Environmental Heterogeneity and Host Genotype Jointly Shape Endophytic Bacterial Community Composition Associated with an Endemic Chinese Sphagnum Species
Abstract
1. Introduction
2. Materials and Methods
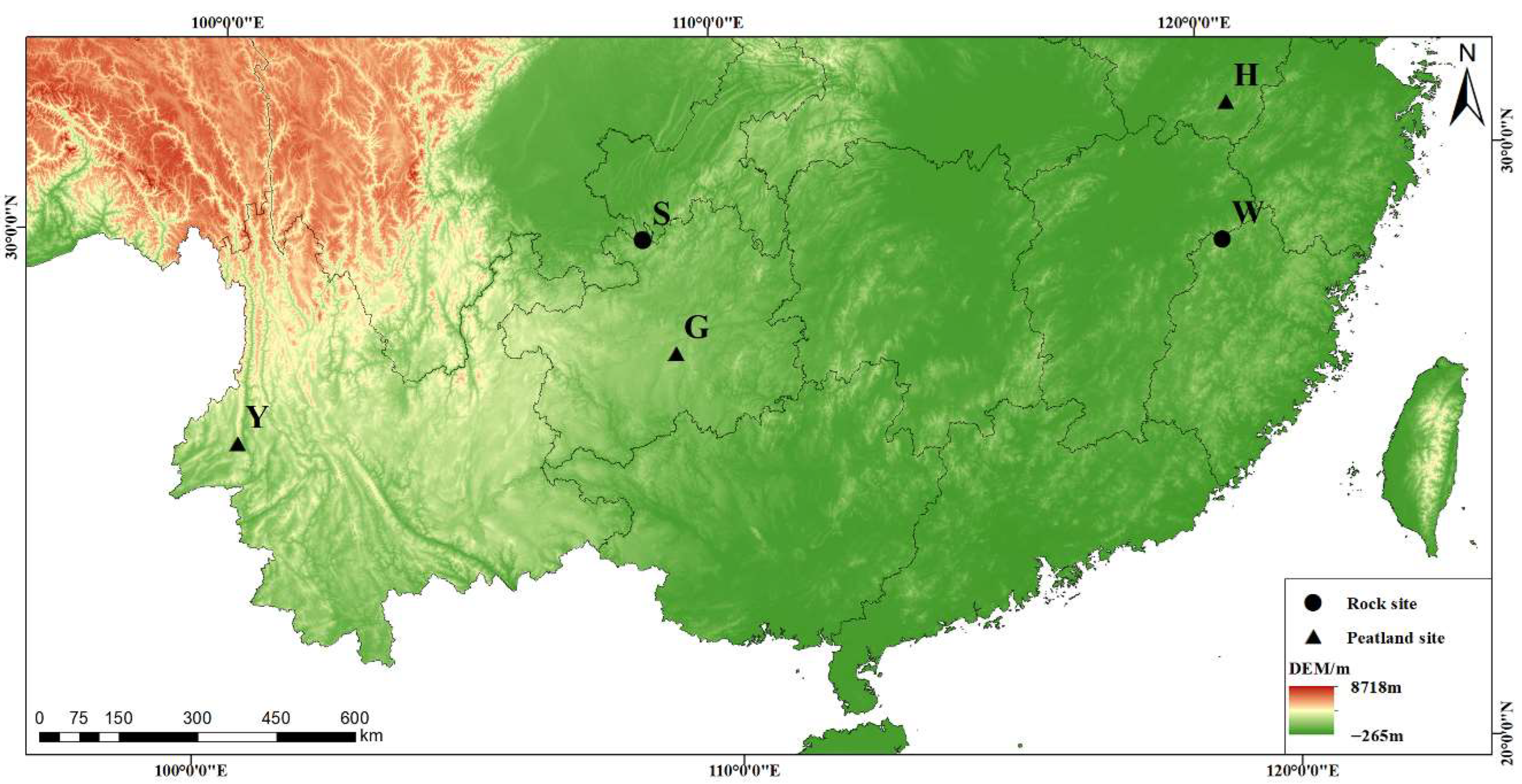
2.1. Sample Collection
2.2. Microsatellite Genotyping of S. multifibrosum
2.3. DNA Extraction, PCR, and High-Throughput Sequencing of Endophytic Bacteria
2.4. Bioinformatic Analyses
2.5. Environmental Variables
2.6. Data Analyses
2.6.1. Diversity Analysis and Indicator Identification
2.6.2. Ordination Analysis
2.6.3. Co-Occurrence Network Analysis
3. Results
3.1. Bacterial Community Composition
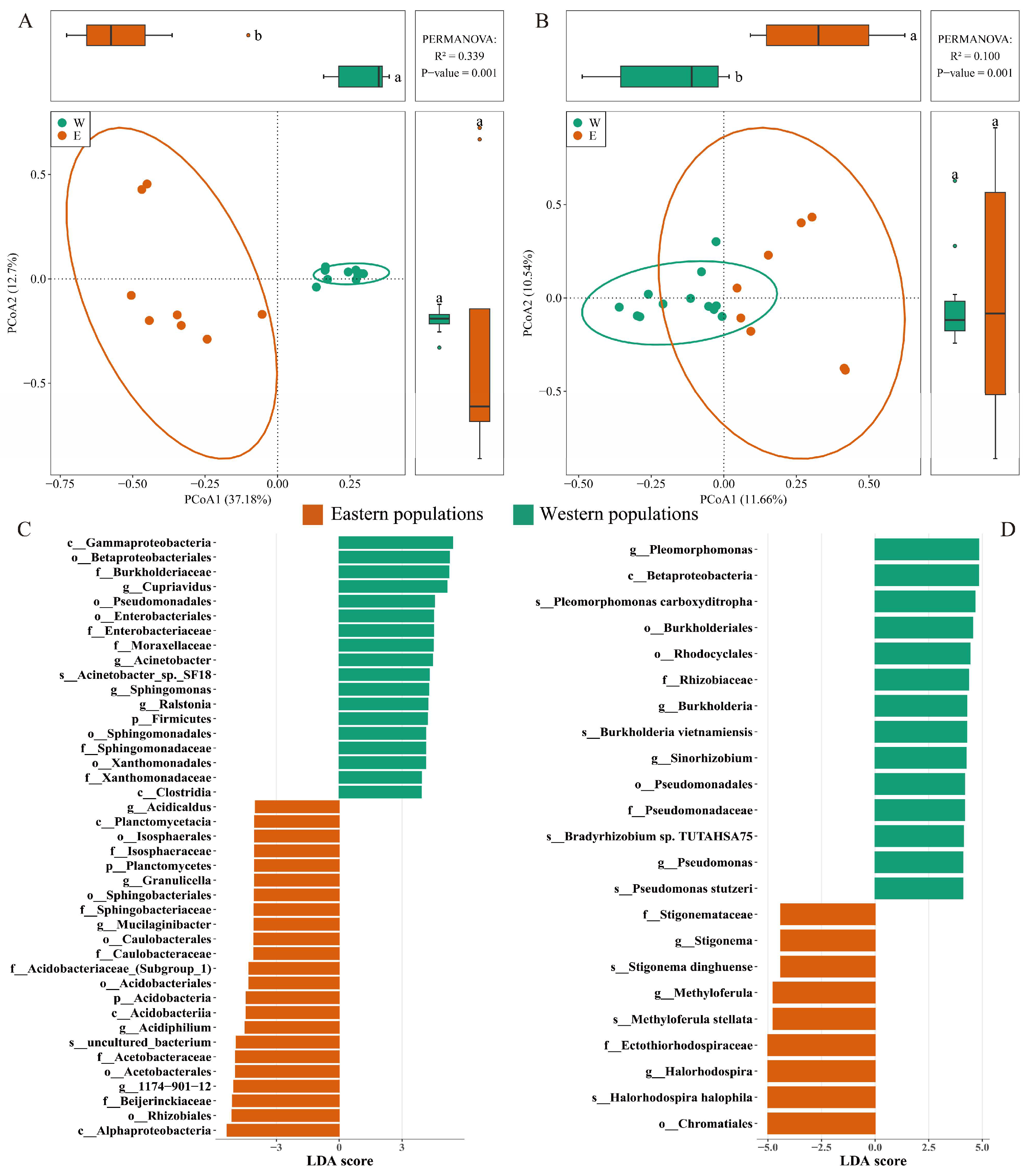
3.2. Distributions of Bacterial Communities and Indicator Taxa
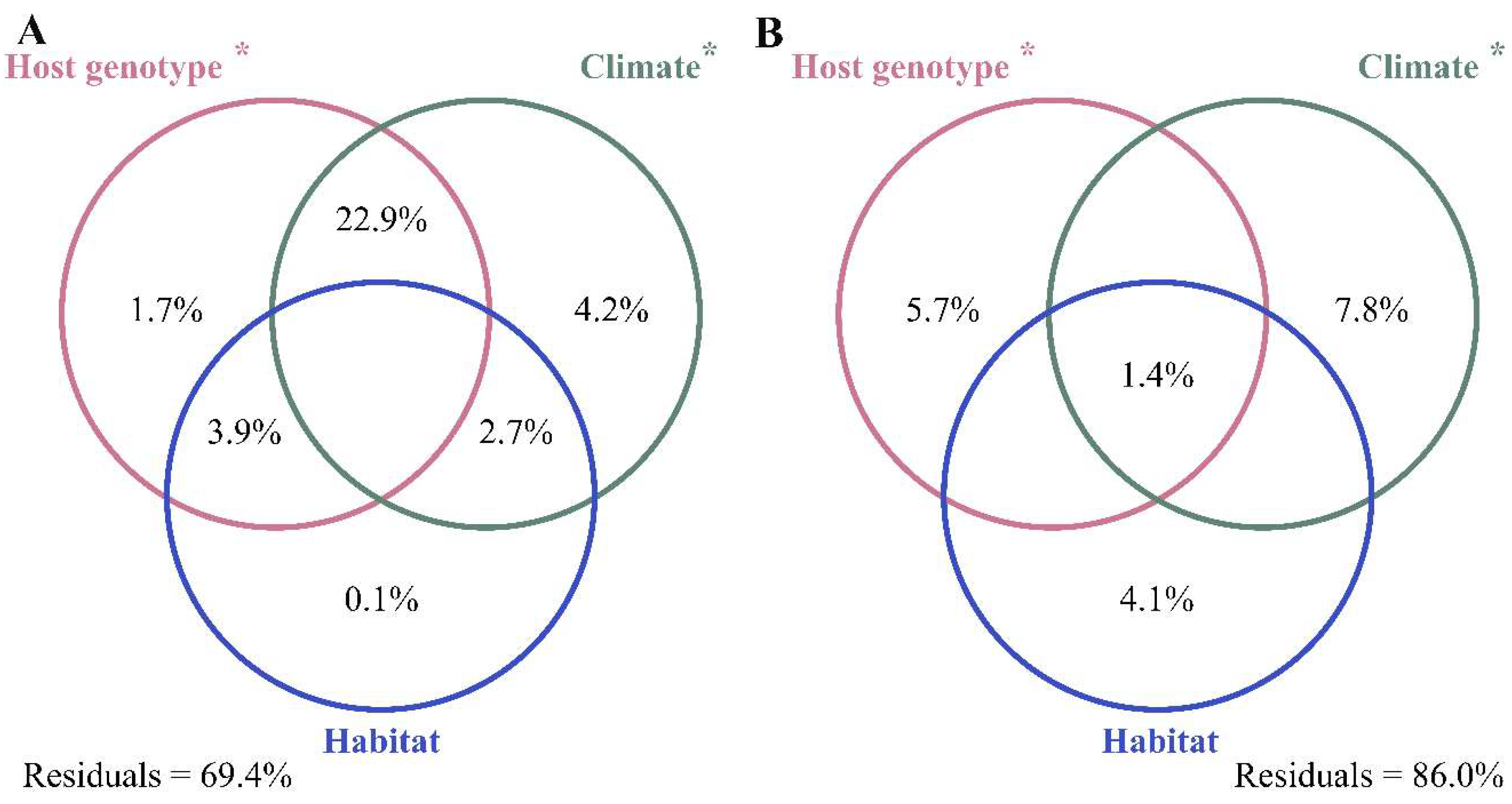
3.3. Abiotic and Biotic Effects on Bacterial Communities
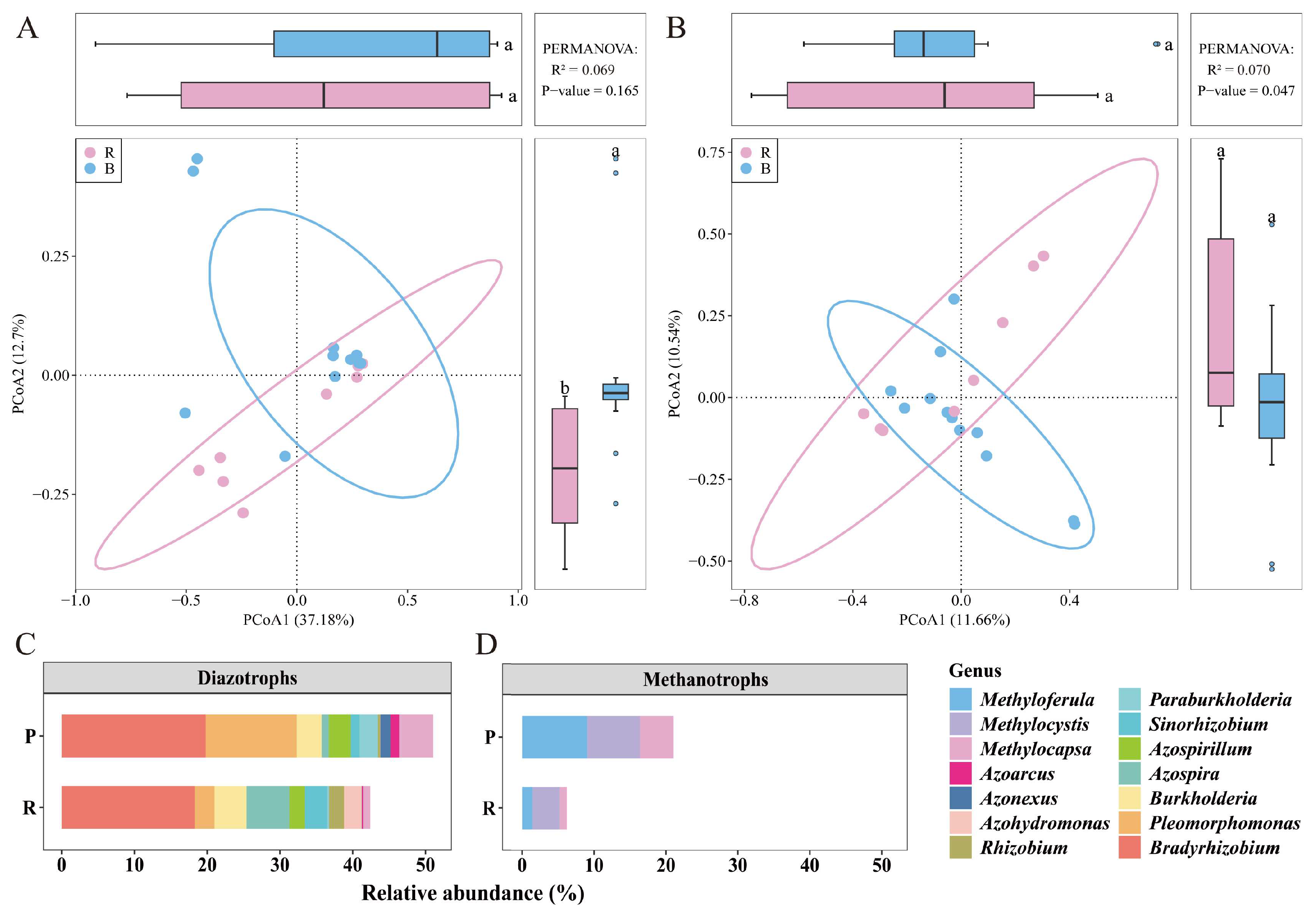
3.4. Differences in Bacterial Communities Between Two Habitats
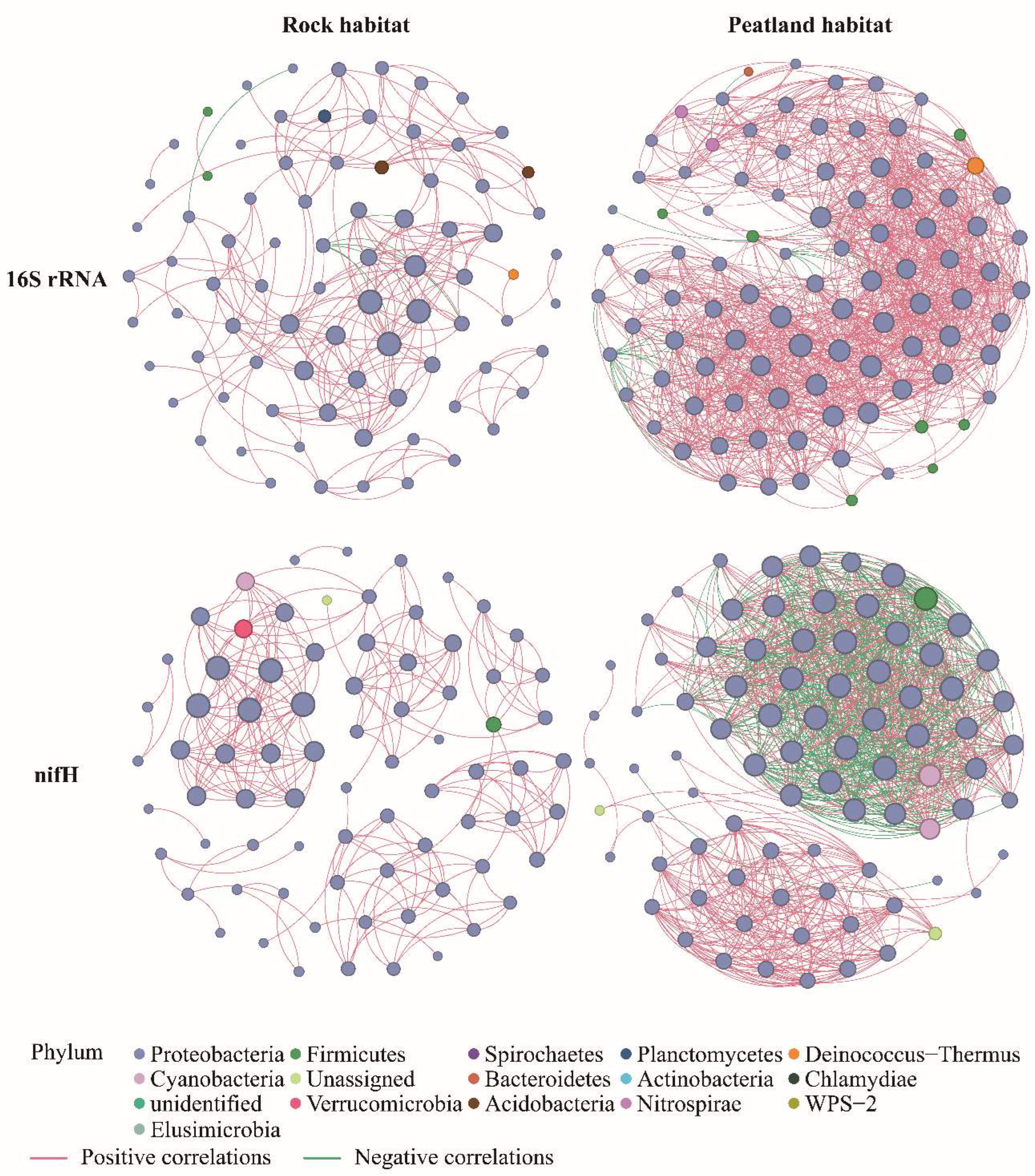
3.5. Bacterial Co-Occurrence Networks
4. Discussion
4.1. Endophytic Bacterial Composition and Their Ecological Functions
4.2. Abiotic and Biotic Influences on Bacterial Community Composition
4.3. Bacterial Functions Differ Between RH and PH Communities: Conservation Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gorham, E. Northern peatlands: Role in the carbon cycle and probable responses to climatic warming. Ecol. Appl. 1991, 1, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Jassey, V.E.; Chiapusio, G.; Binet, P.; Buttler, A.; Laggoun-Défarge, F.; Delarue, F.; Bernard, N.; Mitchell, E.A.; Toussaint, M.L.; Francez, A.J.; et al. Above- and belowground linkages in Sphagnum peatland: Climate warming affects plant-microbial interactions. Glob. Change Biol. 2013, 19, 811–823. [Google Scholar] [CrossRef]
- Kolton, M.; Weston, D.J.; Mayali, X.; Weber, P.K.; McFarlane, K.J.; PettRidge, J.; Somoza, M.M.; Lietard, J.; Glass, J.B.; Lilleskov, E.A.; et al. Defining the Sphagnum core microbiome across the North American Continent reveals a central role for diazotrophic methanotrophs in the nitrogen and carbon cycles of boreal peatland ecosystems. mBio 2022, 13, e03714–e03721. [Google Scholar] [CrossRef]
- Norby, R.J.; Childs, J.; Hanson, P.J.; Warren, J.M. Rapid loss of an ecosystem engineer: Sphagnum decline in an experimentally warmed bog. Ecol. Evol. 2019, 9, 12571–12585. [Google Scholar] [CrossRef]
- Kostka, J.E.; Weston, D.J.; Glass, J.B.; Lilleskov, E.A.; Shaw, A.J.; Turetsky, M.R. The Sphagnum microbiome: New insights from an ancient plant lineage. New Phytol. 2016, 211, 57–64. [Google Scholar] [CrossRef]
- Rousk, K.; Jones, D.L.; DeLuca, T.H. Moss-cyanobacteria associations as biogenic sources of nitrogen in boreal forest ecosystems. Front. Microbiol. 2013, 4, 150. [Google Scholar] [CrossRef]
- Bragina, A.; Berg, C.; Müller, H.; Moser, D.; Berg, G. Insights into functional bacterial diversity and its effects on Alpine bog ecosystem functioning. Sci. Rep. 2013, 3, 1955. [Google Scholar] [CrossRef] [PubMed]
- Kip, N.; van Winden, J.F.; Pan, Y.; Bodrossy, L.; Reichart, G.-J.; Smolders, A.J.P.; Jetten, M.S.M.; Damsté, J.S.S.; Op den Camp, H.J.M. Global prevalence of methane oxidation by symbiotic bacteria in peat-moss ecosystems. Nat. Geosci. 2010, 3, 617–621. [Google Scholar] [CrossRef]
- Larmola, T.; Tuittila, E.-S.; Tiirola, M.; Nykanen, H.; Martikainen, P.J.; Yrjala, K.; Tuomivirta, T.; Fritze, H. The role of Sphagnum mosses in the methane cycling of a boreal mire. Ecology 2010, 91, 2356–2365. [Google Scholar] [CrossRef] [PubMed]
- Larmola, T.; Leppänen, S.M.; Tuittila, E.-S.; Aarva, M.; Merilä, P.; Fritze, H.; Tiirola, M. Methanotrophy induces nitrogen fixation during peatland development. Proc. Natl. Acad. Sci. USA 2014, 111, 734–739. [Google Scholar] [CrossRef]
- Lindo, Z.; Nilsson, M.C.; Gundale, M.J. Bryophyte-cyanobacteria associations as regulators of the northern latitude carbon balance in response to global change. Glob. Change Biol. 2013, 19, 2022–2035. [Google Scholar] [CrossRef]
- Opelt, K.; Chobot, V.; Hadacek, F.; Schönmann, S.; Eberl, L.; Berg, G. Investigations of the structure and function of bacterial communities associated with Sphagnum mosses. Environ. Microbiol. 2007, 9, 2795–2809. [Google Scholar] [CrossRef] [PubMed]
- Shcherbakov, A.V.; Bragina, A.V.; Kuzmina, E.Y.; Berg, C.; Muntyan, A.N.; Makarova, N.M.; Malfanova, N.V.; Cardinale, M.; Berg, G.; Chebotar, V.K.; et al. Endophytic bacteria of Sphagnum mosses as promising objects of agricultural microbiology. Microbiology 2013, 82, 306–315. [Google Scholar] [CrossRef]
- Weston, D.J.; Timm, C.M.; Walker, A.P.; Gu, L.; Muchero, M.; Schmutz, J.; Shaw, A.J.; Tuskan, G.A.; Warren, J.M.; Wullschleger, S.D. Sphagnum physiology in the context of changing climate: Emergent influences of genomics, modelling and host-microbiome interactions on understanding ecosystem function. Plant Cell Environ. 2015, 38, 1737–1751. [Google Scholar] [CrossRef]
- Bragina, A.; Oberauner-Wappis, L.; Zachow, C.; Halwachs, B.; Thallinger, G.G.; Müller, H.; Berg, G. The Sphagnum microbiome supports bog ecosystem functioning under extreme conditions. Mol. Ecol. 2014, 23, 4498–4510. [Google Scholar] [CrossRef]
- Turetsky, M.R.; Weston, D.J.; Cox, W.D.; Petro, C.; Shaw, A.J. The challenging but unique eco-evolutionary aspects of Sphagnum moss. New Phytol. 2025, 247, 1608–1621. [Google Scholar] [CrossRef]
- Bragina, A.; Berg, C.; Cardinale, M.; Shcherbakov, A.; Chebotar, V.; Berg, G. Sphagnum mosses harbour highly specific bacterial diversity during their whole lifecycle. ISME J. 2012, 6, 802–813. [Google Scholar] [CrossRef]
- Bragina, A.; Berg, C.; Berg, G. The core microbiome bonds the Alpine bog vegetation to a transkingdom metacommunity. Mol. Ecol. 2015, 24, 4795–4807. [Google Scholar] [CrossRef]
- Holland-Moritz, H.; Stuart, J.E.M.; Lewis, L.R.; Miller, S.N.; Mack, M.C.; Ponciano, J.M.; McDaniel, S.F.; Fierer, N. The bacterial communities of Alaskan mosses and their contributions to N2-fixation. Microbiome 2021, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Carrell, A.A.; Kolton, M.; Glass, J.B.; Pelletier, D.A.; Warren, M.J.; Kostka, J.E.; Iversen, C.M.; Hanson, P.J.; Weston, D.J. Experimental warming alters the community composition, diversity, and N2 fixation activity of peat moss (Sphagnum fallax) microbiomes. Glob. Change Biol. 2019, 25, 2993–3004. [Google Scholar] [CrossRef] [PubMed]
- Tveit, A.T.; Kiss, A.; Winkel, M.; Horn, F.; Hájek, T.; Svenning, M.M.; Wagner, D.; Liebner, S. Environmental patterns of brown moss- and Sphagnum-associated microbial communities. Sci. Rep. 2020, 10, 22412. [Google Scholar] [CrossRef]
- Živković, T.; Carrell, A.A.; Granath, G.; Shaw, A.J.; Pelletier, D.A.; Schadt, C.W.; Klingeman, D.M.; Nilsson, M.B.; Helbig, M.; Warshan, D.; et al. Host species-microbiome interactions contribute to Sphagnum moss growth acclimation to warming. Glob. Change Biol. 2025, 31, e70066. [Google Scholar] [CrossRef]
- Kox, M.A.R.; Lüke, C.; Fritz, C.; van den Elzen, E.; van Alen, T.; Op den Camp, H.J.M.; Lamers, L.P.M.; Jetten, M.S.M.; Ettwig, K.F. Effects of nitrogen fertilization on diazotrophic activity of microorganisms associated with Sphagnum magellanicum. Plant Soil 2016, 406, 83–100. [Google Scholar] [CrossRef]
- Yang, X.; Chen, H.; Gao, Y.; Wu, N.; Xue, D. Endophytic microbiome of Sphagnum palustre as indicators of ecosystem response to nitrogen deposition. Environ. Technol. Innov. 2025, 38, 104122. [Google Scholar] [CrossRef]
- Carrell, A.A.; Veličković, D.; Lawrence, T.J.; Bowen, B.P.; Louie, K.B.; Carper, D.L.; Chu, R.K.; Mitchell, H.D.; Orr, G.; Markillie, L.M. Novel metabolic interactions and environmental conditions mediate the boreal peatmoss–cyanobacteria mutualism. ISME J. 2021, 16, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Kox, M.A.R.; Aalto, S.L.; Penttila, T.; Ettwig, K.F.; Jetten, M.S.M.; van Kessel, M.A.H.J. The influence of oxygen and methane on nitrogen fixation in subarctic Sphagnum mosses. AMB Express 2018, 8, 76. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xue, D.; Chen, X.; Qiu, Q.; Chen, H. Structure and functions of endophytic bacterial communities associated with Sphagnum mosses and their drivers in two different nutrient types of peatlands. Microb. Ecol. 2024, 87, 47. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014, 10, e1004283. [Google Scholar] [CrossRef]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I.; et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef] [PubMed]
- Karasov, T.L.; Neumann, M.; Leventhal, L.; Symeonidi, E.; Shirsekar, G.; Hawks, A.; Monroe, G.; Pathodopsis Team Exposito-Alonso, M.; Bergelson, J.; Weigel, D.; et al. Continental-scale associations of Arabidopsis thaliana phyllosphere members with host genotype and drought. Nat. Microbiol. 2024, 9, 2748–2758. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2023: Synthesis Report; IPCC: Geneva, Switzerland, 2023. [Google Scholar]
- Petro, C.; Carrell, A.A.; Wilson, R.M.; Duchesneau, K.; Noble-Kuchera, S.; Song, T.; Iversen, C.M.; Childs, J.; Schwaner, G.; Chanton, J.P.; et al. Climate drivers alter nitrogen availability in surface peat and decouple N2 fixation from CH4 oxidation in the Sphagnum moss microbiome. Glob. Change Biol. 2023, 29, 3159–3176. [Google Scholar] [CrossRef]
- Le Geay, M.; Lauga, B.; Walcker, R.; Jassey, V.E.J. A meta-analysis of peatland microbial diversity and function responses to climate change. Soil Biol. Biochem. 2024, 189, 109287. [Google Scholar] [CrossRef]
- Ma, X.; Song, Y.; Song, C.; Wang, X.; Wang, N.; Gao, S.; Cheng, X.; Liu, Z.; Gao, J.; Du, Y. Effect of nitrogen addition on soil microbial functional gene abundance and community diversity in permafrost peatland. Microorganisms 2021, 9, 2498. [Google Scholar] [CrossRef]
- Bragazza, L.; Buttler, A.; Habermacher, J.; Brancaleoni, L.; Gerdol, R.; Fritze, H.; Hanajík, P.; Laiho, R.; Johnson, D. High nitrogen deposition alters the decomposition of bog plant litter and reduces carbon accumulation. Glob. Change Biol. 2011, 18, 1163–1172. [Google Scholar] [CrossRef]
- Basińska, A.M.; Reczuga, M.K.; Gąbka, M.; Stróżecki, M.; Łuców, D.; Samson, M.; Urbaniak, M.; Leśny, J.; Chojnicki, B.H.; Gilbert, D.; et al. Experimental warming and precipitation reduction affect the biomass of microbial communities in a Sphagnum peatland. Ecol. Indic. 2020, 112, 106059. [Google Scholar] [CrossRef]
- Slate, M.L.; Antoninka, A.; Bailey, L.; Berdugo, M.B.; Callaghan, D.A.; Cárdenas, M.; Chmielewski, M.W.; Fenton, N.J.; Holland-Moritz, H.; Hopkins, S.; et al. Impact of changing climate on bryophyte contributions to terrestrial water, carbon, and nitrogen cycles. New Phytol. 2024, 242, 2411–2429. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Crosby, M.R.; He, S. Moss Flora of China; English Version 1; Missouri Botanical Garden Press: St. Louis, MO, USA, 1999. [Google Scholar]
- Cao, T.; Zhu, R.; Tan, B.C.; Guo, S.; Gao, C.; Wu, P.; Li, X. A report of the first national red list of Chinese endangered bryophytes. J. Hattori Bot. Lab. 2006, 99, 275–295. [Google Scholar]
- Shaw, A.J.; Cao, T.; Wang, L.S.; Flatberg, K.I.; Flatberg, B.; Shaw, B.; Zhou, P.; Boles, S.B.; Terraccino, S. Genetic variation in three Chinese peat mosses (Sphagnum) based on microsatellite markers, with primer information and analysis of ascertainment bias. Bryologist 2008, 111, 271–281. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb. Ecol. 2001, 41, 252–263. [Google Scholar]
- Engelbrektson, A.; Kunin, V.; Wrighton, K.C.; Zvenigorodsky, N.; Chen, F.; Ochman, H.; Hugenholtz, P. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010, 4, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Poly, F.; Ranjard, L.; Nazaret, S.; Gourbieère, F.; Monrozier, L.J. Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl. Environ. Microbiol. 2001, 67, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2024, 42, 715–718, Erratum in Nat. Biotechnol. 2024, 42, 813. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Gao, Y.; Zhou, F.; Ciais, P.; Miao, C.; Yang, T.; Jia, Y.L.; Zhou, X.D.; Klaus, B.-B.; Yang, T.T.; Yu, G. Dataset of Nitrogen Deposition in China (1980–2013); National Ecosystem Science Data Center: Beijing, China, 2021. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, Vienna, Austria. 2024. Available online: https://www.R-project.org/ (accessed on 5 April 2025).
- McMurdie, P.J.; Holmes, S. An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.; Blanchet, F.G.; Kindt, R. Vegan Community Ecology Package Version 2.62. 2022. Available online: http://cran.r-project.org (accessed on 12 June 2025).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar]
- Graves, S.; Piepho, H.; Selzer, L.; Dorai-Raj, S. multcompView: Visualizations of Paired Comparisons. R Package Version 0.1-10. 2024. Available online: https://CRAN.R-project.org/package=multcompView (accessed on 5 April 2025).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Hijmans, R. Geosphere: Spherical Trigonometry. R Package Version 1.5-20. 2024. Available online: https://CRAN.R-project.org/package=geosphere (accessed on 12 June 2025).
- Frichot, E.; François, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. InterJournal, Complex Systems, 1695. 2006. Available online: https://igraph.org (accessed on 15 July 2025).
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Web Soc. Media 2009, 3, 361–362. [Google Scholar] [CrossRef]
- Leppänen, S.M.; Rissanen, A.J.; Tiirola, M. Nitrogen fixation in Sphagnum mosses is affected by moss species and water table level. Plant Soil 2015, 389, 185–196. [Google Scholar] [CrossRef]
- Holland-Moritz, H.; Stuart, J.; Lewis, L.R.; Miller, S.; Mack, M.C.; McDaniel, S.F.; Fierer, N. Novel bacterial lineages associated with boreal moss species. Environ. Microbiol. 2018, 20, 2625–2638. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.; Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2004, 2, 621–631. [Google Scholar] [CrossRef]
- Khadem, A.F.; Pol, A.; Jetten, M.S.M.; Op den Camp, H.J.M. Nitrogen fixation by the verrucomicrobial methanotroph ‘Methylacidiphilum fumariolicum’ SolV. Microbiology 2010, 156, 1052–1059. [Google Scholar] [CrossRef]
- Zehr, J.P.; Jenkins, B.D.; Short, S.M.; Steward, G.F. Nitrogenase gene diversity and microbial community structure: A cross-system comparison. Environ. Microbiol. 2003, 5, 539–554. [Google Scholar] [CrossRef]
- Andersen, R.; Chapman, S.J.; Artz, R.R.E. Microbial communities in natural and disturbed peatlands: A review. Soil Biol. Biochem. 2013, 57, 979–994. [Google Scholar] [CrossRef]
- Berg, A.; Danielsson, Å.; Svensson, B.H. Transfer of fixed-N from N2-fixing cyanobacteria associated with the moss Sphagnum riparium results in enhanced growth of the moss. Plant Soil 2013, 362, 271–278. [Google Scholar] [CrossRef]
- Hemond, H.F. The nitrogen budget of Thoreau’s bog. Ecology 1983, 64, 99–109. [Google Scholar] [CrossRef]
- Dedysh, S.N. Cultivating uncultured bacteria from northern wetlands: Knowledge gained and remaining gaps. Front. Microbiol. 2011, 2, 184. [Google Scholar] [CrossRef]
- Kolton, M.; Marks, A.; Wilson, R.M.; Chanton, J.P.; Kotska, J.E. Impact of warming on greenhouse gas production and microbial diversity in anoxic peat from a Sphagnum-dominated bog (Grand Rapids, Minnesota, United States). Front. Microbiol. 2019, 10, 870. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, B.; Pelikan, C.; Herbold, C.W.; Köstlbacher, S.; Albertsen, M.; Eichorst, S.A.; Glavina del Rio, T.; Huemer, M.; Nielsen, P.H.; Rattei, T.; et al. Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 2018, 12, 1729–1742. [Google Scholar] [CrossRef]
- Kalam, S.; Basu, A.; Ahmad, I.; Sayyed, R.Z.; El-Enshasy, H.A.; Dailin, D.J.; Suriani, N.L. Recent understanding of soil Acidobacteria and their ecological significance: A critical review. Front. Microbiol. 2020, 11, 580024. [Google Scholar] [CrossRef]
- Op den Camp, H.J.M.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.H.; Yokota, A. Pleomorphomonas oryzae gen. nov, sp. nov, a nitrogen-fixing bacterium isolated from paddy soil of Oryza sativa. Int. J. Syst. Evol. Microbiol. 2005, 55, 1233–1237. [Google Scholar] [CrossRef]
- Bragina, A.; Maier, S.; Berg, C.; Müller, H.; Chobot, V.; Hadacek, F.; Berg, G. Similar diversity of Alphaproteobacteria and nitrogenase gene amplicons on two related Sphagnum mosses. Front. Microbiol. 2012, 2, 275. [Google Scholar] [CrossRef]
- Nazaries, L.; Murrell, J.C.; Millard, P.; Baggs, L.; Singh, B.K. Methane, microbes and models: Fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 2013, 15, 2395–2417. [Google Scholar] [CrossRef] [PubMed]
- Belova, S.E.; Kulichevskaya, I.S.; Bodelier, P.L.E.; Dedysh, S.N. Methylocystis bryophila sp. nov, a facultatively methanotrophic bacterium from acidic Sphagnum peat, and emended description of the genus Methylocystis (ex Whittenbury et al. 1970) Bowman et al. 1993. Int. J. Syst. Evol. Microbiol. 2013, 63, 1096–1104. [Google Scholar] [CrossRef]
- Dedysh, S.N.; Khmelenina, V.N.; Suzina, N.E.; Trotsenko, Y.A.; Semrau, J.D.; Liesack, W.; Tiedje, J.M. Methylocapsa acidiphila gen. nov, sp. nov, a novel methane-oxidizing and dinitrogen-fixing acidophilic bacterium from Sphagnum bog. Int. J. Syst. Evol. Microbiol. 2002, 52, 251–261. [Google Scholar] [CrossRef]
- Ho, A.; Bodelier, P.L.E. Diazotrophic methanotrophs in peatlands: The missing link? Plant Soil 2015, 389, 419–423. [Google Scholar] [CrossRef]
- Duan, Y.; Siegenthaler, A.; Skidmore, A.K.; Chariton, A.A.; Laros, I.; Rousseau, M.; De Groot, G.A. Forest top canopy bacterial communities are influenced by elevation and host tree traits. Environ. Microbiome 2024, 19, 21. [Google Scholar] [CrossRef]
- Long, J.; Luo, W.; Xie, J.; Yuan, Y.; Wang, J.; Kang, L.; Li, Y.; Zhang, Z.; Hong, M. Environmental factors influencing phyllosphere bacterial communities in giant pandas’ staple food bamboos. Front. Microbiol. 2021, 12, 748141. [Google Scholar] [CrossRef]
- Lyu, X.; Li, P.; Jin, L.; Yang, F.; Pucker, B.; Wang, C.; Liu, L.; Zhao, M.; Shi, L.; Zhang, Y.; et al. Tracing the evolutionary and genetic footprints of atmospheric tillandsioids transition from land to air. Nat. Commun. 2024, 15, 9599. [Google Scholar] [CrossRef] [PubMed]
- Marín, I.; Arahal, D.R. The Family Beijerinckiaceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 115–133. [Google Scholar]
- Ares, A.; Pereira, J.; Garcia, E.; Costa, J.; Tiago, I. The leaf bacterial microbiota of female and male kiwifruit plants in distinct seasons: Assessing the impact of Pseudomonas syringae pv. actinidiae. Phytobiomes J. 2021, 5, 275–287. [Google Scholar] [CrossRef]
- Jo, Y.; Jung, D.R.; Park, T.H.; Lee, D.; Park, M.K.; Lim, K.; Shin, J.H. Changes in microbial community structure in response to gummosis in peach tree bark. Plants 2022, 11, 2834. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621, Erratum in Nat. Rev. Microbiol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Tack, A.J.M.; Wasserman, B.; Liu, J.; Berg, G.; Norelli, J.; Droby, S.; Wisniewski, M. Evidence for host-microbiome co-evolution in apple. New Phytol. 2022, 234, 2088–2100. [Google Scholar] [CrossRef]
- Delaux, P.-M.; Schornack, S. Plant evolution driven by interactions with symbiotic and pathogenic microbes. Science 2021, 371, eaba6605. [Google Scholar] [CrossRef]
- Wicaksono, W.A.; Cernava, T.; Berg, C.; Berg, G. Bog ecosystems as a playground for plant-microbe coevolution: Bryophytes and vascular plants harbour functionally adapted bacteria. Microbiome 2021, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Duan, G.-L.; García-Palacios, P.; Yang, G.; Cui, H.-L.; Yan, M.; Yin, Y.; Yi, X.-Y.; Li, L.; Delgado-Baquerizo, M.; et al. Environmental factors and host genotype control foliar epiphytic microbial community of wild soybeans across China. Front. Microbiol. 2023, 14, 1065302. [Google Scholar] [CrossRef]
- Gomes, W.D.S.; Partelli, F.L.; Veloso, T.G.R.; da Silva, M.C.S.; Moreli, A.P.; Moreira, T.R.; Pereira, L.L. Efects of Coffea canephora genotypes on the microbial community of soil and fruit. Sci. Rep. 2024, 14, 29035. [Google Scholar] [CrossRef] [PubMed]
- Carrell, A.A.; Lawrence, T.J.; Cabugao, K.G.M.; Carper, D.L.; Pelletier, D.A.; Lee, J.H.; Jawdy, S.S.; Grimwood, J.; Schmutz, J.; Hanson, P.J.; et al. Habitat-adapted microbial communities mediate Sphagnum peatmoss resilience to warming. New Phytol. 2022, 234, 2111–2125. [Google Scholar] [CrossRef]
- Song, Y.; Liu, C.; Song, C.; Wang, X.; Ma, X.; Gao, J.; Gao, S.; Wang, L. Linking soil organic carbon mineralization with soil microbial and substrate properties under warming in permafrost peatlands of Northeastern China. Catena 2021, 203, 105348. [Google Scholar] [CrossRef]
- Roux, F.; Frachon, L.; Bartoli, C. The genetic architecture of adaptation to leaf and root bacterial microbiota in Arabidopsis thaliana. Mol. Biol. Evol. 2023, 40, msad093. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, Y.; Zhang, M.; Chu, C.; Chen, Y.; Fang, S.; Jin, G.; Jiang, M.; Lian, J.Y.; Li, Y.; et al. Diversity and biogeography of plant phyllosphere bacteria are governed by latitude-dependent mechanisms. New Phytol. 2023, 240, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Xiong, C.; Wei, Z.; Chen, Q.L.; Ma, B.; Zhou, S.Y.D.; Tan, J.; Zhang, L.M.; Cui, H.L.; Duan, G.L. Impacts of global change on the phyllosphere microbiome. New Phytol. 2022, 234, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Keane, B.; Shuttleworth, E.L.; Evans, M.G.; Ritson, J.P.; Harris, A.; Johnston, A.; Alderson, D.M.; Clay, G.D. Recovery of Sphagnum from drought is controlled by species-specific moisture thresholds. Sci. Rep. 2025, 15, 22167. [Google Scholar] [CrossRef]
- Kox, M.A.R.; Smolders, A.J.P.; Speth, D.R.; Lamers, L.P.M.; Op den Camp, H.J.M.; Jetten, M.S.M.; van Kessel, M.A.H.J. A novel laboratory-scale mesocosm setup to study methane emission mitigation by Sphagnum mosses and associated methanotrophs. Front. Microbiol. 2021, 12, 652486. [Google Scholar] [CrossRef]
- Raghoebarsing, A.A.; Smolders, A.J.; Schmid, M.C.; Rijpstra, W.I.; Wolters-Arts, M.; Derksen, J.; Jetten, M.S.; Schouten, S.; Sinninghe Damsté, J.S.; Lamers, L.P.; et al. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature 2005, 436, 1153–1156. [Google Scholar] [CrossRef]
- Cao, W.; Xiong, Y.; Zhao, D.; Tan, H.; Qu, J. Bryophytes and the symbiotic microorganisms, the pioneers of vegetation restoration in karst rocky desertification areas in southwestern China. Appl. Microbiol. Biotechnol. 2020, 104, 873–891. [Google Scholar] [CrossRef]
- Vorobev, A.V.; Baani, M.; Doronina, N.V.; Brady, A.L.; Liesack, W.; Dunfield, P.F.; Dedysh, S.N. Methyloferula stellata gen. nov, sp. nov, an acidophilic, obligately methanotrophic bacterium that possesses only a soluble methane monooxygenase. Int. J. Syst. Evol. Microbiol. 2011, 61, 2456–2463. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.; Hergoualc’h, K.; Kauffman, J.B.; Murdiyarso, D.; Kolka, R. An appraisal of Indonesia’s immense peat carbon stock using national peatland maps: Uncertainties and potential losses from conversion. Carbon. Balance Manag. 2017, 12, 12. [Google Scholar] [CrossRef]
- Kox, M.A.R.; Kop, L.F.; Elzen, E.; Alen, T.; Lamers, L.; Kessel, M.V.; Jetten, M. Functional redundancy of the methane-oxidising and nitrogen-fixing microbial community associated with Sphagnum fallax and Sphagnum palustre in two Dutch fens. Mires Peat 2020, 26, 16. [Google Scholar]
- Frolking, S.; Roulet, N.; Fuglestvedt, J. How northern peatlands influence the Earth’s radiative budget: Sustained methane emission versus sustained carbon sequestration. J. Geophys. Res.-Biogeosci. 2006, 111, G01008. [Google Scholar] [CrossRef]
- Bragazza, L.; Buttler, A.; Robroek, B.J.; Albrecht, R.; Zaccone, C.; Jassey, V.E.; Signarbieux, C. Persistent high temperature and low precipitation reduce peat carbon accumulation. Glob. Change Biol. 2016, 22, 4114–4123. [Google Scholar] [CrossRef]
- Ma, X.-Y.; Xu, H.; Cao, Z.-Y.; Shu, L.; Zhu, R.L. Will climate change cause the global peatland to expand or contract? Evidence from the habitat shift pattern of Sphagnum mosses. Glob. Change Biol. 2022, 28, 6419–6432. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Sala, A.V.; Charman, D.J.; Brewer, S.; Page, S.E.; Prentice, I.C.; Friedlingstein, P.; Moreton, S.; Amesbury, M.J.; Beilman, D.W.; Björck, S.; et al. Latitudinal limits to the predicted increase of the peatland carbon sink with warming. Nat. Clim. Change 2018, 8, 907–913. [Google Scholar] [CrossRef]
- Loisel, J.; Gallego-Sala, A.V.; Amesbury, M.J.; Magnan, G.; Anshari, G.; Beilman, D.W.; Benavides, J.C.; Blewett, J.; Camill, P.; Charman, D.J.; et al. Expert assessment of future vulnerability of the global peatland carbon sink. Nat. Clim. Change 2021, 11, 70–77, Erratum in Nat. Clim. Chang. 2021, 11, 362. [Google Scholar] [CrossRef]
- Qiu, C.; Ciais, P.; Zhu, D.; Guenet, B.; Chang, J.; Chaudhary, N.; Kleinen, T.; Li, X.; Müller, J.; Xi, Y.; et al. A strong mitigation scenario maintains climate neutrality of northern peatlands. One Earth 2022, 5, 86–97. [Google Scholar] [CrossRef]

| Gene | Variable | CCA1 | CCA2 | r2 | p |
|---|---|---|---|---|---|
| 16S rRNA | BIO5 | 0.678 | 0.735 | 0.917 | 0.001 *** |
| BIO13 | −0.996 | 0.089 | 0.912 | 0.001 *** | |
| BIO10 | 0.721 | 0.693 | 0.559 | 0.003 ** | |
| ND | −0.990 | −0.143 | 0.418 | 0.015 * | |
| SR | −0.997 | −0.075 | 0.035 | 0.750 | |
| nifH | BIO5 | −0.372 | −0.928 | 0.969 | 0.001 *** |
| ND | 0.950 | 0.313 | 0.683 | 0.001 *** | |
| BIO13 | 0.951 | 0.311 | 0.679 | 0.001 *** | |
| BIO10 | −0.223 | −0.975 | 0.557 | 0.004 ** | |
| SR | −0.470 | 0.883 | 0.028 | 0.800 |
| Habitat | Rock Habitat | Peatland Habitat | |||
|---|---|---|---|---|---|
| Population | W | S | H | G | Y |
| 16S rRNA gene | 6.41 | 5.14 | 6.18 | 5.14 | 5.79 |
| nifH gene | 5.28 | 4.75 | 4.97 | 5.28 | 4.80 |
| Metric | 16S rRNA Gene | nifH Gene | ||
|---|---|---|---|---|
| Rock Habitat | Peatland Habitat | Rock Habitat | Peatland Habitat | |
| Number of nodes | 82 | 95 | 77 | 76 |
| Number of edges | 238 | 1184 | 242 | 305 |
| Average degree | 5.805 | 24.926 | 6.286 | 8.026 |
| Graph density | 0.072 | 0.265 | 0.083 | 0.107 |
| Modularity | 0.633 | 0.383 | 0.732 | 0.696 |
| Average clustering coefficient | 0.687 | 0.672 | 0.846 | 0.768 |
| Average path length | 4.800 | 2.137 | 2.644 | 3.663 |
| Eigenvector centrality | 0.840 | 0.526 | 0.842 | 0.825 |
| Average betweenness centrality | 100.939 | 54.453 | 14.130 | 43.842 |
| Negative correlations (%) | 2.940 | 2.280 | 0.000 | 0.980 |
| Positive correlations (%) | 97.060 | 97.720 | 100.000 | 99.020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Sun, X.; Deng, H.; Zhao, Z. Environmental Heterogeneity and Host Genotype Jointly Shape Endophytic Bacterial Community Composition Associated with an Endemic Chinese Sphagnum Species. Microorganisms 2025, 13, 2538. https://doi.org/10.3390/microorganisms13112538
Liu Y, Sun X, Deng H, Zhao Z. Environmental Heterogeneity and Host Genotype Jointly Shape Endophytic Bacterial Community Composition Associated with an Endemic Chinese Sphagnum Species. Microorganisms. 2025; 13(11):2538. https://doi.org/10.3390/microorganisms13112538
Chicago/Turabian StyleLiu, Yan, Xuechun Sun, Hongping Deng, and Zhengwu Zhao. 2025. "Environmental Heterogeneity and Host Genotype Jointly Shape Endophytic Bacterial Community Composition Associated with an Endemic Chinese Sphagnum Species" Microorganisms 13, no. 11: 2538. https://doi.org/10.3390/microorganisms13112538
APA StyleLiu, Y., Sun, X., Deng, H., & Zhao, Z. (2025). Environmental Heterogeneity and Host Genotype Jointly Shape Endophytic Bacterial Community Composition Associated with an Endemic Chinese Sphagnum Species. Microorganisms, 13(11), 2538. https://doi.org/10.3390/microorganisms13112538






