Sulfur Cycling and Life Strategies in Successional Biocrusts Link to Biomass Carbon in Dryland Ecosystems
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection Description
2.2. Environmental Factor Measurement and Nucleic Acid Extraction
2.3. Metagenome Sequencing, Assembly, and Binning
2.4. Functional Annotations and Geochip Analysis
2.5. Classification of Microbial Lifestyle Strategies
2.6. Statistical Analysis
3. Results
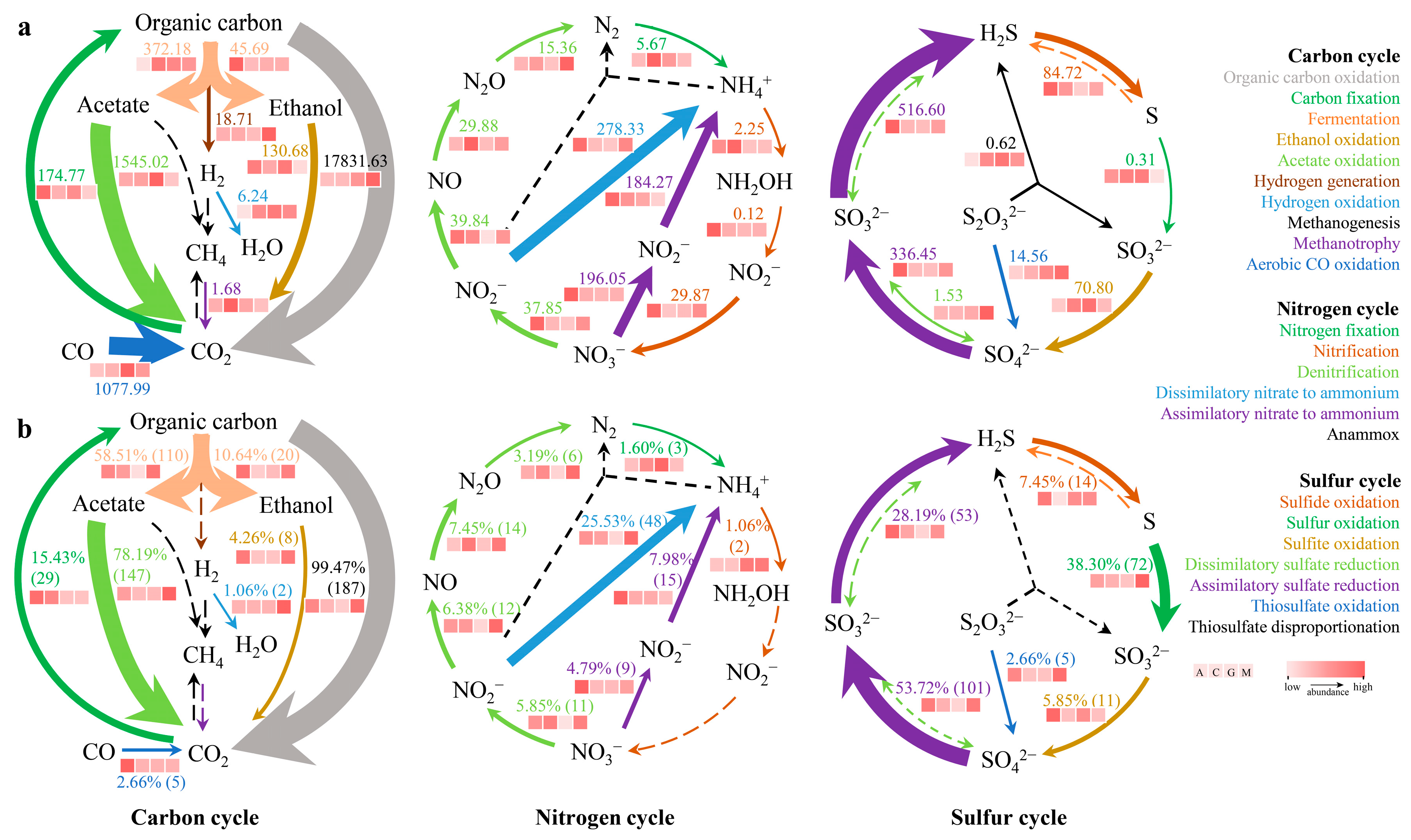
3.1. Sulfur Cycle Patterns
3.2. Carbon, Nitrogen, and Sulfur Cycling Patterns
3.3. Coupling of the Sulfur Cycle with Carbon Metabolism
3.4. Life Strategies
3.5. Relationship Between the Sulfur Cycle and Life Strategies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pufahl, P.K.; Squires, A.D.; Murphy, J.B.; Quesada, C.; Lokier, S.W.; Álvaro, J.J.; Hatch, J. Ordovician Ironstone of the Iberian Margin: Coastal Upwelling, Ocean Anoxia and Palaeozoic Biodiversity. Depos. Rec. 2020, 6, 581–604. [Google Scholar] [CrossRef]
- Beulig, F.; Røy, H.; McGlynn, S.E.; Jørgensen, B.B. Cryptic CH4 Cycling in the Sulfate–Methane Transition of Marine Sediments Apparently Mediated by ANME-1 Archaea. ISME J. 2019, 13, 250–262. [Google Scholar] [CrossRef]
- Balk, M.; Keuskamp, J.A.; Laanbroek, H.J. Potential Activity, Size, and Structure of Sulfate-Reducing Microbial Communities in an Exposed, Grazed and a Sheltered, Non-Grazed Mangrove Stand at the Red Sea Coast. Front. Microbiol. 2015, 6, 1478. [Google Scholar] [CrossRef] [PubMed]
- Raven, M.R.; Keil, R.G.; Webb, S.M. Microbial Sulfate Reduction and Organic Sulfur Formation in Sinking Marine Particles. Science 2021, 371, 178–181. [Google Scholar] [CrossRef]
- Gomez-Saez, G.V.; Dittmar, T.; Holtappels, M.; Pohlabeln, A.M.; Lichtschlag, A.; Schnetger, B.; Boetius, A.; Niggemann, J. Sulfurization of Dissolved Organic Matter in the Anoxic Water Column of the Black Sea. Sci. Adv. 2021, 7, eabf6199. [Google Scholar] [CrossRef]
- Montoya, L.; Celis, L.B.; Razo-Flores, E.; Alpuche-Solís, Á.G. Distribution of CO2 Fixation and Acetate Mineralization Pathways in Microorganisms from Extremophilic Anaerobic Biotopes. Extremophiles 2012, 16, 805–817. [Google Scholar] [CrossRef]
- Boyle, N.R.; Morgan, J.A. Computation of Metabolic Fluxes and Efficiencies for Biological Carbon Dioxide Fixation. Metab. Eng. 2011, 13, 150–158. [Google Scholar] [CrossRef]
- Meier, D.V.; Pjevac, P.; Bach, W.; Hourdez, S.; Girguis, P.R.; Vidoudez, C.; Amann, R.; Meyerdierks, A. Niche Partitioning of Diverse Sulfur-Oxidizing Bacteria at Hydrothermal Vents. ISME J. 2017, 11, 1545–1558. [Google Scholar] [CrossRef] [PubMed]
- Grote, J.; Jost, G.; Labrenz, M.; Herndl, G.J.; Jürgens, K. Epsilonproteobacteria Represent the Major Portion of Chemoautotrophic Bacteria in Sulfidic Waters of Pelagic Redoxclines of the Baltic and Black Seas. Appl. Environ. Microbiol. 2008, 74, 7546–7551. [Google Scholar] [CrossRef]
- Nakagawa, S.; Shtaih, Z.; Banta, A.; Beveridge, T.J.; Sako, Y.; Reysenbach, A.-L. Sulfurihydrogenibium yellowstonense sp. nov., an Extremely Thermophilic, Facultatively Heterotrophic, Sulfur-Oxidizing Bacterium from Yellowstone National Park, and Emended Descriptions of the Genus Sulfurihydrogenibium, Sulfurihydrogenibium subterraneum and Sulfurihydrogenibium azorense. Int. J. Syst. Evol. Microbiol. 2005, 55, 2263–2268. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jiang, L.; Xie, S.; Alain, K.; Wang, Z.; Wang, J.; Liu, D.; Shao, Z. Disproportionation of Inorganic Sulfur Compounds by Mesophilic Chemolithoautotrophic Campylobacterota. mSystems 2023, 8, e00954-22. [Google Scholar] [CrossRef]
- Tabatabai, M.A. Chemistry of Sulfur in Soils. In Chemical Processes in Soils; SSSA Book Series; Soil Science Society of America: Madison, WI, USA, 2018; pp. 193–226. [Google Scholar]
- Kawano, Y.; Suzuki, K.; Ohtsu, I. Current Understanding of Sulfur Assimilation Metabolism to Biosynthesize L-Cysteine and Recent Progress of Its Fermentative Overproduction in Microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 8203–8211. [Google Scholar] [CrossRef]
- Hausmann, B.; Pelikan, C.; Herbold, C.W.; Köstlbacher, S.; Albertsen, M.; Eichorst, S.A.; Glavina del Rio, T.; Huemer, M.; Nielsen, P.H.; Rattei, T.; et al. Peatland Acidobacteria with a Dissimilatory Sulfur Metabolism. ISME J. 2018, 12, 1729–1742. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, B.B. The Sulfur Cycle of a Coastal Marine Sediment (Limfjorden, Denmark). Limnol. Oceanogr. 1977, 22, 814–832. [Google Scholar] [CrossRef]
- Schad, M.; Konhauser, K.O.; Sánchez-Baracaldo, P.; Kappler, A.; Bryce, C. How Did the Evolution of Oxygenic Photosynthesis Influence the Temporal and Spatial Development of the Microbial Iron Cycle on Ancient Earth? Free Radic. Biol. Med. 2019, 140, 154–166. [Google Scholar] [CrossRef]
- Lovley, D.R.; Phillips, E.J.P. Organic Matter Mineralization with Reduction of Ferric Iron in Anaerobic Sediments. Appl. Environ. Microbiol. 1986, 51, 683–689. [Google Scholar] [CrossRef] [PubMed]
- Odum, E.P. The Strategy of Ecosystem Development. Science 1969, 164, 262–270. [Google Scholar] [CrossRef]
- Malik, A.A.; Martiny, J.B.H.; Brodie, E.L.; Martiny, A.C.; Treseder, K.K.; Allison, S.D. Defining Trait-Based Microbial Strategies with Consequences for Soil Carbon Cycling under Climate Change. ISME J. 2020, 14, 1–9. [Google Scholar] [CrossRef]
- Li, C.; Liao, H.; Xu, L.; Wang, C.; He, N.; Wang, J.; Li, X. The Adjustment of Life History Strategies Drives the Ecological Adaptations of Soil Microbiota to Aridity. Mol. Ecol. 2022, 31, 2920–2934. [Google Scholar] [CrossRef]
- Ho, A.; Di Lonardo, D.P.; Bodelier, P.L.E. Revisiting Life Strategy Concepts in Environmental Microbial Ecology. FEMS Microbiol. Ecol. 2017, 93, fix006. [Google Scholar] [CrossRef]
- Chorover, J.; Kretzschmar, R.; Garcia-Pichel, F.; Sparks, D.L. Soil Biogeochemical Processes within the Critical Zone. Elements 2007, 3, 321–326. [Google Scholar] [CrossRef]
- Bowker, M.A.; Maestre, F.T.; Eldridge, D.; Belnap, J.; Castillo-Monroy, A.; Escolar, C.; Soliveres, S. Biological Soil Crusts (Biocrusts) as a Model System in Community, Landscape and Ecosystem Ecology. Biodivers. Conserv. 2014, 23, 1619–1637. [Google Scholar] [CrossRef]
- Garcia-Pichel, F. The Microbiology of Biological Soil Crusts. Annu. Rev. Microbiol. 2023, 77, 149–171. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Liu, Y.; Wang, Z.; Zhao, L.; Zhang, W.; Wang, Y.; Li, X. Variations in Microbial Functional Potential Associated with Phosphorus and Sulfur Cycling in Biological Soil Crusts of Different Ages at the Tengger Desert, China. Appl. Soil Ecol. 2021, 165, 104022. [Google Scholar] [CrossRef]
- Meier, D.V.; Imminger, S.; Gillor, O.; Woebken, D. Distribution of Mixotrophy and Desiccation Survival Mechanisms across Microbial Genomes in an Arid Biological Soil Crust Community. mSystems 2021, 6, e00786-20. [Google Scholar] [CrossRef]
- Tang, K.; Yuan, B.; Jia, L.; Pan, X.; Feng, F.; Jin, K. Spatial and Temporal Distribution of Aerobic Anoxygenic Phototrophic Bacteria: Key Functional Groups in Biological Soil Crusts. Environ. Microbiol. 2021, 23, 3211–3225. [Google Scholar] [CrossRef]
- Cordero, P.R.F.; Bayly, K.; Leung, P.M.; Huang, C.; Islam, Z.F.; Schittenhelm, R.B.; King, G.M.; Greening, C. Atmospheric Carbon Monoxide Oxidation Is a Widespread Mechanism Supporting Microbial Survival. ISME J. 2019, 13, 2868–2881. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Q.; Han, Y.; Zhang, D.; Zhang, C.-C.; Hu, C. Carbon Cycle in the Microbial Ecosystems of Biological Soil Crusts. Soil Biol. Biochem. 2022, 171, 108729. [Google Scholar] [CrossRef]
- Bay, S.K.; Waite, D.W.; Dong, X.; Gillor, O.; Chown, S.L.; Hugenholtz, P.; Greening, C. Chemosynthetic and Photosynthetic Bacteria Contribute Differentially to Primary Production across a Steep Desert Aridity Gradient. ISME J. 2021, 15, 3339–3356. [Google Scholar] [CrossRef] [PubMed]
- Leung, P.M.; Bay, S.K.; Meier, D.V.; Chiri, E.; Cowan, D.A.; Gillor, O.; Woebken, D.; Greening, C. Energetic Basis of Microbial Growth and Persistence in Desert Ecosystems. mSystems 2020, 5, e00495-19. [Google Scholar] [CrossRef] [PubMed]
- Klatt, J.M.; Meyer, S.; Häusler, S.; Macalady, J.L.; de Beer, D.; Polerecky, L. Structure and Function of Natural Sulphide-Oxidizing Microbial Mats under Dynamic Input of Light and Chemical Energy. ISME J. 2016, 10, 921–933. [Google Scholar] [CrossRef]
- Ngosong, C.; Buse, T.; Ewald, M.; Richter, A.; Glaser, K.; Schöning, I.; Ruess, L. Influence of Management Intensity and Environmental Conditions on Microbiota in Biological Soil Crust and Crust-Free Soil Habitats of Temperate Forests. Soil Biol. Biochem. 2020, 144, 107761. [Google Scholar] [CrossRef]
- Ghosh, W.; Dam, B. Biochemistry and Molecular Biology of Lithotrophic Sulfur Oxidation by Taxonomically and Ecologically Diverse Bacteria and Archaea. FEMS Microbiol. Rev. 2009, 33, 999–1043. [Google Scholar] [CrossRef]
- Brocks, J.J.; Love, G.D.; Summons, R.E.; Knoll, A.H.; Logan, G.A.; Bowden, S.A. Biomarker Evidence for Green and Purple Sulphur Bacteria in a Stratified Palaeoproterozoic Sea. Nature 2005, 437, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Bar-Even, A.; Flamholz, A.; Noor, E.; Milo, R. Thermodynamic Constraints Shape the Structure of Carbon Fixation Pathways. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1646–1659. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Dou, Y.; Wang, B.; Xue, Z.; Wang, Y.; An, S.; Chang, S.X. Deciphering Factors Driving Soil Microbial Life-history Strategies in Restored Grasslands. iMeta 2022, 2, e66. [Google Scholar] [CrossRef]
- Treseder, K.K. Ecological Strategies of Microbes: Thinking Outside the Triangle. J. Ecol. 2023, 111, 1625–1628. [Google Scholar] [CrossRef]
- Han, Y.; Wang, Q.; Li, Q.; Hu, C. Active Metabolism and Biomass Dynamics of Biocrusts Are Shaped by Variation in Their Successional State and Seasonal Energy Sources. Sci. Total Environ. 2022, 831, 154756. [Google Scholar] [CrossRef]
- Schimel, J.; Balser, T.C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef]
- Wang, Q.; Han, Y.; Lan, S.; Hu, C. Metagenomic Insight Into Patterns and Mechanism of Nitrogen Cycle During Biocrust Succession. Front. Microbiol. 2021, 12, 633428. [Google Scholar] [CrossRef]
- Lan, S.; Wu, L.; Zhang, D.; Hu, C. Successional Stages of Biological Soil Crusts and Their Microstructure Variability in Shapotou Region (China). Environ. Earth Sci. 2012, 65, 77–88. [Google Scholar] [CrossRef]
- Lan, S.; Ouyang, H.; Wu, L.; Zhang, D.; Hu, C. Biological Soil Crust Community Types Differ in Photosynthetic Pigment Composition, Fluorescence and Carbon Fixation in Shapotou Region of China. Appl. Soil Ecol. 2017, 111, 9–16. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An Extraction Method for Measuring Soil Microbial Biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.M.; Yiu, S.M.; Chin, F.Y.L. IDBA-UD: A de Novo Assembler for Single-Cell and Metagenomic Sequencing Data with Highly Uneven Depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Noguchi, H.; Park, J.; Takagi, T. MetaGene: Prokaryotic Gene Finding from Environmental Genome Shotgun Sequences. Nucleic Acids Res. 2006, 34, 5623–5630. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-Hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Li, R.; Li, Y.; Kristiansen, K.; Wang, J. SOAP: Short Oligonucleotide Alignment Program. Bioinformatics 2008, 24, 713–714. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Qiu, Z.; Mu, R.; Qiao, X.; Zhang, L.; Lian, C.-A.; Deng, C.; Wu, Y.; Xu, Z.; Li, B.; et al. Recovery of High-Qualitied Genomes from a Deep-Inland Salt Lake Using BASALT. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An Adaptive Binning Algorithm for Robust and Efficient Genome Reconstruction from Metagenome Assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An Automated Binning Algorithm to Recover Genomes from Multiple Metagenomic Datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning Metagenomic Contigs by Coverage and Composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Olm, M.R.; Brown, C.T.; Brooks, B.; Banfield, J.F. dRep: A Tool for Fast and Accurate Genomic Comparisons That Enables Improved Genome Recovery from Metagenomes through de-Replication. ISME J. 2017, 11, 2864–2868. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the Quality of Microbial Genomes Recovered from Isolates, Single Cells, and Metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Breister, A.M.; Liu, Y.; Kieft, K.; Cowley, E.S.; Karaoz, U.; Anantharaman, K. METABOLIC: High-Throughput Profiling of Microbial Genomes for Functional Traits, Metabolism, Biogeochemistry, and Community-Scale Functional Networks. Microbiome 2022, 10, 2. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar] [CrossRef]
- Hahn, C.R.; Farag, I.F.; Murphy, C.L.; Podar, M.; Elshahed, M.S.; Youssef, N.H. Microbial Diversity and Sulfur Cycling in an Early Earth Analogue: From Ancient Novelty to Modern Commonality. mBio 2022, 13, e00016-22. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.L.; MacLeod, F.I.; White, R.A.; Visscher, P.T.; Burns, B.P. Microbial Dark Matter Filling the Niche in Hypersaline Microbial Mats. Microbiome 2020, 8, 159. [Google Scholar] [CrossRef] [PubMed]
- Bell, E.; Lamminmäki, T.; Alneberg, J.; Andersson, A.F.; Qian, C.; Xiong, W.; Hettich, R.L.; Frutschi, M.; Bernier-Latmani, R. Active Sulfur Cycling in the Terrestrial Deep Subsurface. ISME J. 2020, 14, 1260–1272. [Google Scholar] [CrossRef] [PubMed]
- Probst, A.J.; Castelle, C.J.; Singh, A.; Brown, C.T.; Anantharaman, K.; Sharon, I.; Hug, L.A.; Burstein, D.; Emerson, J.B.; Thomas, B.C.; et al. Genomic Resolution of a Cold Subsurface Aquifer Community Provides Metabolic Insights for Novel Microbes Adapted to High CO2 Concentrations. Environ. Microbiol. 2017, 19, 459–474. [Google Scholar] [CrossRef]
- Tran, P.Q.; Bachand, S.C.; McIntyre, P.B.; Kraemer, B.M.; Vadeboncoeur, Y.; Kimirei, I.A.; Tamatamah, R.; McMahon, K.D.; Anantharaman, K. Depth-Discrete Metagenomics Reveals the Roles of Microbes in Biogeochemical Cycling in the Tropical Freshwater Lake Tanganyika. ISME J. 2021, 15, 1971–1986. [Google Scholar] [CrossRef]
- Vigneron, A.; Cruaud, P.; Culley, A.I.; Couture, R.-M.; Lovejoy, C.; Vincent, W.F. Genomic Evidence for Sulfur Intermediates as New Biogeochemical Hubs in a Model Aquatic Microbial Ecosystem. Microbiome 2021, 9, 16. [Google Scholar] [CrossRef]
- Cai, R.; He, W.; Liu, R.; Zhang, J.; Zhang, X.; Sun, C. Deep-Sea In Situ Insights into the Formation of Zero-Valent Sulfur Driven by a Bacterial Thiosulfate Oxidation Pathway. mBio 2022, 13, e00143-22. [Google Scholar] [CrossRef]
- Anantharaman, K.; Brown, C.T.; Hug, L.A.; Sharon, I.; Castelle, C.J.; Probst, A.J.; Thomas, B.C.; Singh, A.; Wilkins, M.J.; Karaoz, U.; et al. Thousands of Microbial Genomes Shed Light on Interconnected Biogeochemical Processes in an Aquifer System. Nat. Commun. 2016, 7, 13219. [Google Scholar] [CrossRef] [PubMed]
- Vikram, S.; Guerrero, L.D.; Makhalanyane, T.P.; Le, P.T.; Seely, M.; Cowan, D.A. Metagenomic Analysis Provides Insights into Functional Capacity in a Hyperarid Desert Soil Niche Community. Environ. Microbiol. 2016, 18, 1875–1888. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ou, P.; Zhao, B.; Zhang, W.; Yu, K.; Xie, K.; Zhuang, W.-Q. Assimilatory and Dissimilatory Sulfate Reduction in the Bacterial Diversity of Biofoulant from a Full-Scale Biofilm-Membrane Bioreactor for Textile Wastewater Treatment. Sci. Total Environ. 2021, 772, 145464. [Google Scholar] [CrossRef]
- Houghton, J.L.; Foustoukos, D.I.; Flynn, T.M.; Vetriani, C.; Bradley, A.S.; Fike, D.A. Thiosulfate Oxidation by Thiomicrospira thermophila: Metabolic Flexibility in Response to Ambient Geochemistry. Environ. Microbiol. 2016, 18, 3057–3072. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, B.B.; Findlay, A.J.; Pellerin, A. The Biogeochemical Sulfur Cycle of Marine Sediments. Front. Microbiol. 2019, 10, 849. [Google Scholar] [CrossRef]
- Hu, X.; Liu, J.; Liu, H.; Zhuang, G.; Xun, L. Sulfur Metabolism by Marine Heterotrophic Bacteria Involved in Sulfur Cycling in the Ocean. Sci. China Earth Sci. 2018, 61, 1369–1378. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, H.; Cui, F.; Liu, H.; Xun, L. Recombinant Escherichia coli with Sulfide:Quinone Oxidoreductase and Persulfide Dioxygenase Rapidly Oxidises Sulfide to Sulfite and Thiosulfate via a New Pathway. Environ. Microbiol. 2016, 18, 5123–5136. [Google Scholar] [CrossRef]
- Toohey, J.; Cooper, A. Thiosulfoxide (Sulfane) Sulfur: New Chemistry and New Regulatory Roles in Biology. Molecules 2014, 19, 12789–12813. [Google Scholar] [CrossRef]
- Wong, B.; Brown, L.; Buckingham, R.; Sweet, W.; Russ, B.; Gorensek, M. Sulfur Dioxide Disproportionation for Sulfur Based Thermochemical Energy Storage. Sol. Energy 2015, 118, 134–144. [Google Scholar] [CrossRef]
- Sousa, F.M.; Pereira, J.G.; Marreiros, B.C.; Pereira, M.M. Taxonomic Distribution, Structure/Function Relationship and Metabolic Context of the Two Families of Sulfide Dehydrogenases: SQR and FCSD. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 742–753. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Adams, A.M.; Kieft, K.; Breier, J.A.; Fortunato, C.S.; Sheik, C.S.; Huber, J.A.; Li, M.; Dick, G.J.; et al. Sulfur Cycling Connects Microbiomes and Biogeochemistry in Deep-Sea Hydrothermal Plumes. ISME J. 2023, 17, 1604–1616. [Google Scholar] [CrossRef]
- Klatt, J.M.; Al-Najjar, M.A.A.; Yilmaz, P.; Lavik, G.; de Beer, D.; Polerecky, L. Anoxygenic Photosynthesis Controls Oxygenic Photosynthesis in a Cyanobacterium from a Sulfidic Spring. Appl. Environ. Microbiol. 2015, 81, 2025–2031. [Google Scholar] [CrossRef]
- Voorhies, A.A.; Biddanda, B.A.; Kendall, S.T.; Jain, S.; Marcus, D.N.; Nold, S.C.; Sheldon, N.D.; Dick, G.J. Cyanobacterial Life at Low O2: Community Genomics and Function Reveal Metabolic Versatility and Extremely Low Diversity in a Great Lakes Sinkhole Mat. Geobiology 2012, 10, 250–267. [Google Scholar] [CrossRef]
- Alef, K.; Kleiner, D. Rapid and Sensitive Determination of Microbial Activity in Soils and in Soil Aggregates by Dimethylsulfoxide Reduction. Biol. Fertil. Soils 1989, 8, 349–355. [Google Scholar] [CrossRef]
- Spiese, C.E.; Kieber, D.J.; Nomura, C.T.; Kiene, R.P. Reduction of Dimethylsulfoxide to Dimethylsulfide by Marine Phytoplankton. Limnol. Oceanogr. 2009, 54, 560–570. [Google Scholar] [CrossRef]
- Li, C.-Y.; Cao, H.-Y.; Wang, Q.; Carrión, O.; Zhu, X.; Miao, J.; Wang, P.; Chen, X.-L.; Todd, J.D.; Zhang, Y.-Z. Aerobic Methylation of Hydrogen Sulfide to Dimethylsulfide in Diverse Microorganisms and Environments. ISME J. 2023, 17, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Asher, E.C.; Dacey, J.W.H.; Stukel, M.; Long, M.C.; Tortell, P.D. Processes Driving Seasonal Variability in DMS, DMSP, and DMSO Concentrations and Turnover in Coastal Antarctic Waters. Limnol. Oceanogr. 2017, 62, 104–124. [Google Scholar] [CrossRef]
- Maier, S.; Tamm, A.; Wu, D.; Caesar, J.; Grube, M.; Weber, B. Photoautotrophic Organisms Control Microbial Abundance, Diversity, and Physiology in Different Types of Biological Soil Crusts. ISME J. 2018, 12, 1032–1046. [Google Scholar] [CrossRef]
- Rajeev, L.; da Rocha, U.N.; Klitgord, N.; Luning, E.G.; Fortney, J.; Axen, S.D.; Shih, P.M.; Bouskill, N.J.; Bowen, B.P.; Kerfeld, C.A.; et al. Dynamic Cyanobacterial Response to Hydration and Dehydration in a Desert Biological Soil Crust. ISME J. 2013, 7, 2178–2191. [Google Scholar] [CrossRef]
- Hunt, K.A.; Jennings, R.d.; Inskeep, W.P.; Carlson, R.P. Stoichiometric Modelling of Assimilatory and Dissimilatory Biomass Utilisation in a Microbial Community. Environ. Microbiol. 2016, 18, 4946–4960. [Google Scholar] [CrossRef]
- Smith, A.R.; Kieft, B.; Mueller, R.; Fisk, M.R.; Mason, O.U.; Popa, R.; Colwell, F.S. Carbon Fixation and Energy Metabolisms of a Subseafloor Olivine Biofilm. ISME J. 2019, 13, 1737–1749. [Google Scholar] [CrossRef]
- Li, Y.; Hu, C. Biogeographical Patterns and Mechanisms of Microbial Community Assembly That Underlie Successional Biocrusts across Northern China. npj Biofilms Microbiomes 2021, 7, 13. [Google Scholar] [CrossRef]
- Kristensen, E.; Holmer, M.; Bussarawit, N. Benthic Metabolism and Sulfate Reduction in a Southeast Asian Mangrove Swamp. Mar. Ecol. Prog. Ser. 1991, 73, 93–103. [Google Scholar] [CrossRef]
- Guo, W.; Wen, Y.; Chen, Y.; Zhou, Q. Sulfur Cycle as an Electron Mediator between Carbon and Nitrate in a Constructed Wetland Microcosm. Front. Environ. Sci. Eng. 2020, 14, 52. [Google Scholar] [CrossRef]
- Ouyang, H.; Hu, C. Insight into Climate Change from the Carbon Exchange of Biocrusts Utilizing Non-Rainfall Water. Sci. Rep. 2017, 7, 41525. [Google Scholar] [CrossRef] [PubMed]
- Naidoo, Y.; Valverde, A.; Pierneef, R.E.; Cowan, D.A. Differences in Precipitation Regime Shape Microbial Community Composition and Functional Potential in Namib Desert Soils. Microb. Ecol. 2022, 83, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Daebeler, A.; Kitzinger, K.; Koch, H.; Herbold, C.W.; Steinfeder, M.; Schwarz, J.; Zechmeister, T.; Karst, S.M.; Albertsen, M.; Nielsen, P.H.; et al. Exploring the Upper pH Limits of Nitrite Oxidation: Diversity, Ecophysiology, and Adaptive Traits of Haloalkalitolerant Nitrospira. ISME J. 2020, 14, 2967–2979. [Google Scholar] [CrossRef]
- Malik, A.A.; Puissant, J.; Buckeridge, K.M.; Goodall, T.; Jehmlich, N.; Chowdhury, S.; Gweon, H.S.; Peyton, J.M.; Mason, K.E.; van Agtmaal, M.; et al. Land Use Driven Change in Soil pH Affects Microbial Carbon Cycling Processes. Nat. Commun. 2018, 9, 3890. [Google Scholar] [CrossRef]
- Baumann, K.; Marschner, P. Effects of Salinity on Microbial Tolerance to Drying and Rewetting. Biogeochemistry 2013, 112, 71–80. [Google Scholar] [CrossRef]
- Thompson, L.R.; Williams, G.J.; Haroon, M.F.; Shibl, A.; Larsen, P.; Shorenstein, J.; Knight, R.; Stingl, U. Metagenomic Covariation along Densely Sampled Environmental Gradients in the Red Sea. ISME J. 2017, 11, 138–151. [Google Scholar] [CrossRef]
- Louca, S.; Hawley, A.K.; Katsev, S.; Torres-Beltran, M.; Bhatia, M.P.; Kheirandish, S.; Michiels, C.C.; Capelle, D.; Lavik, G.; Doebeli, M.; et al. Integrating Biogeochemistry with Multiomic Sequence Information in a Model Oxygen Minimum Zone. Proc. Natl. Acad. Sci. USA 2016, 113, E5925–E5933. [Google Scholar] [CrossRef]
- Sherman, R.E.; Fahey, T.J.; Howarth, R.W. Soil-Plant Interactions in a Neotropical Mangrove Forest: Iron, Phosphorus and Sulfur Dynamics. Oecologia 1998, 115, 553–563. [Google Scholar] [CrossRef]
- Cotner, J.B.; Anderson, N.J.; Osburn, C. Accumulation of Recalcitrant Dissolved Organic Matter in Aerobic Aquatic Systems. Limnol. Oceanogr. Lett. 2022, 7, 401–409. [Google Scholar] [CrossRef]
- Mooshammer, M.; Hofhansl, F.; Frank, A.H.; Wanek, W.; Hämmerle, I.; Leitner, S.; Schnecker, J.; Wild, B.; Watzka, M.; Keiblinger, K.M.; et al. Decoupling of Microbial Carbon, Nitrogen, and Phosphorus Cycling in Response to Extreme Temperature Events. Sci. Adv. 2017, 3, e1602781. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The Microbial Nitrogen-Cycling Network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Moran, M.A. The Global Ocean Microbiome. Science 2015, 350, aac8455. [Google Scholar] [CrossRef] [PubMed]
- Cernava, T.; Aschenbrenner, I.A.; Soh, J.; Sensen, C.W.; Grube, M.; Berg, G. Plasticity of a Holobiont: Desiccation Induces Fasting-like Metabolism within the Lichen Microbiota. ISME J. 2019, 13, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Seah, B.K.B.; Antony, C.P.; Huettel, B.; Zarzycki, J.; Schada von Borzyskowski, L.; Erb, T.J.; Kouris, A.; Kleiner, M.; Liebeke, M.; Dubilier, N.; et al. Sulfur-Oxidizing Symbionts without Canonical Genes for Autotrophic CO2 Fixation. mBio 2019, 10, e01112-19. [Google Scholar] [CrossRef]
- Lange, O.L.; Belnap, J.; Reichenberger, H. Photosynthesis of the Cyanobacterial Soil-Crust Lichen Collema tenax from Arid Lands in Southern Utah, USA: Role of Water Content on Light and Temperature Responses of CO2 Exchange. Funct. Ecol. 1998, 12, 195–202. [Google Scholar] [CrossRef]
- Kiene, R.; Bates, T. Biological Removal of Dimethyl Sulphide from Sea Water. Nature 1990, 345, 702–705. [Google Scholar] [CrossRef]
- Bay, S.K.; Dong, X.; Bradley, J.A.; Leung, P.M.; Grinter, R.; Jirapanjawat, T.; Arndt, S.K.; Cook, P.L.M.; LaRowe, D.E.; Nauer, P.A.; et al. Trace Gas Oxidizers Are Widespread and Active Members of Soil Microbial Communities. Nat. Microbiol. 2021, 6, 246–256. [Google Scholar] [CrossRef]
- Muyzer, G.; Stams, A.J.M. The Ecology and Biotechnology of Sulphate-Reducing Bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Ohtsu, I.; Kawano, Y.; Suzuki, M.; Morigasaki, S.; Saiki, K.; Yamazaki, S.; Nonaka, G.; Takagi, H. Uptake of L-Cystine via an ABC Transporter Contributes Defense of Oxidative Stress in the L-Cystine Export-Dependent Manner in Escherichia coli. PLoS ONE 2015, 10, e0120619. [Google Scholar] [CrossRef]
- Hwang, J.-H.; Kim, H.-C.; Choi, J.-A.; Abou-Shanab, R.A.I.; Dempsey, B.A.; Regan, J.M.; Kim, J.R.; Song, H.; Nam, I.-H.; Kim, S.-N.; et al. Photoautotrophic Hydrogen Production by Eukaryotic Microalgae under Aerobic Conditions. Nat. Commun. 2014, 5, 4234. [Google Scholar] [CrossRef] [PubMed]
- Greening, C.; Berney, M.; Hards, K.; Cook, G.M.; Conrad, R. A Soil Actinobacterium Scavenges Atmospheric H2 Using Two Membrane-Associated, Oxygen-Dependent [NiFe] Hydrogenases. Proc. Natl. Acad. Sci. USA 2014, 111, 4257–4261. [Google Scholar] [CrossRef] [PubMed]
- Constant, P.; Chowdhury, S.P.; Pratscher, J.; Conrad, R. Streptomycetes Contributing to Atmospheric Molecular Hydrogen Soil Uptake Are Widespread and Encode a Putative High-Affinity [NiFe]-Hydrogenase. Environ. Microbiol. 2010, 12, 821–829. [Google Scholar] [CrossRef]
- McKinlay, J.B.; Harwood, C.S. Carbon Dioxide Fixation as a Central Redox Cofactor Recycling Mechanism in Bacteria. Proc. Natl. Acad. Sci. USA 2010, 107, 11669–11675. [Google Scholar] [CrossRef]
- Paulo, L.M.; Stams, A.J.M.; Sousa, D.Z. Methanogens, Sulphate and Heavy Metals: A Complex System. Rev. Environ. Sci. Biotechnol. 2015, 14, 537–553. [Google Scholar] [CrossRef]
- Bothe, H.; Schmitz, O.; Yates, M.G.; Newton, W.E. Nitrogen Fixation and Hydrogen Metabolism in Cyanobacteria. Microbiol. Mol. Biol. Rev. 2010, 74, 529–551. [Google Scholar] [CrossRef]
- Williamson, M.; MacArthur, R.H.; Wilson, E.O. The Theory of Island Biogeography. J. Anim. Ecol. 1969, 38, 464. [Google Scholar] [CrossRef]
- Pianka, E.R. On R- and K-Selection. Am. Nat. 1970, 104, 592–597. [Google Scholar] [CrossRef]
- Busi, S.B.; Bourquin, M.; Fodelianakis, S.; Michoud, G.; Kohler, T.J.; Peter, H.; Pramateftaki, P.; Styllas, M.; Tolosano, M.; De Staercke, V.; et al. Genomic and Metabolic Adaptations of Biofilms to Ecological Windows of Opportunity in Glacier-Fed Streams. Nat. Commun. 2022, 13, 1729. [Google Scholar] [CrossRef]
- Song, H.-K.; Song, W.; Kim, M.; Tripathi, B.M.; Kim, H.; Jablonski, P.; Adams, J.M. Bacterial Strategies along Nutrient and Time Gradients, Revealed by Metagenomic Analysis of Laboratory Microcosms. FEMS Microbiol. Ecol. 2017, 93, fix114. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Neilson, J.W.; Kushwaha, P.; Maier, R.M.; Barberán, A. Life-History Strategies of Soil Microbial Communities in an Arid Ecosystem. ISME J. 2021, 15, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- Soong, J.L.; Fuchslueger, L.; Marañon-Jimenez, S.; Torn, M.S.; Janssens, I.A.; Penuelas, J.; Richter, A. Microbial Carbon Limitation: The Need for Integrating Microorganisms into Our Understanding of Ecosystem Carbon Cycling. Glob. Chang. Biol. 2020, 26, 1953–1961. [Google Scholar] [CrossRef]
- Beck, A.; Bernstein, H.; Carlson, R. Stoichiometric Network Analysis of Cyanobacterial Acclimation to Photosynthesis-Associated Stresses Identifies Heterotrophic Niches. Processes 2017, 5, 32. [Google Scholar] [CrossRef]
- de Vries, F.T.; Shade, A. Controls on Soil Microbial Community Stability under Climate Change. Front. Microbiol. 2013, 4, 265. [Google Scholar] [CrossRef]
- Mackelprang, R.; Vaishampayan, P.; Fisher, K. Adaptation to Environmental Extremes Structures Functional Traits in Biological Soil Crust and Hypolithic Microbial Communities. mSystems 2022, 7, e01419-21. [Google Scholar] [CrossRef] [PubMed]
- Kearns, P.J.; Shade, A. Trait-Based Patterns of Microbial Dynamics in Dormancy Potential and Heterotrophic Strategy: Case Studies of Resource-Based and Post-Press Succession. ISME J. 2018, 12, 2575–2581. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Onishi, F.; Shiroyama, M.; Miura, M.; Tanaka, N.; Oshiro, S.; Nonaka, G.; Nakanishi, T.; Ohtsu, I. Improved Fermentative L-Cysteine Overproduction by Enhancing a Newly Identified Thiosulfate Assimilation Pathway in Escherichia coli. Appl. Microbiol. Biotechnol. 2017, 101, 6879–6889. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, M.; Li, Q.; Han, Y.; Wang, Q.; Yang, H.; Li, H.; Hu, C. Sulfur Cycling and Life Strategies in Successional Biocrusts Link to Biomass Carbon in Dryland Ecosystems. Microorganisms 2025, 13, 2594. https://doi.org/10.3390/microorganisms13112594
Zhou M, Li Q, Han Y, Wang Q, Yang H, Li H, Hu C. Sulfur Cycling and Life Strategies in Successional Biocrusts Link to Biomass Carbon in Dryland Ecosystems. Microorganisms. 2025; 13(11):2594. https://doi.org/10.3390/microorganisms13112594
Chicago/Turabian StyleZhou, Maocheng, Qi Li, Yingchun Han, Qiong Wang, Haijian Yang, Hua Li, and Chunxiang Hu. 2025. "Sulfur Cycling and Life Strategies in Successional Biocrusts Link to Biomass Carbon in Dryland Ecosystems" Microorganisms 13, no. 11: 2594. https://doi.org/10.3390/microorganisms13112594
APA StyleZhou, M., Li, Q., Han, Y., Wang, Q., Yang, H., Li, H., & Hu, C. (2025). Sulfur Cycling and Life Strategies in Successional Biocrusts Link to Biomass Carbon in Dryland Ecosystems. Microorganisms, 13(11), 2594. https://doi.org/10.3390/microorganisms13112594





