculture 2021, 11, 1046. [Google Scholar] [CrossRef]
- Barbe, V.; Cruveiller, S.; Kunst, F.; Lenoble, P.; Meurice, G.; Sekowska, A.; Vallenet, D.; Wang, T.; Moszer, I.; Medigue, C.; et al. From a consortium sequence to a unified sequence: The Bacillus subtilis 168 reference genome a decade later. Microbiology 2009, 155 Pt 6, 1758–1775. [Google Scholar] [CrossRef] [PubMed]
- Veith, B.; Herzberg, C.; Steckel, S.; Feesche, J.; Maurer, K.H.; Ehrenreich, P.; Bäumer, S.; Henne, A.; Liesegang, H.; Merkl, R.; et al. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 2004, 7, 204–211. [Google Scholar] [CrossRef]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef] [PubMed]
- Bohm, M.E.; Huptas, C.; Krey, V.M.; Scherer, S. Massive horizontal gene transfer, strictly vertical inheritance and ancient duplications differentially shape the evolution of Bacillus cereus enterotoxin operons hbl, cytK and nhe. BMC Evol. Biol. 2015, 15, 246. [Google Scholar] [CrossRef] [PubMed]
- Cazalet, C.; Rusniok, C.; Brüggemann, H.; Zidane, N.; Magnier, A.; Ma, L.; Tichit, M.; Jarraud, S.; Bouchier, C.; Vandenesch, F.; et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004, 36, 1165–1173. [Google Scholar] [CrossRef]
- Parulekar, R.S.; Barale, S.S.; Sonawane, K.D. Antibiotic resistance and inhibition mechanism of novel aminoglycoside phosphotransferase APH(5) from, B. subtilis subsp. subtilis strain, R.K. Braz. J. Microbiol. 2019, 50, 887–898. [Google Scholar] [CrossRef]
- Urban-Chmiel, R.; Marek, A.; Wieczorek, K.; Dec, M.; Stepien-Pysniak, D.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef]
- Piña-Iturbe, A.; Suazo, I.D.; Hoppe-Elsholz, G.; Ulloa-Allendes, D.; González, P.A.; Kalergis, A.M.; Bueno, S.M. Horizontally Acquired Homologs of Xenogeneic Silencers: Modulators of Gene Expression Encoded by Plasmids, Phages and Genomic Islands. Genes 2020, 11, 142. [Google Scholar] [CrossRef]
- Zhang, R.; Zhang, C.T. Identification of genomic islands in the genome of Bacillus cereus by comparative analysis with Bacillus anthracis. Physiol. Genom. 2003, 16, 19–23. [Google Scholar] [CrossRef]
- Botelho, J.; Grosso, F.; Peixe, L. Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updates 2019, 44, 26–47. [Google Scholar] [CrossRef] [PubMed]
- Millan, A.S.; Maclean, R.C. Fitness Costs of Plasmids: A Limit to Plasmid Transmission. Microbiol. Spectr. 2017, 5, 10. [Google Scholar] [CrossRef]
- Saidumohamed, B.E.; Bhat, S.G. Indian oil sardine (Sardinella longiceps) gut derived Bacillus safensis SDG14 with enhanced probiotic competence for food and feed applications. Food Res. Int. 2021, 150 Pt A, 110475. [Google Scholar] [CrossRef]
- Stephen, J.; Salam, F.; Lekshmi, M.; Kumar, S.H.; Varela, M.F. The Major Facilitator Superfamily and Antimicrobial Resistance Efflux Pumps of the ESKAPEE Pathogen Staphylococcus aureus. Antibiotics 2023, 12, 343. [Google Scholar] [CrossRef]
- Anthony, T.; Chellappa, G.S.; Rajesh, T.; Gunasekaran, P. Functional analysis of a putative holin-like peptide-coding gene in the genome of Bacillus licheniformis AnBa9. Arch. Microbiol. 2010, 192, 51–56. [Google Scholar] [CrossRef]
- Sutcliffe, S.G.; Shamash, M.; Hynes, A.P.; Maurice, C.F. Common Oral Medications Lead to Prophage Induction in Bacterial Isolates from the Human Gut. Viruses 2021, 13, 455, Erratum in Viruses 2022, 15, 25. [Google Scholar] [CrossRef]
- Kim, M.S.; Bae, J.W. Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J. 2018, 12, 1127–1141. [Google Scholar] [CrossRef] [PubMed]
- Dragos, A.; Priyadarshini, B.; Hasan, Z.; Strube, M.L.; Kempen, P.J.; Maróti, G.; Kaspar, C.; Bose, B.; Burton, B.M.; Bischofs, I.B.; et al. Pervasive prophage recombination occurs during evolution of spore-forming Bacilli. Isme J. 2021, 15, 1344–1358. [Google Scholar] [CrossRef]
- Abe, K.; Yoshinari, A.; Aoyagi, T.; Hirota, Y.; Iwamoto, K.; Sato, T. Regulated DNA rearrangement during sporulation in Bacillus weihenstephanensis KBAB4. Mol. Microbiol. 2013, 90, 415–427. [Google Scholar] [CrossRef]
- Bondy-Denomy, J.; Qian, J.; Westra, E.R.; Buckling, A.; Guttman, D.S.; Davidson, A.R.; Maxwell, K.L. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016, 10, 2854–2866. [Google Scholar] [CrossRef] [PubMed]
- Grossman, A.S.; Mauer, T.J.; Forest, K.T.; Goodrich-Blair, H. A Widespread Bacterial Secretion System with Diverse Substrates. mBio 2021, 12, e0195621. [Google Scholar] [CrossRef] [PubMed]
- El-Aouar, R.A.; Nicolas, A.; Castro, T.L.D.; Deplanche, M.; Azevedo, V.A.D.; Goossens, P.L.; Taieb, F.; Lina, G.; Le Loir, Y.; Berkova, N. Heterogeneous Family of Cyclomodulins: Smart Weapons That Allow Bacteria to Hijack the Eukaryotic Cell Cycle and Promote Infections. Front. Cell. Infect. Microbiol. 2017, 7, 208, Erratum in Front. Cell. Infect. Microbiol. 2017, 7, 364. [Google Scholar]
- Hassan, M.U.; Williamson, M.P. Bioinformatic analysis of WxL domain proteins. Saudi J. Biol. Sci. 2023, 30, 103526. [Google Scholar] [CrossRef]
- Hamouda, H.I.; Ali, N.; Su, H.; Feng, J.; Lu, M.; Li, F.L. Exploration of Two Pectate Lyases from Caldicellulosiruptor bescii Reveals that the CBM66 Module Has a Crucial Role in Pectic Biomass Degradation. Appl. Environ. Microbiol. 2020, 86, e00787-20. [Google Scholar] [CrossRef]
- Monica, P.; Kapoor, M. Alkali-stable GH11 endo-β-1,4 xylanase (XynB) from Bacillus subtilis strain CAM 21: Application in hydrolysis of agro-industrial wastes, fruit/vegetable peels and weeds. Prep. Biochem. Biotechnol. 2021, 51, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Wang, Y.; Li, J.; Shen, Q.; Zhang, R. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 2013, 97, 8823–8830. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Q.; Liu, D.; Zhang, H.; Dong, X.; Zhang, G.; Liu, Y.; Zhang, R. Quorum sensing signal autoinducer-2 promotes root colonization of Bacillus velezensis SQR9 by affecting biofilm formation and motility. Appl. Microbiol. Biotechnol. 2020, 104, 7177–7185. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, H.; Sun, X.; Liu, Y.; Yan, W.; Xun, W.; Shen, Q.; Zhang, R. Bacillus velezensis Wall Teichoic Acids Are Required for Biofilm Formation and Root Colonization. Appl. Environ. Microbiol. 2019, 85, e02116-18. [Google Scholar] [CrossRef]
- Slamti, L.; Perchat, S.; Huillet, E.; Lereclus, D. Quorum sensing in Bacillus thuringiensis is required for completion of a full infectious cycle in the insect. Toxins 2014, 6, 2239–2255. [Google Scholar] [CrossRef] [PubMed]
- Huynh-Phuoc, V.; Ly, T.Q.; Purbiantoro, W.; Ngo, H.V.T.; Afonso, F.; Vu, N.U.; Cheng, T.C. Bacillus safensis isolated from white-leg shrimp, Penaeus vannamei in Taiwan with antagonistic activity against common Vibrio pathogens. Biocatal. Agric. Biotechnol. 2022, 44, 102477. [Google Scholar] [CrossRef]
- Wu, P.S.; Liu, C.H.; Hu, S.Y. Probiotic Bacillus safensis NPUST1 Administration Improves Growth Performance, Gut Microbiota, and Innate Immunity against Streptococcus iniae in Nile tilapia (Oreochromis niloticus). Microorganisms 2021, 9, 2494. [Google Scholar] [CrossRef]
- Mahapatra, S.; Yadav, R.; Ramakrishna, W. Bacillus subtilis impact on plant growth, soil health and environment: Dr. Jekyll and Mr. Hyde. J. Appl. Microbiol. 2022, 132, 3543–3562. [Google Scholar] [CrossRef] [PubMed]
- Hascoët, A.S.; Ripolles-Avila, C.; Cervantes-Huamán, B.R.H.; Rodríguez-Jerez, J.J. In Vitro Preformed Biofilms of Bacillus safensis Inhibit the Adhesion and Subsequent Development of Listeria monocytogenes on Stainless-Steel Surfaces. Biomolecules 2021, 11, 475. [Google Scholar] [CrossRef]
- Ajawatanawong, P. Molecular Phylogenetics: Concepts for a Newcomer. Adv. Biochem. Eng. Biotechnol. 2017, 160, 185–196. [Google Scholar]
- Yang, Z.; Rannala, B. Molecular phylogenetics: Principles and practice. Nat. Rev. Genet. 2012, 13, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, T.; Sugano, M. 16S rRNA gene sequence analysis for bacterial identification in the clinical laboratory. Rinsho Byori Jpn. J. Clin. Pathol. 2013, 61, 1107–1115. [Google Scholar]
- Fischer, S.; Krause, T.; Lederer, F.; Merroun, M.L.; Shevchenko, A.; Hübner, R.; Firkala, T.; Stumpf, T.; Jordan, N.; Jain, R. Bacillus safensis JG-B5T affects the fate of selenium by extracellular production of colloidally less stable selenium nanoparticles. J. Hazard. Mater. 2020, 384, 121146. [Google Scholar] [CrossRef]
- Abril, A.G.; Rama, J.L.R.; Feijoo-Siota, L.; Calo-Mata, P.; Salazar, S.; Peix, A.; Velázquez, E.; Villa, T.G. Bacillus safensis subsp. osmophilus subsp. nov., isolated from condensed milk, and description of Bacillus safensis subsp. safensis subsp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Ran, X.Q.; Zhu, Z.M.; Long, H.; Tian, Q.; You, L.J.; Wu, X.D.; Liu, Q.; Huang, S.H.; Li, S.; Niu, X.; et al. Manganese Stress Adaptation Mechanisms of Bacillus safensis Strain ST7 From Mine Soil. Front. Microbiol. 2021, 12, 758889. [Google Scholar] [CrossRef]
- Rodionova, I.A.; Li, X.Q.; Thiel, V.; Stolyar, S.; Stanton, K.; Fredrickson, J.K.; Bryant, D.A.; Osterman, A.L.; Best, A.A.; Rodionov, D.A. Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria. Front. Microbiol. 2013, 4, 407. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.H. Bacillus velezensis: A Valuable Member of Bioactive Molecules within Plant Microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).