Abstract
Bacteria utilize chemotaxis to sense the surrounding chemical signals to seek a more favorable survival environment. The chemotaxis process includes signal sensing, signal transduction, and signal response (i.e., regulating flagellar rotation to control motility). Agrobacterium fabrum, as a soil-born facultative phytopathogen, can survive in diverse environments from bulk soil, the rhizosphere, to the plant-associated niches, and needs to cope with diverse challenges from various survival environments. It must recognize a variety of environmental signals and thus has evolved a chemosensory signaling system more complicated than the prototypical chemotaxis system. The chemosensory system of A. fabrum possesses one histidine kinase, but more chemoreceptors, coupling proteins (2 CheWs), and response regulators (2 CheYs and 1 CheB) than the well-studied prototypical system in the model bacterium Escherichia coli, which has only one CheW, one CheY, and fewer chemoreceptors. More response regulators imply that the chemosensory system may involve other physiological functions beyond chemotaxis. In this review, we outline the recent advances in the prototypical chemosensory signaling system and discuss the functions of protein components in A. fabrum’s chemosensory system by comparing those proteins with the homologous proteins observed in the paradigm and other closely related species. Meanwhile, we place particular emphasis on reviewing the data about the chemosensory system of A. fabrum, propose a “one-system two-pathways” model depicting that A. fabrum possibly utilizes one histidine kinase to assemble two chemosensory signaling pathways, and envision future directions for studying this system. The insights provided will aid in understanding the diversity of chemosensory signaling pathways in other organisms and the molecular mechanism mediating the signal crosstalk among chemosensory signaling pathways.
1. Introduction
Bacterial chemotaxis is defined as the behavior of motile bacteria and archaea in which they track a variety of environmental cues and move toward more favorable environment [1]. Through sensing the environmental attractants or repellents, bacterial cells can efficiently locate optimal growth environments [2,3,4]. Bacterial chemotaxis utilizes a highly conserved chemosensory signaling system to transduce signals. Chemotaxis is essential not only for bacteria to locate more favorable environments under nutrient stress [5], but also for various human [6], animal [7,8], and plant pathogens to invade their hosts [9,10,11]. Chemotaxis also significantly affects biofilm formation [12,13,14], adhesion ability [15], polymicrobial communication [16,17,18], bacterial symbiosis with plants [19,20,21,22], and the regulation of second messenger levels [23,24,25]. In addition, chemotaxis is very important in enabling bacteria to move toward biodegradable pollutants, and thus plays a crucial role in the biodegradation of some pollutants that may pose serious environmental and health risks [2,26].
Molecular study of bacterial chemotaxis began in the 1960s, and was pioneered by Adler’s work [1,27]. For over six decades, the molecular mechanism of bacterial chemotaxis has been well understood. The chemosensory pathway begins with the sensing of chemical gradients through chemoreceptors, and then the sensory signals are processed by the core signaling unit (CSU) and transmitted to the flagellar motor by cytoplasmic proteins, causing the bacteria to “run” or “tumble” via the change in flagellar rotation. The chemosensory pathway used by chemotaxis is highly conserved and has been identified in more than half of the sequenced bacterial genomes [28,29]. The chemosensory pathway represents a major signal transduction mechanism in bacteria and has become a paradigmatic model for the study of bacterial signal transduction [30]. What is more important is that the chemosensory system exhibits significant diversity in terms of the proportional composition of core proteins and the involvement of auxiliary proteins [31]. Moreover, the phenomenon of multiple chemosensory systems occurring in a single bacterium is also very common [29,32,33].
Agrobacterium fabrum (formerly known as Agrobacterium tumefaciens) is a remarkable and well-studied bacterium that has significantly influenced the fields of plant biology and agricultural biotechnology [34,35]. This Gram-negative soil bacterium is renowned for its unique ability to transfer the segments of its DNA, known as the T-DNA (transferred DNA), into the cells of plants [36]. In nature, the transfer and integration of the T-DNA into the plant host genome leads to the production of abnormal plant growths, or tumors, which is how Agrobacterium fabrum earned its former name “tumefaciens” (meaning tumor-forming) [20,37]. However, this ability to transfer DNA has also made it an invaluable tool in genetic engineering to create transgenic plants with improved traits [38,39]. A. fabrum is a facultative phytopathogen and able to live in diverse environments, such as soil, the rhizosphere, and the plant-associated niches, which are challenged by multiple stressors, such as diverse nutritional sources, microbial competition, and plant defenses [40,41]. It is required to sense various environmental signals. In fact, A. fabrum has chemotaxis responses to a variety of chemical substances [2,42]. Over twenty genome sequences derived from A. fabrum are publicly available [40]. All the published genomes of A. fabrum strains are annotated to encode a single complete set of core chemotaxis proteins—without the presence of multiple sets or variations in the component proportions of core proteins among different strains [43], demonstrating that neither multiple sets nor compositional variations in core chemotaxis proteins occur in different A. fabrum strains, though the number of chemoreceptors may differ among strains from different ecological environments. Such composition of core chemotaxis proteins is also found in other plant-associated bacteria, such as Sinorhizobium meliloti [44,45], Azorhizobium caulinodans [46], Rhodobacter sphaeroides, and Caulobacter crescentus [47]. Therefore, A. fabrum’s chemosensory system may potentially represent a distinct class of chemosensory systems prevalent in plant–microbe symbionts. A review of this chemosensory system will be helpful for understanding the diversity of chemosensory pathways and the ecological functions of the chemosensory system in plant-associated bacteria. Notably, while numerous reviews have addressed prototypical chemosensory systems, none have dealt with the recent advances in A. fabrum’s chemosensory system.
2. Prototype of Chemosensory Signaling System and Co-Existence of Multiple Systems
Most bacterial signaling transduction adopts a two-component system, which consists of a sensory kinase and a response regulator. Typically, the sensory kinase is a transmembrane protein that senses the extracellular signal and transmits the signal to the cytoplasmic response regulator through phosphorylating the latter [2]. The phosphorylated response regulator regulates the corresponding physiological functions [48]. The chemotaxis signaling pathway is a special case of a two-component system [2,31]. The sensory kinase of the chemotaxis pathway is divided into three different proteins: chemoreceptor (or methyl-accepting chemotaxis protein, MCP), coupling (or adaptor) protein, and histidine kinase. Such functional segregation of signal sensing (chemoreceptor) and phosphorylation activation (histidine kinase) might be beneficial to the evolutionary adaptation of the chemotaxis system to sense different chemoeffectors in new environmental niches via the acquisition or loss of specific chemoreceptors without affecting the function of the histidine kinase [49,50].
Besides chemotaxis, this chemosensory signaling system is reported to be involved in other physiological functions [8,11]. According to their functions, chemosensory signaling systems can be classified into three groups: those regulating the flagellar motility (Fla), type IV pilus-based motility (Tfp), and those involved in non-motility functions (or alternative cellular functions, ACF) (such as biofilm formation, cell morphology, and the control of second messengers) [11,31]. The Fla group is the most diverse and further classed into 17 subclasses (F1–F17), while Tfp and ACF groups contain a single system class [11,29,31]. The core of the chemosensory signaling system contains at least four essential components, namely the chemoreceptor (MCP), coupling protein (CheW), histidine kinase (CheA), and response regulator (CheY).
2.1. Prototype of Chemosensory Signaling System
The signaling pathway of the chemosensory system is highly conserved across prokaryotes and best understood in Escherichia coli [28,31]. The chemosensory signaling system consists of two signaling modules: one for rapid signal transduction to regulate the physiological functions and another for slower adaptation to the new environmental signal strength [49]. The rapid signal transduction module comprises the MCP–CheW–CheA ternary complex (Figure 1A) and CheY. The binding of ligand to the chemoreceptor MCP causes the conformation change that is transmitted to the histidine kinase CheA and modulates the kinase activity. The kinase CheA can phosphorylate the response regulator CheY. Therefore, CheA, together with a phosphatase CheZ that can specifically dephosphorylate the phosphorylated CheY (CheY-P), controls the cytoplasmic level of CheY-P. The diffusible CheY-P regulates the corresponding functions. For chemotaxis, CheY-P binds to the flagellar motor to change the flagellar rotation. The slower adaptation module is achieved by a methyltransferase CheR and a methylesterase CheB [51]. CheR is an MCP-specific methyltransferase that can methylate the glutamyl residues of chemoreceptors. Due to its ability to accept methyl groups, the chemoreceptor is named as the methyl-accepting chemotaxis protein (MCP). CheB can also be phosphorylated by CheA. The phosphorylated CheB (CheB-P) removes methyl groups from the MCPs and controls the methylation state of the MCPs in conjunction with CheR. The methylation state of the MCPs affects the MCPs’ ability to activate CheA and thus adjusts MCPs’ sensitivity to the ligand to adapt the new ligand concentration [52,53].
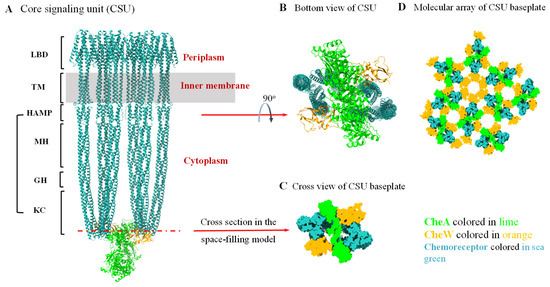
Figure 1.
Structure of core signaling unit (CSU) and architecture of the chemosensory array baseplate in the prototypical chemosensory system of E. coli. (A) All-atom ribbon model of the native E. coli CSU (PDB 8C5V). Specific regions of the chemoreceptors are labeled in the left side. LBD: the ligand-binding domain; TM: transmembrane helical region; CSD: cytoplasmic signaling domain; HAMP: domain existing in histidine kinases, adenylate cyclases, methyl-accepting chemotaxis proteins, and phosphatases; MH: methylation helix bundle; GH: glycine hinge; KC: kinase control region. (B) The bottom view of the CSU showing the baseplate structure of the CSU. (C) Cross sectional view of the CSU baseplate in the space-filling CSU model showing the roughly hexagonal baseplate. (D) The chemosensory array baseplate model assembled from the cross-sectional views of the repeated CSU baseplate. CheW and CheA P5-domain form a hexameric ring in an alternating order. Additional CheW molecules may change the ratio of CheA P5-domain to CheW molecules in the array and possibly form a hexameric CheW ring without CheA P5-domain.
Typical chemoreceptors, which sense the environmental signal outside the bacterial cell, are generally transmembrane proteins containing a periplasmic ligand-binding domain (LBD), a transmembrane (TM) helical region, and a cytoplasmic signaling domain (CSD), which comprise four regions (or sub-domains), the HAMP (found in histidine kinase, adenylyl cyclase, methyl-binding protein, and phosphatase) domain, methylation helix (MH) bundle, flexible region (or glycine hinge, GH), and protein-interacting region (or kinase control region, KC) [54]. Two chemoreceptors form a homodimer. Three chemoreceptor dimers further make up a trimer-of-dimers. Two trimers-of-dimers, together with one CheA dimer and two CheW monomers, constitute the smallest functional unit, called the core signaling unit (CSU) (Figure 1A). CheA and CheW bind to the kinase control region of the chemoreceptor to form the baseplate of the CSU (Figure 1B,C) [51,55,56]. Many CSUs then form into larger structures, in which the trimers-of-dimers of the chemoreceptors are packed into a supramolecular array across the bacterial inner membrane at the cell pole. The interaction between the CheW and CheA P5-domain forms the alternating hexameric rings that link the CSUs together to assemble the baseplate of the supramolecular array. Variation in the ratio of CheA to CheW can change the composition of the hexameric ring. Additional CheW molecules can form hexameric CheW rings in the supramolecular array (Figure 1D). Recent results have demonstrated that the hexameric CheW rings enhance the response sensitivity and cooperativity of the chemosensory array, but are not indispensable for the assembly of CheA-containing arrays [57].
In recent years, studies on the detailed structure of the CSU and the signaling mechanism by which the transmembrane chemoreceptor propagates the periplasmic ligand-binding signal to its membrane-distal tip, which is 200 Å away, to control the kinase CheA have made great strides. The sub-nanometer resolution three-dimensional map of a complete CSU was acquired by using cryo-electron tomography, which reveals the structure of the CSU in unprecedented detail [30,58,59,60]. More recently, the combination of cryo-electron tomography and molecular simulation presents the complete atomic structure of the native CSU, which provides new insight into the interactions between neighboring chemoreceptor ligand-binding domains, and elucidates previously unresolved interactions between individual CheA domains [56]. Hydrogen deuterium exchange mass spectrometry was employed to measure the signaling-related allosteric changes in the chemoreceptors, CheW and CheA within functional complexes, revealing that the cytoplasmic domain of the chemoreceptor remains disordered even within functional complexes and this intrinsic disorder plays a crucial mechanistic role in controlling CheA kinase activity through long-range allosterically stabilizing the catalytic domain of CheA [61,62]. Protein in vivo cross-linking and Förster resonance energy transfer (FRET)-based kinase assays were used to distinguish the transitional changes in each subunit in the structure and interacting interface, which are caused by various dynamic stimulus signals [63,64,65]. All these recent results are very useful for understanding the details of how the signals are mechanically transmitted to CheA.
2.2. Co-Existence of Multiple Chemosensory Systems
Comparative analysis of bacterial genome sequences demonstrated that the chemosensory system presents in most prokaryotic species and that more than half of all motile bacteria have multiple chemosensory signaling systems in their genomes [29,66]. Experimental evidence from both genetic investigation and cryo-EM observation also demonstrated that bacteria may express multiple chemoreceptor arrays segregated into distinguishable assemblies [11,67]. An “average” bacterial genome has 14 chemoreceptor genes [68]. The cytoplasmic domains of various chemoreceptors have different lengths due to the number of seven-residue repeats inserted in the cytoplasmic signaling domain although they are highly conserved [69]. Arrays are only formed among chemoreceptors with the same physical length, resulting in distinct chemoreceptor arrays in many bacterial cells [70,71]. Recently, more and more experimental observations have demonstrated that different types of chemoreceptor arrays present in a single bacterial cell [32,72,73,74,75]. Moreover, protein components encoded by the genes of different chemotaxis system clusters can assemble into promiscuous chemosensory signaling arrays [76,77]. The signals transduced by various chemosensory signaling arrays may be involved in different physiological functions. The coexistence of multiple chemosensory signaling pathways within a cell results in the crosstalk between chemosensory systems [13,72,76] and raises the questions of how these signals from different systems are coordinated in the signal transduction and functionally separated at the level of response regulators [78].
3. Chemosensory-Related Proteins Encoded by Agrobacterium fabrum Genome
Comparative genomics analysis of 22 A. fabrum genomes showed that all A. fabrum genomes encode only one chemosensory histidine kinase CheA, two CheWs, and two CheYs, signifying that A. fabrum has only one complete set of core chemosensory protein components and that the chemosensory systems in different strains are highly similar, although different A. fabrum genomes encode different numbers of chemoreceptors [43]. In this review, we will only discuss the chemosensory system in the model strain C58 of A. fabrum due to the differences in some accessory chemosensory proteins and chemoreceptors in different strains. The genome of A. fabrum C58 carries one chemotaxis gene cluster, which encodes most of the main chemosensory protein components, including one copy of CheA, CheB, CheR, CheD, CheS, and MCP and two copies of CheY. Besides the chemotaxis gene cluster, A. fabrum C58 has two CheW-encoding genes and an additional nineteen MCP-encoding genes scattered on different locations of the genome [20,43].
3.1. Chemoreceptor (Or Methyl-Accepting Chemotaxis Protein, MCP)
Bacterial chemoreceptors detect a variety of signals. In fact, they vary widely in sensing mode, cellular location, protein topology, and above all, the type of LBD, even though their CSDs are highly conserved. Over a hundred different types of LBD have been identified among chemoreceptors, but only a few of them are prevalent [79]. Based on LBD and membrane topology, chemoreceptors can be classified into four major classes: class I, the typical transmembrane chemoreceptor class with a periplasmic LBD; class II, the transmembrane chemoreceptors with an N-terminal cytoplasmic LBD; class III, the transmembrane chemoreceptors with a cytoplasmic LBD after the last transmembrane helix; and class IV, the cytoplasmic (soluble) chemoreceptors [80]. Some LBDs of the same type recognize structurally very different ligands, whereas some ligands can be recognized by a number of different LBD types [81]. The lack of a structure–function correlation in LBDs prevents a reliable prediction of the function of chemoreceptors by extrapolation from the experimentally studied homologs. Moreover, some chemoreceptors, especially those without LBDs, indirectly recognize their ligands through the interaction partner proteins [3,82].
A. fabrum C58 has a total of 20 chemoreceptor-encoding genes. Only one gene is located on the chemotaxis gene cluster, while 19 genes are scattered in various locations of the genome. In addition to 13 on the circular chromosome and 5 on the linear chromosome, the Ti and At plasmids each carry one chemoreceptor-encoding gene [20]. The LBDs and categories of all 20 chemoreceptors are summarized in Figure 2. Among the 20 chemoreceptors, 14 contain at least one transmembrane region: 12 are typical transmembrane chemoreceptors with periplasmic LBDs, and 2 are transmembrane chemoreceptors with cytosolic LBDs. The remaining six chemoreceptors lack transmembrane regions and are classified as cytosolic chemoreceptors [43]. Twelve chemoreceptors carry four known LBD types (calcium channels and chemotaxis, CACHE; cyclases/histidine kinases associated sensory extracellular fold, CHASE; found in Per-Arnt-Sim protein, PAS; Protoglobin), while the LBD types of the remaining eight are unknown [83]. The functions of seven A. fabrum chemoreceptors have been experimentally verified: (1) The atu0514-encoded chemoreceptor affects the chemotactic responses of A. fabrum to a broad range of chemoeffectors [84]; (2) The atu0526-encoded chemoreceptor is the only chemoreceptor that recognizes formic acid [85]; (3) The atu1912-encoded chemoreceptor senses pyruvate and propionate [86]; (4) Deletion of the atu1027 gene abolishes the aerotactic response of A. fabrum to atmospheric air [87]; (5) The chemoreceptor encoded by atu0373 is the primary chemoreceptor for choline, acetylcholine, betaine, and L-carnitine in A. fabrum [88]; (6) The atu0872-encoded chemoreceptor is a phenolic acid-sensing one [89]; and (7) The atu2173-encoded chemoreceptor broadly mediates amino acid chemotaxis, responding to all 17 proteinic amino acids except Glu, Asp, and Gly [90].
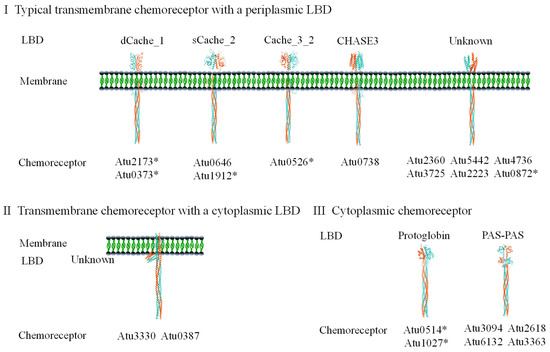
Figure 2.
The chemoreceptors annotated in the A. fabrum C58 genome. Chemoreceptors are named after the genes that encode them. Chemoreceptors marked with an asterisk (*) has been experimentally characterized. LBDs were predicted based on Pfam database annotations [83]. Categories were assigned based on predicted transmembrane region number and localization in each chemoreceptor.
According to the classification of chemoreceptors based on the number of heptad repeats in the flexible region, all of the A. fabrum chemoreceptors belong to the category of 36H [70]. Sequence alignment of 20 A. fabrum chemoreceptors showed that their sequences in the protein interaction region, which interacts with CheA and CheW, are highly conserved with 95% identical residues. However, individual key residues, which are directly involved in the interaction with CheA and CheW, show divergence among 20 chemoreceptors, implying that different chemoreceptors possibly possess different affinities to interact with CheA and CheW. Sequence analysis of 20 A. fabrum chemoreceptors also showed that only 9 chemoreceptors possess a C-terminal pentapeptide for tethering two chemotaxis-adaptational proteins, CheB and CheR.
3.2. Coupling Protein, CheW
CheW is an adaptor protein that couples the histidine kinase CheA to chemoreceptors to form the CSU [91,92]. The genome of A. fabrum C58 carries two CheW-encoding genes, atu2075 and atu2617. Neither is located on the chemotaxis gene cluster, but downstream of atu2617 lies a cytosolic chemoreceptor-encoding gene, atu2618 [93]. We are not sure whether this gene organization indicates that CheW encoded by the atu2617 gene preferentially couples CheA to the cytoplasmic chemoreceptors rather than the transmembrane chemoreceptors. These two CheWs share 47% identical residues, but key residues involved in the interactions with the chemoreceptor and CheA show high divergence, suggesting that they may have distinct affinities for coupling CheA to different chemoreceptors. Reverse genetic experiments via gene deletion verified that CheW2617 (encoded by atu2617, CheW2) had a significantly more severe impact on A. fabrum chemotaxis than CheW2075 (encoded by atu2075, CheW1), even when both CheWs were expressed by using the same system and thus had identical molecular counts [93]. However, both CheWs are very important for the chemotaxis of A. fabrum. Only when both CheWs are deficient does A. fabrum chemotaxis completely disappear. Experimental results also showed that the molecular counts of the two CheWs differ in the wild-type cells [93].
3.3. Histidine Kinase, CheA
CheA is the key player in the chemosensory signaling system and responsible for transducing the allosterically mechanistic signal from the chemoreceptors into the chemical signal of phosphorylated CheY (CheY-P). In the prototypical CSU, CheA exists in dimeric form and one CheA subunit uses ATP to trans-phosphorylate the other. Phosphorylated CheA then transfers the phosphoryl group to the response regulators, such as CheY or CheB [2]. CheA proteins from E. coli and Thermotoga maritima consist of five domains. From the N-terminus to the C-terminus, the five domains of CheA are labeled P1 through P5 (P1: histidine-containing phosphotransfer domain, Hpt; P2: P2/CheY-binding domain; P3: dimerization domain, H-kinase_dim; P4: histidine kinase domain, HATPase_c; P5: CheW-like domain, homologous to the CheW protein) [94]. These five CheA domains were recognized as members of conserved domain families. However, comparative analysis of 13,673 CheA protein sequences from 7367 genomes showed that only 46% of the CheA protein sequences have the classical five domain architecture [95].
The genome of A. fabrum C58 encodes only one CheA. A. fabrum CheA is encoded by the atu0517 gene located on the chemotaxis gene cluster. We compared A. fabrum CheA with E. coli CheA and found that A. fabrum CheA has the classical five domain architecture, but contains 98 additional residues inserted between the P2 and P3 domains as compared with E. coli CheA. It is unknown whether the long residue insertion is very important for the function of A. fabrum CheA. Moreover, only 30.83% of the residues of A. fabrum CheA are identical to those of E. coli CheA. In particular, when the corresponding domains of A. fabrum CheA and E. coli CheA are individually compared, the P2/CheY-binding domain shows the greatest sequence divergence, with all other domains exhibiting less divergence than this domain, which implies that the binding between CheA and CheY in A. fabrum may adopt a mechanism different from that in E. coli.
3.4. Response Regulators, CheY and CheB
Response regulators belong to a super protein family that regulates diverse signal relays via the phosphorylation of one of their conserved aspartate residues. They are characterized by the presence of a conserved receiver (REC, or Response_reg in Pfam) domain with a β5α5 fold structure, a central five-stranded parallel β sheet surrounded by five α helices [96]. The conserved aspartate residue in the receiver domain of the response regulator accepts the phosphoryl group from auto-phosphorylated histidine kinase, resulting in response regulator activation. In most cases, the response arising from the activated response regulator should be terminated in a timely manner. Therefore, all response regulators are able to auto-dephosphorylate. However, auto-dephosphorylation is often unregulated and thus, in some cases, bacteria have to evolve a specific phosphatase to remove the phosphoryl group from the activated response regulator [97]. Usually, a typical chemosensory system contains two types of response regulators, CheY and CheB.
In general, CheY is used to regulate the flagellar motor [98]. However, many bacteria have multiple CheYs and some CheYs do not regulate the flagellar motor [99]. For example, both Azorhizobium caulinodans [46,97] and Rhizobium meliloti [45] encode only one CheA, but two CheYs that have a clearly different influence on the chemotactic response. The chemotaxis gene cluster of A. fabrum C58 has two CheY-encoding genes, atu0516 and atu0520. Both A. fabrum CheY variants possess the typical β5α5 fold structure, but they share only 35.66% identical amino acid residues and their residues corresponding to the E. coli CheY residues that were identified to interact with CheA and motor protein FliM are divergent. CheA-dependent cellular CheY localization assays and in vitro pull-down experiments showed that the affinity of atu0516-encoded CheY (CheY516, CheY1) to CheA is approximately 1.6-fold higher than that of atu0520-encoded CheY (CheY520, CheY2), but the affinity of CheY2 to FliM is approximately 5-fold higher than that of CheY1 [100]. These results suggest that two A. fabrum CheYs have distinct functions. The function of regulating flagellar motility is likely performed by CheY2, whereas CheY1 probably has additional physiological functions beyond flagellar motility regulation.
CheB is the other type of response regulator in the chemosensory system. Unlike CheY, which consists solely of a REC domain, CheB is a typical response regulator comprising two domains (an N-terminal REC domain and a C-terminal methylesterase/deamidase effector domain) connected by a long linker region [101,102]. In fact, over three-fourths of response regulators (approximately 77%) are two-domain proteins containing a REC domain and an effector (or output) domain. The REC domains in various response regulators are highly conserved, whereas the effector domains, which are responsible for outputting the response, vary widely in structure and function depending on the specific response regulator [96]. CheB’s effector domain is a methylesterase/deamidase enzyme domain that can catalyze the demethylation of specific methyl glutamate residues formed by methyltransferase CheR on the chemoreceptors and the deamidation of specific glutamine residues in the methyl-accepting region of the chemoreceptors to create some methyl-accepting sites [103]. However, some CheBs, for example, Bacillus subtilis CheB, may not have the deamidase activity. Similarly to CheY, CheB’s catalytic activity is activated through phosphorylation via CheA. Phosphorylated CheB (CheB-P) is tethered to the C-terminal pentapeptides of chemoreceptors and then recruited to the chemoreceptor substrate [33,104,105]. CheB is recruited to the chemoreceptor array as stimulated by a repellent and dissociates from the array once adaptation is complete [106].
A. fabrum CheB is encoded by the atu0519 gene in the chemotaxis gene cluster and has two CheB-characterized domains. CheB is highly conserved. A. fabrum CheB has 46.76% of its residues identical to those of E. coli CheB, 46.91% of its residues identical to those of Pectobacterium atrosepticum CheB, and 47.61% of its residues identical to those of Salmonella typhimurium CheB. The crystal structures of both P. atrosepticum CheB and S. typhimurium CheB were solved. According to the comparison of the sequences and structures of these CheBs, the residues in the long linker between the two domains are more divergent than the residues in the REC and effector domains. However, the chemoreceptor-tethering region of CheB is located on the N-terminal part of the linker with high residue divergency. Therefore, the high residue divergency in this region results in the fact that CheBs from various bacteria have significantly different affinities to tether to the C-terminal pentapeptide of the chemoreceptor. For example, CheBs from E. coli and S. typhimurium can tether to the C-terminal pentapeptide of the chemoreceptor, but P. atrosepticum CheB cannot [107]. Nine A. fabrum chemoreceptors possess the C-terminal pentapeptide and thus A. fabrum CheB should have the capacity to recognize the C-terminal pentapeptide.
3.5. Methyltransferase and Deamidase for Chemoreceptor Modification, CheR and CheD
CheR is highly conserved and exists in almost all chemosensory systems [29]. A. fabrum CheR was annotated to be encoded by the atu0518 gene. CheR is an S-adenosylmethionine-dependent methyltransferase, which belongs to a subclass of the super methyltransferase family that catalyzes the methylation of a variety of substrates and ubiquitously exists across all life forms [108]. CheR catalyzes the methylation of specific glutamyl residues in chemoreceptors and, together with phosphorylated CheB, regulates the methylation level of the chemoreceptors to adapt the new signal strength. Like CheB, different CheRs have different specificities and affinities to tether to the C-terminal pentapeptide of the chemoreceptor and to methylate the chemoreceptors [105,109,110]. The structure of CheR has been solved in some bacteria. For example, CheRs from S. typhimurium and B. subtilis possess a similar structure with a smaller N-terminal helical domain linked to a larger C-terminal α/β domain via a single polypeptide linker and a small antiparallel β sheet subdomain appended to the C-terminal domain [108]. Sequence comparison and structure prediction showed that A. fabrum CheR is highly similar to these experimentally studied CheRs. Therefore, A. fabrum CheR should have functions highly similar to other homologous CheRs, although no experimental study on A. fabrum CheR is reported.
CheD homologs are found in most nonenteric chemotactic bacteria, although they are not present in enterobacteria like E. coli. The best-studied CheD is B. subtilis CheD [111,112]. Other relatively well-characterized CheDs are the CheDs from Thermotoga maritima [113], Borrelia burgdorferi [114], and Pseudomonas aeruginosa [115]. CheD not only deamidates the glutamine residues in a conserved structural motif of the chemoreceptors to create some methyl-accepting sites, but also hydrolyzes glutamyl-methylesters at the modification sites of the chemoreceptors [113]. However, all these experimentally studied CheDs possess specificity to the chemoreceptor. They can only bind and deamidate some specific chemoreceptors. The A. fabrum atu0521 gene was annotated to encode CheD. A. fabrum CheD shows relatively low sequence identity to other CheD homologs (14.36% identity with B. subtilis CheD, 29.12% identity with T. maritima CheD, 26.5% identity with P. aeruginosa CheD, 22.16% identity with B. burgdorferi CheD). Therefore, more experimental data are required to verify the functions of A. fabrum CheD.
3.6. The Other Chemosensory-Related Proteins Encoded by the A. faberum Genome
Besides the above-mentioned chemotaxis proteins, the chemotaxis gene cluster in A. fabrum has two genes, atu0515 and atu0522, which are annotated to encode conserved hypothetical proteins, respectively. According to the locations of these two genes in the gene cluster and the sequence analysis of their encoded proteins, atu0515-encoded proteins and atu0522-encoded proteins are, respectively, very similar to the CheS and CheT that were identified in Sinorhizobium meliloti.
The atu0515-encoded protein has a sequence highly homologous to S. meliloti CheS, whose homolog is not present in the model chemotaxis system, but is found in other members of α-proteobacteria, such as Sinorhizobium medicae, A. fabrum, Rhizobium leguminosarum, Rhodobacter sphaeroides, and Caulobacter crescentus [47]. CheS is a single-domain protein containing the STAS (sulfate transporter and anti-sigma factor antagonist) domain. Most of the small STAS domain-only proteins are anti-sigma factor antagonists, which are involved in the regulation of sigma factors and anti-sigma factors [116]. S. meliloti CheS can bind with CheA to form a CheA/CheS complex and affect the binding of CheA to CheY. Furthermore, S. meliloti CheS helps CheA to differentiate two CheYs and results in the transfer of the phosphoryl group from one CheY to another CheY via CheA [47]. A. fabrum CheS exhibits a sequence identity of 58.59% to S. meliloti CheS, demonstrating that A. fabrum CheS should have functions similar to S. meliloti CheS.
The protein encoded by the atu0522 gene is highly homologous to the most recently reported CheT that was identified in S. meliloti [44]. The genes encoding CheT homologs are also found in other closely related α-proteobacteria such as S. medicae, R. leguminosarum, C. crescentus, and Hoeflea sp. 10. The sequences of CheT homologs do not show significant homology with any known chemotactic protein in the chemotaxis paradigm model. Sequence analysis shows that CheT has a phosphatase motif (DXXXQ sequence). CheT structure predicted by AlphaFold is highly similar to the crystal structure of E. coli CheZ, which is a CheY-specific phosphatase terminating the CheY response signal in the paradigm model. S. meliloti CheT can differentiate two CheYs and enhance the dephosphorylation of only one phosphorylated CheY. S. meliloti CheT can form a complex with CheR and both the CheT and CheR in the CheT/CheR complex can maintain their original activities [44]. The protein encoded by the A. fabrum atu0522 gene exhibits a sequence identity of 70% to S. meliloti CheT. Therefore, the functions of atu0522-encoded CheT should be very similar to those of S. meliloti CheT.
4. The Possible Signaling Pathways in the Agrobacterium fabrum Chemosensory System
The chemosensory signaling system is particularly important to phytopathogenic bacteria [117,118,119]. Up to 90% of phytopathogenic bacteria have chemotaxis-related genes [120], whereas such genes are found in only 47% of all bacteria [79]. Phytopathogenic bacteria often have multiple chemosensory systems and a high number of chemoreceptors, almost twice the number in those species not classified as plant-associated bacteria [120]. However, the A. fabrum genome encodes only one histidine kinase CheA although it contains twenty chemoreceptor-encoding genes, two CheW-encoding genes, and two CheY-encoding genes. CheA is the central component of the chemosensory signaling system. Based on the understanding of the established chemosensory system, A. fabrum can only assemble one complete set of the chemosensory system due to the presence of only one histidine kinase CheA, but one chemosensory system cannot illustrate why A. fabrum needs two CheWs and two CheYs and how A. fabrum possibly utilizes one chemosensory system to regulate various other physiological processes except for the flagellar motility. A. fabrum also has inducible type IV pili [34]. However, it remains unknown whether and how the chemosensory system mediates type IV pilus-based motility. Moreover, the A. fabrum genome encodes three additional chemosensory-related proteins (CheD, CheS, and CheT) that are not found in the prototypical model chemosensory system. The molecular mechanism of signal transduction and regulation unveiled in the model chemosensory system is also unable to rationally illustrate why the A. fabrum chemosensory signaling system requires CheS, CheT, and CheD. Therefore, the A. fabrum chemosensory signaling system should be more complicated than the established chemosensory systems.
It should be pointed out that the characteristic composition of A. fabrum chemosensory-related protein components is not unique in bacteria. As mentioned above, many bacteria, such as Azorhizobium caulinodans [46,97], Rhizobium meliloti [45], Sinorhizobium medicae, Rhizobium leguminosarum, Rhodobacter sphaeroides, and Caulobacter crescentus [47], also have the composition of one histidine kinase for two coupling proteins and multiple response regulators, as well as the additional proteins (CheD, CheS, and CheT) that are not found in the prototypical chemosensory system. Therefore, the molecular mechanism of signal transduction and regulation in the A. fabrum chemosensory system may become a typical representative of a large group of plant-associated bacteria.
By integrating all published experimental data on the A. fabrum chemosensory signaling system and comparing its chemosensory protein components with homologs, we propose a model to describe two signaling pathways in the chemosensory system of A. fabrum and the possible regulation between the two pathways (Figure 3). In this model, since two CheWs have different affinities to couple CheA to different chemoreceptors [93], each CheW can preferentially couple CheA to the chemoreceptors that have high affinities to the corresponding CheW and form the MCP–CheW–CheA CSU without another CheW. As a result, two CheWs are possibly able to assemble two types of CSUs: the MCP–CheW2617–CheA CSU and the MCP–CheW2075–CheA CSU. One CheW can be partially complemented by the other only when the molecular concentration of one CheW is too small to compete with the other. These two CSUs may have distinct affinities to bind two CheYs due to the different CheWs [100] and accordingly each CSU can only phosphorylate the CheY possessing high affinity. According to the effects of two CheWs and two CheYs on A. fabrum chemotaxis [93,100], the CSU with CheW2617 may preferentially phosphorylate CheY520, whereas the CSU with CheW2075 may mainly phosphorylate CheY516. Two A. fabrum CheYs may be used to regulate different physiological functions. Only CheY520 was supposed to directly regulate flagellar motor. Under normal conditions, each CSU phosphorylates its cognate CheY to regulate specific downstream functions. Consequently, the A. fabrum chemosensory system may form two signaling pathways (Figure 3). Cross-phosphorylation may occur only when the cognate CheY is limiting.
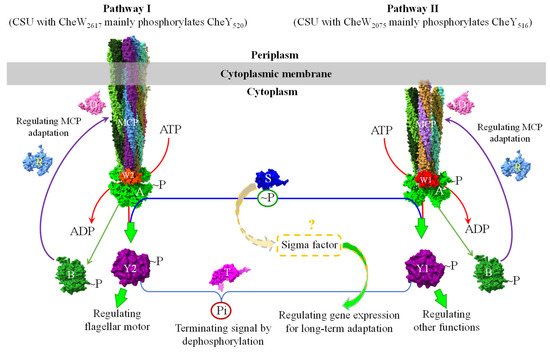
Figure 3.
A schematic model describing the chemosensory signaling system of A. fabrum. Two CheWs couple CheA to different chemoreceptors depending on their affinities to the chemoreceptors to assemble two different CSUs (core signaling units) and thus possibly form two chemosensory signaling pathways, Pathway I and Pathway II. Two CheWs may affect the affinities of CheA to two CheYs. CSU with CheW2617 mainly (or preferentially) phosphorylates CheY520. CSU with CheW2075 mainly (or preferentially) phosphorylates CheY516. CheY520 mainly regulates flagellar motor, whereas CheY516 is supposed to mainly regulate other functions. CheS is supposed to regulate the distribution of the phosphoryl group between two CheYs via CheA and the gene expression via interacting with some sigma factors. CheT possesses different capabilities to dephosphorylate two phosphorylated CheYs. Thus, CheS and CheT possibly mediate the signal interchange between two pathways. CheD, CheR and the phosphorylated CheB regulate the chemoreceptors’ sensitivities to adapt the changing ligand concentration. For the detailed functions of these proteins, please see the text. Abbreviations: A, CheA; B, CheB; CSU, core signaling unit; D, CheD; MCP, chemoreceptor; R, CheR; S, CheS (encoded by atu0515); T, CheT (encoded by atu0522); W1, CheW1 (encoded by atu2075; CheW2075); W2, CheW2 (encoded by atu2617; CheW2617); Y1, CheY1 (encoded by atu0516; CheY516); Y2, CheY2 (encoded by atu0520; CheY520); ~P, phosphoryl group; Pi, inorganic phosphate.
CheS in S. meliloti (a species closely related to A. fabrum) assists CheA in distinguishing between the two CheYs, enabling CheA to mediate cyclic phosphoryl group transfer between them via a CheA–relay mechanism [47]. CheT directly mediates the differential dephosphorylation of the two phosphorylated CheYs [44]. Through these mechanisms, we speculate that CheS and CheT collectively regulate the transfer and distribution of phosphoryl groups between the two CheYs. Therefore, it is plausible that A. fabrum CheS and CheT modulate signal crosstalk between the two pathways by regulating the CheA-dependent phosphoryl group shuttling between the two CheYs via CheA and the CheT-mediated differential dephosphorylation of the two phosphorylated CheYs.
CheD is hypothesized to regulate the methyl-accepting sites in the chemoreceptor. As in the prototypical chemotaxis system, CheR and phosphorylated CheB modulate chemoreceptor sensitivity in response to changing signal strengths through methylation/demethylation. However, once bacteria find a favorable growth environment through chemotaxis, they must also adjust their metabolism to adapt to the new conditions. For long-term adaptation to a new environment, bacteria need to regulate the expression of certain genes [121]. In addition, A. fabrum CheS is a small protein composed solely of a STAS domain and is encoded by a gene within the chemotaxis gene cluster. Since most small STAS domain-only proteins function as anti-sigma factor antagonists, which regulate gene expression, this model proposes that CheS might also regulate gene expression through interaction with sigma factors.
5. Perspectives on Multiple Chemosensory Pathways Sharing a Histidine Kinase
In summary, the chemosensory systems of bacteria exhibit diversity due to differences in their ecological living conditions. Some bacteria, such as E. coli, inhabit relatively stable environments; their chemosensory systems are correspondingly simple, consisting of only a few chemoreceptors, one CheW, one CheA, and one CheY, and these systems solely regulate chemotaxis. In contrast, bacteria living in more dynamic environments—P. aeruginosa, for instance—possess highly intricate chemosensory systems. Not only have they evolved more chemoreceptors, but they also feature multiple chemosensory systems and use different systems to regulate a variety of physiological functions beyond chemotaxis. However, a group of plant-associated bacteria, represented by A. fabrum, employs a strategy involving more chemoreceptors, one CheA for two (or more) CheWs and two (or more) CheYs. We may call this the “one-system two-pathways” strategy. This allows them to respond to diverse signals in various ecological environments and fulfill the need to regulate multiple physiological functions.
In the chemosensory system of A. fabrum, the two CheWs may couple CheA to different chemoreceptors based on their distinct affinities for different chemoreceptors, forming two types of CSUs. These two CSUs may endow CheA with different affinities for the two CheYs due to differences in the CheWs, thereby enabling discrimination between the two CheYs and regulating their phosphorylation levels, respectively. The two phosphorylated CheYs may independently regulate different physiological functions. Therefore, a single histidine kinase may orchestrate two signaling pathways. However, experimental validation of the model proposed in Figure 3 requires addressing the following key questions:
- How is the chemosensory array assembled in A. fabrum?
It remains unresolved whether the chemosensory supramolecular array is assembled via the initial formation of a chemoreceptor array with CheW later coupling CheA, or via pre-assembled CSUs that pack into the array. It is also unknown whether all 20 chemoreceptors form a single supramolecular array through random combination or assemble into relatively independent multiple supramolecular arrays based on differences in the signals they recognize or their structures.
Given the potential diversity of the 20 chemoreceptors, the assembly process of the chemosensory array is inherently complex. First, since the 20 chemoreceptor-encoding genes are controlled by different operators, some of them may be regulated by A. fabrum’s life cycle and growth niches, making it impossible for all 20 chemoreceptors to be simultaneously and stably expressed. Second, the six cytoplasmic chemoreceptors may form distinct arrays from transmembrane ones. Given the dynamic regulation and spatial distribution of the chemoreceptors, they are unlikely to form identical molecular arrays across different cells or even at different stages within the same cell.
- 2.
- How are the signals transduced in the A. fabrum chemosensory system?
If chemoreceptors recognizing different signals form separate arrays, the two pathways described in Figure 3 would possibly exist in reality. Different signals are basically transduced independently of each other, and signal exchange may occur between different arrays. In this case, CheS and CheT would function as the signal coordinators (or mediators) of the two pathways via shuttling among arrays.
In contrast, if all 20 chemoreceptors assemble into a single array, signals from different chemoreceptors may converge on CheA, which then distributes phosphate groups to different regulators to coordinate distinct physiological functions. In such a scenario, CheS and CheT might localize to the array and function to assist CheA in distributing phosphate groups to different regulators.
It remains unclear whether A. fabrum possesses additional CheA-phosphorylatable response regulators beyond CheB, CheY1, and CheY2 and whether the two CheW proteins influence CheA’s affinities for different response regulators, thereby influencing CheA’s ability to distribute phosphate groups and coordinate signal transduction. Furthermore, no conclusive evidence currently demonstrates whether CheY2 or other potential CheA-phosphorylatable regulators regulate non-chemotactic physiological functions.
- 3.
- Does the A. fabrum chemosensory system play a role in long-term adaptation?
Currently, it remains unclear whether the A. fabrum chemosensory system contributes to the long-term adaptation process. This is because we lack experimental evidence demonstrating that CheS interacts with any sigma factor or anti-sigma factor. Therefore, it is still uncertain whether A. fabrum CheS can regulate gene expression.
Author Contributions
Conceptualization, M.G.; methodology, J.L., M.F. and H.W.; software, N.X. and J.L.; validation, M.G., J.L. and M.F.; formal analysis, M.G. and J.L.; investigation, J.L., M.F., H.W. and N.X.; data curation, J.L. and M.F.; writing—original draft preparation, J.L. and M.G.; writing—review and editing, M.G.; visualization, M.G., J.L. and N.X.; supervision, M.G.; project administration, M.G.; funding acquisition, M.G. and H.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 31870118 and 32300151).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data derived from public domain resources. The data presented in this study are available in [MiST4.0: Microbial Signal Transduction Da-241 tabase (mistdb.com)] at [https://mistdb.com/mist/genomes/GCF_000092025.1/signal-genes?ranks=chemotaxis] accessed on 25 August 2025, reference number (reference [83]). These data were derived from the following resources available in the public domain: [https://alphafold.ebi.ac.uk/], accessed on 25 August 2025.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Adler, J. Chemotaxis in bacteria. Science 1966, 153, 708–716. [Google Scholar] [CrossRef]
- Guo, M.; Huang, Z.; Yang, J. Is there any crosstalk between the chemotaxis and virulence induction signaling in Agrobacterium tumefaciens? Biotechnol. Adv. 2017, 35, 505–551. [Google Scholar] [CrossRef]
- Muok, A.R.; Olsthoorn, F.A.; Briegel, A. Unpacking alternative features of the bacterial chemotaxis system. Annu. Rev. Microbiol. 2024, 78, 169–189. [Google Scholar] [CrossRef]
- Sourjik, V.; Wingreen, N.S. Responding to chemical gradients: Bacterial chemotaxis. Curr. Opin. Cell Biol. 2012, 24, 262–268. [Google Scholar] [CrossRef]
- Matilla, M.A.; Gavira, J.A.; Krell, T. Accessing nutrients as the primary benefit arising from chemotaxis. Curr. Opin. Microbiol. 2023, 75, 102358. [Google Scholar] [CrossRef]
- Zhou, B.; Szymanski, C.M.; Baylink, A. Bacterial chemotaxis in human diseases. Trends Microbiol. 2023, 31, 453–467. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. Targeting motility and chemotaxis as a strategy to combat bacterial pathogens. Microb. Biotechnol. 2023, 16, 2205–2211. [Google Scholar] [CrossRef]
- Xu, Q.; Ali, S.; Afzal, M.; Nizami, A.S.; Han, S.; Dar, M.A.; Zhu, D. Advancements in bacterial chemotaxis: Utilizing the navigational intelligence of bacteria and its practical applications. Sci. Total Environ. 2024, 931, 172967. [Google Scholar] [CrossRef]
- Malvino, M.L. Unraveling the dynamics of Xanthomonas’ flagella: Insights into host-pathogen interactions. Peer J. 2024, 12, e18204. [Google Scholar] [CrossRef]
- Matilla, M.A.; Krell, T. Sensing the environment by bacterial plant pathogens: What do their numerous chemoreceptors recognize? Microb. Biotechnol. 2024, 17, e14368. [Google Scholar] [CrossRef]
- Munar-Palmer, M.; Santamaría-Hernando, S.; Liedtke, J.; Ortega, D.R.; López-Torrejón, G.; Rodríguez-Herva, J.J.; Briegel, A.; López-Solanilla, E. Chemosensory systems interact to shape relevant traits for bacterial plant pathogenesis. mBio 2024, 15, e0087124. [Google Scholar] [CrossRef]
- Bundurus, I.A.; Balta, I.; Pet, I.; Stef, L.; Popescu, C.A.; McCleery, D.; Lemon, J.; Callaway, T.; Douglas, A.; Corcionivoschi, N. Mechanistic concepts involved in biofilm associated processes of Campylobacter jejuni: Persistence and inhibition in poultry environments. Poult. Sci. 2024, 103, 104328. [Google Scholar] [CrossRef]
- Huang, Z.; Wang, Y.-H.; Zhu, H.-Z.; Andrianova, E.P.; Jiang, C.-Y.; Li, D.; Ma, L.; Feng, J.; Liu, Z.-P.; Xiang, H.; et al. Cross talk between chemosensory pathways that modulate chemotaxis and biofilm formation. mBio 2019, 10, e02876-18. [Google Scholar] [CrossRef]
- Oliveira, N.M.; Foster, K.R.; Durham, W.M. Single-cell twitching chemotaxis in developing biofilms. Proc. Natl. Acad. Sci. USA 2016, 113, 6532–6537. [Google Scholar] [CrossRef]
- Xu, X.; Li, H.; Qi, X.; Chen, Y.; Qin, Y.; Zheng, J.; Jiang, X. cheA, cheB, cheR, cheV, and cheY are involved in regulating the adhesion of Vibrio harveyi. Front. Cell. Infect. Microbiol. 2021, 10, 591751. [Google Scholar]
- Jazleena, P.J.; Das, A.; Guiseppi, A.; Debard, F.; Sharma, J.; Yaikhomba, M.; Mignot, T.; Mauriello, E.M.F.; Gayathri, P. Di-HAMP domains of a cytoplasmic chemoreceptor modulate nucleoid array formation and downstream signaling. mBio 2025, 16, e0005725. [Google Scholar] [CrossRef] [PubMed]
- Seymour, J.R.; Brumley, D.R.; Stocker, R.; Raina, J.B. Swimming towards each other: The role of chemotaxis in bacterial interactions. Trends Microbiol. 2024, 32, 640–649. [Google Scholar] [CrossRef]
- Yarrington, K.D.; Shendruk, T.N.; Limoli, D.H. The type IV pilus chemoreceptor PilJ controls chemotaxis of one bacterial species towards another. PLoS Biol. 2024, 22, e3002488. [Google Scholar] [CrossRef] [PubMed]
- Aroney, S.T.N.; Poole, P.S.; Sánchez-Cañizares, C. Rhizobial chemotaxis and motility systems at work in the soil. Front. Plant Sci. 2021, 12, 725338. [Google Scholar] [CrossRef]
- Guo, M.; Ye, J.; Gao, D.; Xu, N.; Yang, J. Agrobacterium-mediated horizontal gene transfer: Mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol. Adv. 2019, 37, 259–270. [Google Scholar] [CrossRef]
- Keegstra, J.M.; Carrara, F.; Stocker, R. The ecological roles of bacterial chemotaxis. Nat. Rev. Microbiol. 2022, 20, 491–504, Erratum in Nat. Rev. Microbiol. 2022, 20, 505. [Google Scholar] [CrossRef]
- O’Neal, L.; Vo, L.; Alexandre, G. Specific root exudate compounds sensed by dedicated chemoreceptors shape Azospirillum brasilense chemotaxis in the rhizosphere. Appl. Environ. Microbiol. 2020, 86, e01026-20. [Google Scholar] [CrossRef]
- Fulcher, N.B.; Holliday, P.M.; Klem, E.; Cann, M.J.; Wolfgang, M.C. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol. Microbiol. 2010, 76, 889–904. [Google Scholar] [CrossRef]
- Huang, W.; Wang, D.; Zhang, X.X.; Zhao, M.; Sun, L.; Zhou, Y.; Guan, X.; Xie, Z. Regulatory roles of the second messenger c-di-GMP in beneficial plant-bacteria interactions. Microbiol. Res. 2024, 285, 127748. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hua, C.; Deng, X. c-di-GMP signaling in Pseudomonas syringae complex. Microbiol. Res. 2023, 275, 127445. [Google Scholar] [CrossRef]
- Zhuang, Z.; Sethupathy, S.; Bajón-Fernández, Y.; Ali, S.; Niu, L.; Zhu, D. Microbial chemotaxis in degradation of xenobiotics: Current trends and opportunities. Microbiol. Res. 2025, 290, 127935. [Google Scholar] [CrossRef] [PubMed]
- Adler, J. Chemoreceptors in bacteria. Science 1969, 166, 1588–1597. [Google Scholar] [CrossRef]
- Gumerov, V.M.; Ortega, D.R.; Adebali, O.; Ulrich, L.E.; Zhulin, I.B. MiST 3.0: An updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res. 2020, 48, D459–D464. [Google Scholar] [CrossRef]
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal 2010, 3, ra50. [Google Scholar] [CrossRef]
- Qin, Z.; Zhang, P. Studying bacterial chemosensory array with CryoEM. Biochem. Soc. Trans. 2021, 49, 2081–2089. [Google Scholar] [CrossRef]
- Gumerov, V.M.; Andrianova, E.P.; Zhulin, I.B. Diversity of bacterial chemosensory systems. Curr. Opin. Microbiol. 2021, 61, 42–50. [Google Scholar] [CrossRef]
- Matilla, M.A.; Martín-Mora, D.; Gavira, J.A.; Krell, T. Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol. Mol. Biol. Rev. 2021, 85, e00151-20. [Google Scholar] [CrossRef]
- Ortega, Á.; Krell, T. Chemoreceptors with C-terminal pentapeptides for CheR and CheB binding are abundant in bacteria that maintain host interactions. Comput. Struct. Biotechnol. J. 2020, 18, 1947–1955. [Google Scholar] [CrossRef]
- Guo, M.; Jin, S.; Sun, D.; Hew, C.L.; Pan, S.Q. Recruitment of conjugative DNA transfer substrate to Agrobacterium type IV secretion apparatus. Proc. Natl. Acad. Sci. USA 2007, 104, 20019–20024. [Google Scholar] [CrossRef]
- Loyola-Vargas, V.M.; Méndez-Hernández, H.A.; Quintana-Escobar, A.O. The history of Agrobacterium rhizogenes: From pathogen to a multitasking platform for biotechnology. Methods Mol. Biol. 2024, 2827, 51–69. [Google Scholar]
- Thomson, G.; Dickinson, L.; Jacob, Y. Genomic consequences associated with Agrobacterium-mediated transformation of plants. Plant J. 2024, 117, 342–363. [Google Scholar] [CrossRef]
- Hooykaas, P.J.J. The Ti plasmid, driver of Agrobacterium pathogenesis. Phytopathology 2023, 113, 594–604. [Google Scholar] [CrossRef]
- Azizi-Dargahlou, S.; Pouresmaeil, M. Agrobacterium tumefaciens-mediated plant transformation: A review. Mol. Biotechnol. 2024, 66, 1563–1580. [Google Scholar] [CrossRef]
- Yang, J.; Pan, X.; Xu, Y.; Li, Y.; Xu, N.; Huang, Z.; Ye, J.; Gao, D.; Guo, M. Agrobacterium tumefaciens ferritins play an important role in full virulence through regulating iron homeostasis and oxidative stress survival. Mol. Plant Pathol. 2020, 21, 1167–1178. [Google Scholar] [CrossRef]
- Du, Y.; Zou, J.; Yin, Z.; Chen, T. Pan-chromosome and comparative analysis of Agrobacterium fabrum reveal important traits concerning the genetic diversity, evolutionary dynamics, and niche adaptation of the species. Microbiol. Spectr. 2023, 11, e0292422. [Google Scholar] [CrossRef]
- Xu, N.; Yang, Q.; Yang, X.; Wang, M.; Guo, M. Reconstruction and analysis of a genome-scale metabolic model for Agrobacterium tumefaciens. Mol. Plant Pathol. 2021, 22, 348–360. [Google Scholar] [CrossRef]
- Subramoni, S.; Nathoo, N.; Klimov, E.; Yuan, Z.C. Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front. Plant Sci. 2014, 5, 322. [Google Scholar] [CrossRef]
- Xu, N.; Wang, M.; Yang, X.; Xu, Y.; Guo, M. In silico analysis of the chemotactic system of Agrobacterium tumefaciens. Microb. Genom. 2020, 6, mgen000460. [Google Scholar] [CrossRef]
- Agbekudzi, A.; Arapov, T.D.; Stock, A.M.; Scharf, B.E. The dual role of a novel Sinorhizobium meliloti chemotaxis protein CheT in signal termination and adaptation. Mol. Microbiol. 2024, 122, 429–446. [Google Scholar] [CrossRef] [PubMed]
- Sourjik, V.; Schmitt, R. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol. Microbiol. 1996, 22, 427–436. [Google Scholar] [CrossRef]
- Liu, W.; Bai, X.; Li, Y.; Min, J.; Kong, Y.; Hu, X. CheY1 and CheY2 of Azorhizobium caulinodans ORS571 regulate chemotaxis and competitive colonization with the host plant. Appl. Environ. Microbiol. 2020, 86, e00599-20. [Google Scholar] [CrossRef]
- Dogra, G.; Purschke, F.G.; Wagner, V.; Haslbeck, M.; Kriehuber, T.; Hughes, J.G.; Van Tassell, M.L.; Gilbert, C.; Niemeyer, M.; Ray, W.K.; et al. Sinorhizobium meliloti CheA complexed with CheS exhibits enhanced binding to CheY1, resulting in accelerated CheY1 dephosphorylation. J. Bacteriol. 2012, 194, 1075–1087. [Google Scholar] [CrossRef]
- Wuichet, K.; Cantwell, B.J.; Zhulin, I.B. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 2010, 13, 219–225. [Google Scholar] [CrossRef]
- Colin, R.; Ni, B.; Laganenka, L.; Sourjik, V. Multiple functions of flagellar motility and chemotaxis in bacterial physiology. FEMS Microbiol. Rev. 2021, 45, fuab038. [Google Scholar] [CrossRef]
- Karmakar, R. State of the art of bacterial chemotaxis. J. Basic Microbiol. 2021, 61, 366–379. [Google Scholar] [CrossRef]
- Parkinson, J.S.; Hazelbauer, J.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef]
- Micali, G.; Endres, R.G. Bacterial chemotaxis: Information processing, thermodynamics, and behavior. Curr. Opin. Microbiol. 2016, 30, 8–15. [Google Scholar] [CrossRef]
- Moore, J.P.; Emonet, T. Physics of bacterial chemotaxis. Curr. Biol. 2024, 34, R972–R977. [Google Scholar] [CrossRef]
- Matilla, M.A.; Gavira, J.A.; Monteagudo-Cascales, E.; Zhulin, I.B.; Krell, T. Structural and functional diversity of sensor domains in bacterial transmembrane receptors. Trends Microbiol. 2025, 33, 796–809. [Google Scholar] [CrossRef]
- Briegel, A.; Li, X.; Bilwes, A.M.; Hughes, K.T.; Jensen, G.J.; Crane, B.R. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc. Natl. Acad. Sci. USA 2012, 109, 3766–3771. [Google Scholar] [CrossRef]
- Cassidy, C.K.; Qin, Z.; Frosio, T.; Gosink, K.; Yang, Z.; Sansom, M.S.P.; Stansfeld, P.J.; Parkinson, J.S.; Zhang, P. Structure of the native chemotaxis core signaling unit from phage E-protein lysed E. coli cells. mBio 2023, 14, e0079323. [Google Scholar] [CrossRef]
- Piñas, G.E.; DeSantis, M.D.; Cassidy, C.K.; Parkinson, J.S. Hexameric rings of the scaffolding protein CheW enhance response sensitivity and cooperativity in Escherichia coli chemoreceptor arrays. Sci. Signal. 2022, 15, eabj1737. [Google Scholar] [CrossRef]
- Burt, A.; Cassidy, C.K.; Ames, P.; Bacia-Verloop, M.; Baulard, M.; Huard, K.; Luthey-Schulten, Z.; Desfosses, A.; Stansfeld, P.J.; Margolin, W.; et al. Complete structure of the chemosensory array core signalling unit in an E. coli minicell strain. Nat. Commun. 2020, 11, 743. [Google Scholar] [CrossRef]
- Cassidy, C.K.; Himes, B.A.; Sun, D.; Ma, J.; Zhao, G.; Parkinson, J.S.; Stansfeld, P.J.; Luthey-Schulten, Z.; Zhang, P. Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations. Commun. Biol. 2020, 3, 24. [Google Scholar] [CrossRef]
- Riechmann, C.; Zhang, P. Recent structural advances in bacterial chemotaxis signalling. Curr. Opin. Struct. Biol. 2023, 79, 102565. [Google Scholar] [CrossRef]
- Li, X.; Eyles, S.J.; Thompson, L.K. Hydrogen exchange of chemoreceptors in functional complexes suggests protein stabilization mediates long-range allosteric coupling. J. Biol. Chem. 2019, 294, 16062–16079. [Google Scholar] [CrossRef]
- Tran, T.; Karunanayake Mudiyanselage, A.P.K.K.; Eyles, S.J.; Thompson, L.K. Bacterial chemoreceptor signaling complexes control kinase activity by stabilizing the catalytic domain of CheA. Proc. Natl. Acad. Sci. USA 2023, 120, e2218467120. [Google Scholar] [CrossRef]
- Flack, C.E.; Parkinson, J.S. Structural signatures of Escherichia coli chemoreceptor signaling states revealed by cellular crosslinking. Proc. Natl. Acad. Sci. USA 2022, 119, e2204161119. [Google Scholar] [CrossRef]
- Piñas, G.E.; DeSantis, M.D.; Parkinson, J.S. Noncritical signaling role of a kinase-receptor interaction surface in the Escherichia coli chemosensory core complex. J. Mol. Biol. 2018, 430, 1051–1064. [Google Scholar] [CrossRef]
- Reyes, G.I.; Flack, C.E.; Parkinson, J.S. The structural logic of dynamic signaling in the Escherichia coli serine chemoreceptor. Protein Sci. 2024, 33, e5209. [Google Scholar] [CrossRef]
- Yang, W.; Briegel, A. Diversity of bacterial chemosensory arrays. Trends Microbiol. 2020, 28, 68–80. [Google Scholar] [CrossRef]
- Briegel, A.; Wong, M.L.; Hodges, H.L.; Oikonomou, C.M.; Piasta, K.N.; Harris, M.J.; Fowler, D.J.; Thompson, L.K.; Falke, J.J.; Kiessling, L.L.; et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry 2014, 53, 1575–1585, Erratum in Biochemistry 2014, 53, 6624. [Google Scholar] [CrossRef]
- Lacal, J.; García-Fontana, C.; Muñoz-Martínez, F.; Ramos, J.L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884. [Google Scholar] [CrossRef] [PubMed]
- Herrera Seitz, M.K.; Frank, V.; Massazza, D.A.; Vaknin, A.; Studdert, C.A. Bacterial chemoreceptors of different length classes signal independently. Mol. Microbiol. 2014, 93, 814–822. [Google Scholar] [CrossRef]
- Alexander, R.P.; Zhulin, I.B. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc. Natl. Acad. Sci. USA 2007, 104, 2885–2890. [Google Scholar] [CrossRef]
- Briegel, A.; Ortega, D.R.; Tocheva, E.I.; Wuichet, K.; Li, Z.; Chen, S.; Müller, A.; Iancu, C.V.; Murphy, G.E.; Dobro, M.J.; et al. Universal architecture of bacterial chemoreceptor arrays. Proc. Natl. Acad. Sci. USA 2009, 106, 17181–17186. [Google Scholar] [CrossRef]
- Fortier, E.M.; Bouillet, S.; Infossi, P.; Ali Chaouche, A.; Espinosa, L.; Giudici-Orticoni, M.-T.; Mauriello, E.M.F.; Iobbi-Nivol, C. Defining two chemosensory arrays in Shewanella oneidensis. Biomolecules 2023, 13, 21. [Google Scholar] [CrossRef]
- Omori, F.; Tajima, H.; Asaoka, S.; Nishiyama, S.I.; Sowa, Y.; Kawagishi, I. Chemotaxis and related signaling systems in Vibrio cholerae. Biomolecules 2025, 15, 434. [Google Scholar] [CrossRef]
- Ortega, D.R.; Kjaer, A.; Briegel, A. The chemosensory systems of Vibrio cholerae. Mol. Microbiol. 2020, 114, 367–376. [Google Scholar] [CrossRef]
- Ortega, D.R.; Yang, W.; Subramanian, P.; Mann, P.; Kjær, A.; Chen, S.; Watts, K.J.; Pirbadian, S.; Collins, D.A.; Kooger, R.; et al. Repurposing a chemosensory macromolecular machine. Nat. Commun. 2020, 11, 2041. [Google Scholar] [CrossRef]
- Ganusova, E.E.; Rost, M.; Aksenova, A.; Abdulhussein, M.; Holden, A.; Alexandre, G. Azospirillum brasilense AerC and Tlp4b cytoplasmic chemoreceptors are promiscuous and interact with the two membrane-bound chemotaxis signaling clusters mediating chemotaxis responses. J. Bacteriol. 2023, 205, e0048422. [Google Scholar] [CrossRef]
- O’Neal, L.; Gullett, J.M.; Aksenova, A.; Hubler, A.; Briegel, A.; Ortega, D.; Kjær, A.; Jensen, G.; Alexandre, G. Distinct chemotaxis protein paralogs assemble into chemoreceptor signaling arrays to coordinate signaling output. mBio 2019, 10, e01757-19. [Google Scholar] [CrossRef] [PubMed]
- Boyeldieu, A.; Ali Chaouche, A.; Ba, M.; Honoré, F.A.; Méjean, V.; Jourlin-Castelli, C. The phosphorylated regulator of chemotaxis is crucial throughout biofilm biogenesis in Shewanella oneidensis. NPJ Biofilms Microbiomes. 2020, 6, 54. [Google Scholar] [CrossRef]
- Ortega, Á.; Zhulin, I.B.; Krell, T. Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81, e00033-17. [Google Scholar] [CrossRef]
- Salah Ud-Din, A.I.M.; Roujeinikova, A. Methyl-accepting chemotaxis proteins: A core sensing element in prokaryotes and archaea. Cell Mol. Life Sci. 2017, 74, 3293–3303. [Google Scholar] [CrossRef]
- Matilla, M.A.; Velando, F.; Martín-Mora, D.; Monteagudo-Cascales, E.; Krell, T. A catalogue of signal molecules that interact with sensor kinases, chemoreceptors and transcriptional regulators. FEMS Microbiol. Rev. 2022, 46, fuab043. [Google Scholar] [CrossRef]
- Muok, A.R.; Deng, Y.; Gumerov, V.M.; Chong, J.E.; DeRosa, J.R.; Kurniyati, K.; Coleman, R.E.; Lancaster, K.M.; Li, C.; Zhulin, I.B.; et al. A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species. Proc. Natl. Acad. Sci. USA 2019, 116, 14955–14960. [Google Scholar] [CrossRef]
- Gumerov, V.M.; Ulrich, L.E.; Zhulin, I.B. MiST 4.0: A new release of the microbial signal transduction database, now with a metagenomic component. Nucleic Acids Res. 2024, 52, D647–D653. [Google Scholar] [CrossRef]
- Ye, J.; Gao, M.; Zhou, Q.; Wang, H.; Xu, N.; Guo, M. The only chemoreceptor encoded by che operon affects the chemotactic response of Agrobacterium to various chemoeffectors. Microorganisms 2021, 9, 1923. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Xu, Y.; Zong, R.; Xu, N.; Guo, M. Agrobacterium fabrum atu0526-encoding protein is the only chemoreceptor that regulates chemoattraction toward the broad antibacterial agent formic acid. Biology 2021, 10, 1345. [Google Scholar] [CrossRef]
- Zong, R.; Gao, M.; Zhang, M.; Wang, H.; Xu, N.; Guo, M. Functional identification of Agrobacterium tumefaciens chemoreceptor MCP1912 in regulating chemotactic response. Acta Microbiol. Sin. 2022, 62, 1949–1961. [Google Scholar]
- Huang, Z.; Zou, J.; Guo, M.; Zhang, G.; Gao, J.; Zhao, H.; Yan, F.; Niu, Y.; Wang, G.L. An aerotaxis receptor influences invasion of Agrobacterium tumefaciens into its host. Peer J. 2024, 12, e16898. [Google Scholar] [CrossRef]
- Gavira, J.A.; Gilabert, M.J.; Santamaría-Hernando, S.; Molina-Ollero, A.; Rico-Jiménez, M.; Cabrera, J.J.; López-Solanilla, E.; Matilla, M.A. Acetylcholine chemotaxis in global bacterial plant pathogens. Microbiol. Res. 2025, 300, 128294. [Google Scholar] [CrossRef]
- Xu, N.; Yang, X.; Li, C.; Zhang, C.; Guo, M. Identification and functional characterization of chemoreceptors for phenolic acids in Agrobacterium tumefaciens. Microbiol. Res. 2026, 302, 128348. [Google Scholar] [CrossRef]
- Ye, J. Functional study of chemoreceptor-encoding genes atu0514 and atu2173 in Agrobacterium fabrum C58. Ph.D. Thesis, Yangzhou University, Yangzhou, China, 2022. [Google Scholar]
- Huang, Z.; Pan, X.; Xu, N.; Guo, M. Bacterial chemotaxis coupling protein: Structure, function and diversity. Microbiol. Res. 2019, 219, 40–48. [Google Scholar] [CrossRef]
- Vass, L.R.; Bourret, R.B.; Foster, C.A. Analysis of CheW-like domains provides insights into organization of prokaryotic chemotaxis systems. Proteins 2023, 91, 315–329. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, Q.; Sun, P.; Yang, J.; Guo, M. Two Agrobacterium tumefaciens CheW proteins are incorporated into one chemosensory pathway with different efficiencies. Mol. Plant Microbe Interact. 2018, 31, 460–470. [Google Scholar] [CrossRef]
- Muok, A.R.; Briegel, A.; Crane, B.R. Regulation of the chemotaxis histidine kinase CheA: A structural perspective. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183030. [Google Scholar] [CrossRef]
- Berry, M.A.; Andrianova, E.P.; Zhulin, I.B. Diverse domain architectures of CheA histidine kinase, a central component of bacterial and archaeal chemosensory systems. Microbiol. Spectr. 2024, 12, e0346423. [Google Scholar] [CrossRef]
- Gao, R.; Bouillet, S.; Stock, A.M. Structural basis of response regulator function. Annu. Rev. Microbiol. 2019, 73, 175–197. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, E.N.; Barr, S.A.; Liu, X.; Vass, L.R.; Liu, Y.; Xie, Z.; Bourret, R.B. Azorhizobium caulinodans chemotaxis is controlled by an unusual phosphorelay network. J. Bacteriol. 2022, 204, e0052721. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, P.; Gupta, S.; Pandini, A.; Chen, Y.; Petzold, C.J.; Ralston, C.Y.; Blair, D.F.; Khan, S. Allosteric priming of E. coli CheY by the flagellar motor protein FliM. Biophys. J. 2020, 119, 1108–1122. [Google Scholar] [CrossRef]
- Ganusova, E.E.; Vo, L.T.; Mukherjee, T.; Alexandre, G. Multiple CheY proteins control surface-associated lifestyles of Azospirillum brasilense. Front. Microbiol. 2021, 12, 664826. [Google Scholar] [CrossRef]
- Gao, D.; Zong, R.; Huang, Z.; Ye, J.; Wang, H.; Xu, N.; Guo, M. The divergent key residues of two Agrobacterium fabrum (tumefaciens) CheY paralogs play a key role in distinguishing their functions. Microorganisms 2021, 9, 1134. [Google Scholar] [CrossRef]
- Djordjevic, S.; Goudreau, P.N.; Xu, Q.; Stock, A.M.; West, A.H. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 1998, 95, 1381–1386. [Google Scholar] [CrossRef]
- Li, M.; Xu, X.; Zou, X.; Hazelbauer, G.L. A selective tether recruits activated response regulator CheB to its chemoreceptor substrate. mBio 2021, 12, e0310621. [Google Scholar] [CrossRef]
- Barnakov, A.N.; Barnakova, L.A.; Hazelbauer, G.L. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. USA 1999, 96, 10667–10672. [Google Scholar] [CrossRef]
- Agbekudzi, A.; Scharf, B.E. Chemoreceptors in Sinorhizobium meliloti require minimal pentapeptide tethers to provide adaptational assistance. Mol. Microbiol. 2024, 122, 50–67. [Google Scholar] [CrossRef]
- Velando, F.; Monteagudo-Cascales, E.; Matilla, M.A.; Krell, T. Differential CheR affinity for chemoreceptor C-terminal pentapeptides modulates chemotactic responses. Mol. Microbiol. 2024, 122, 465–476. [Google Scholar] [CrossRef]
- Fukuoka, H.; Nishitani, K.; Deguchi, T.; Oshima, T.; Uchida, Y.; Hamamoto, T.; Che, Y.S.; Ishijima, A. CheB localizes to polar receptor arrays during repellent adaptation. Sci. Adv. 2024, 10, eadp5636. [Google Scholar] [CrossRef]
- Velando, F.; Gavira, J.A.; Rico-Jiménez, M.; Matilla, M.A.; Krell, T. Evidence for pentapeptide-dependent and independent CheB methylesterases. Int. J. Mol. Sci. 2020, 21, 8459. [Google Scholar] [CrossRef] [PubMed]
- Batra, M.; Sharma, R.; Malik, A.; Dhindwal, S.; Kumar, P.; Tomar, S. Crystal structure of pentapeptide-independent chemotaxis receptor methyltransferase (CheR) reveals idiosyncratic structural determinants for receptor recognition. J. Struct. Biol. 2016, 196, 364–374. [Google Scholar] [CrossRef]
- Li, M.; Hazelbauer, G.L. Methyltransferase CheR binds to its chemoreceptor substrates independent of their signaling conformation yet modifies them differentially. Protein Sci. 2020, 29, 443–454. [Google Scholar] [CrossRef]
- Su, Y.; Li, J.; Zhang, W.; Ni, J.; Huang, R.; Wang, Z.; Cheng, S.; Wang, Y.; Tian, Z.; Zhou, Q.; et al. Methylation of PhoP by CheR regulates Salmonella virulence. mBio 2021, 12, e02099-21. [Google Scholar] [CrossRef]
- Glekas, G.D.; Plutz, M.J.; Walukiewicz, H.E.; Allen, G.M.; Rao, C.V.; Ordal, G.W. Elucidation of the multiple roles of CheD in Bacillus subtilis chemotaxis. Mol. Microbiol. 2012, 86, 743–756. [Google Scholar] [CrossRef]
- Walukiewicz, H.E.; Tohidifar, P.; Ordal, G.W.; Rao, C.V. Interactions among the three adaptation systems of Bacillus subtilis chemotaxis as revealed by an in vitro receptor-kinase assay. Mol. Microbiol. 2014, 93, 1104–1118. [Google Scholar] [CrossRef]
- Chao, X.; Muff, T.J.; Park, S.Y.; Zhang, S.; Pollard, A.M.; Ordal, G.W.; Bilwes, A.M.; Crane, B.R. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 2006, 124, 561–571. [Google Scholar] [CrossRef][Green Version]
- Moon, K.H.; Hobbs, G.; Motaleb, M.A. Borrelia burgdorferi CheD promotes various functions in chemotaxis and the pathogenic life cycle of the spirochete. Infect Immun. 2016, 84, 1743–1752. [Google Scholar] [CrossRef]
- Orillard, E.; Watts, K.J. Deciphering the Che2 chemosensory pathway and the roles of individual Che2 proteins from Pseudomonas aeruginosa. Mol. Microbiol. 2021, 115, 222–237. [Google Scholar] [CrossRef]
- Sharma, A.K.; Rigby, A.C.; Alper, S.L. STAS domain structure and function. Cell Physiol. Biochem. 2011, 28, 407–422. [Google Scholar] [CrossRef]
- Gálvez-Roldán, C.; Cerna-Vargas, J.P.; Rodríguez-Herva, J.J.; Krell, T.; Santamaría-Hernando, S.; López-Solanilla, E. A nitrate-sensing domain-containing chemoreceptor is required for successful entry and virulence of Dickeya dadantii 3937 in potato plants. Phytopathology 2023, 113, 390–399. [Google Scholar] [CrossRef]
- Santamaría-Hernando, S.; López-Maroto, Á.; Galvez-Roldán, C.; Munar-Palmer, M.; Monteagudo-Cascales, E.; Rodríguez-Herva, J.J.; Krell, T.; López-Solanilla, E. Pseudomonas syringae pv. tomato infection of tomato plants is mediated by GABA and l-Pro chemoperception. Mol. Plant Pathol. 2022, 23, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Tumewu, S.A.; Matsui, H.; Yamamoto, M.; Noutoshi, Y.; Toyoda, K.; Ichinose, Y. Requirement of γ-aminobutyric acid chemotaxis for virulence of Pseudomonas syringae pv. tabaci 6605. Microbes Environ. 2020, 35, ME20114. [Google Scholar] [CrossRef]
- Sanchis-López, C.; Cerna-Vargas, J.P.; Santamaría-Hernando, S.; Ramos, C.; Krell, T.; Rodríguez-Palenzuela, P.; López-Solanilla, E.; Huerta-Cepas, J.; Rodríguez-Herva, J.J. Prevalence and specificity of chemoreceptor profiles in plant-associated bacteria. mSystems 2021, 6, e0095121. [Google Scholar] [CrossRef]
- Xu, N.; Wang, W.; Cheng, S.; Zuo, J.; Guo, M. Function and regulation of pob genes for 4-hydroxybenzoate catabolism in Agrobacterium tumefaciens. Appl. Environ. Microbiol. 2025, 91, e0025525. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).