Characterization of Bacterial Communities in Volcanic Soil from Northern Patagonian Area of Chile
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Collection of Volcanic Soil Samples
2.2. Physico-Chemical Soil Analysis
2.3. DNA Extraction from Volcanic Soil Samples
2.4. 16S-rRNA Amplicon Sequencing
2.5. Sequencing Data Analysis
2.6. Microbial Diversity Analysis
2.7. Statistical Analysis
2.8. Network of the Microbial Communities
3. Results
3.1. Soil Characteristics
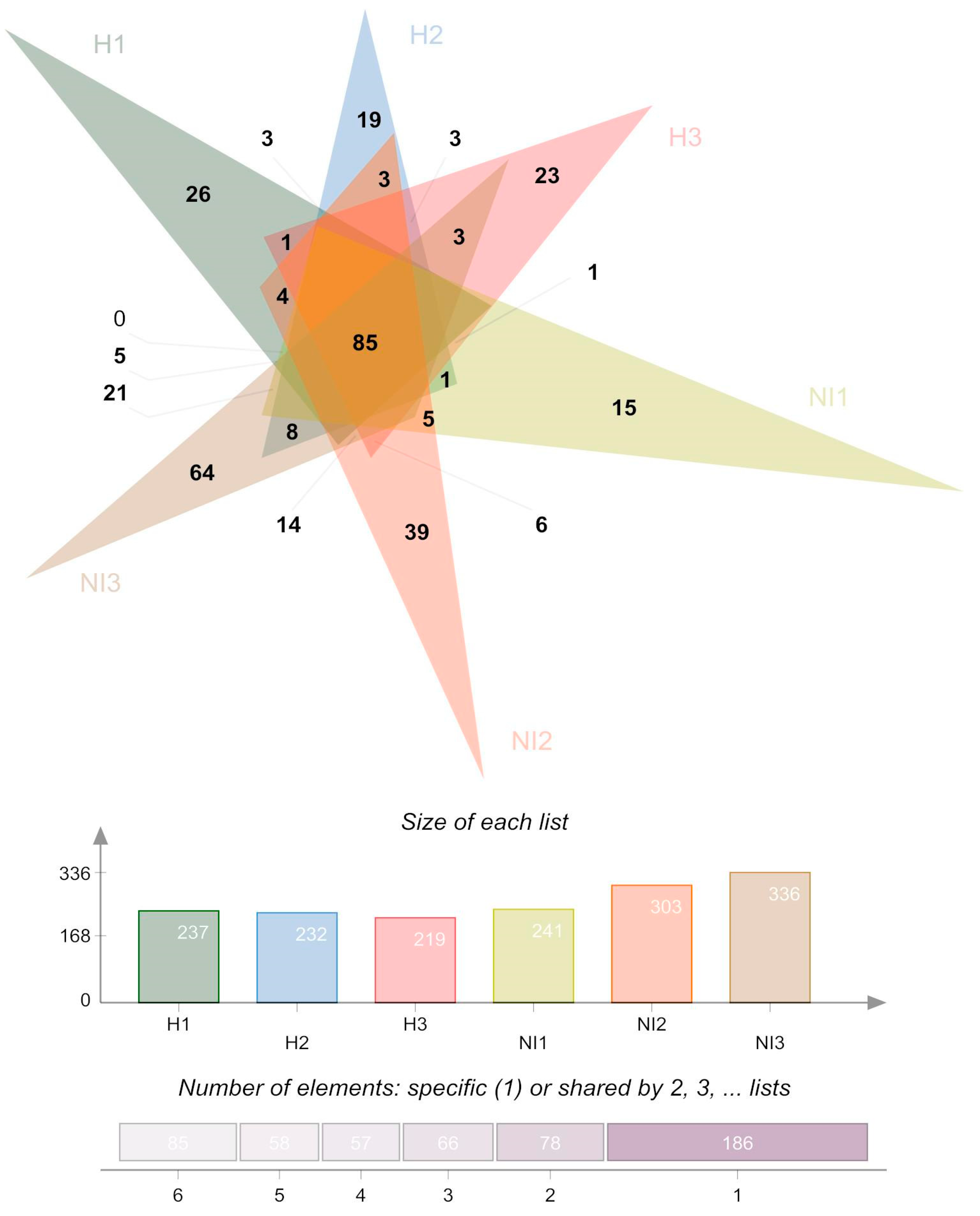
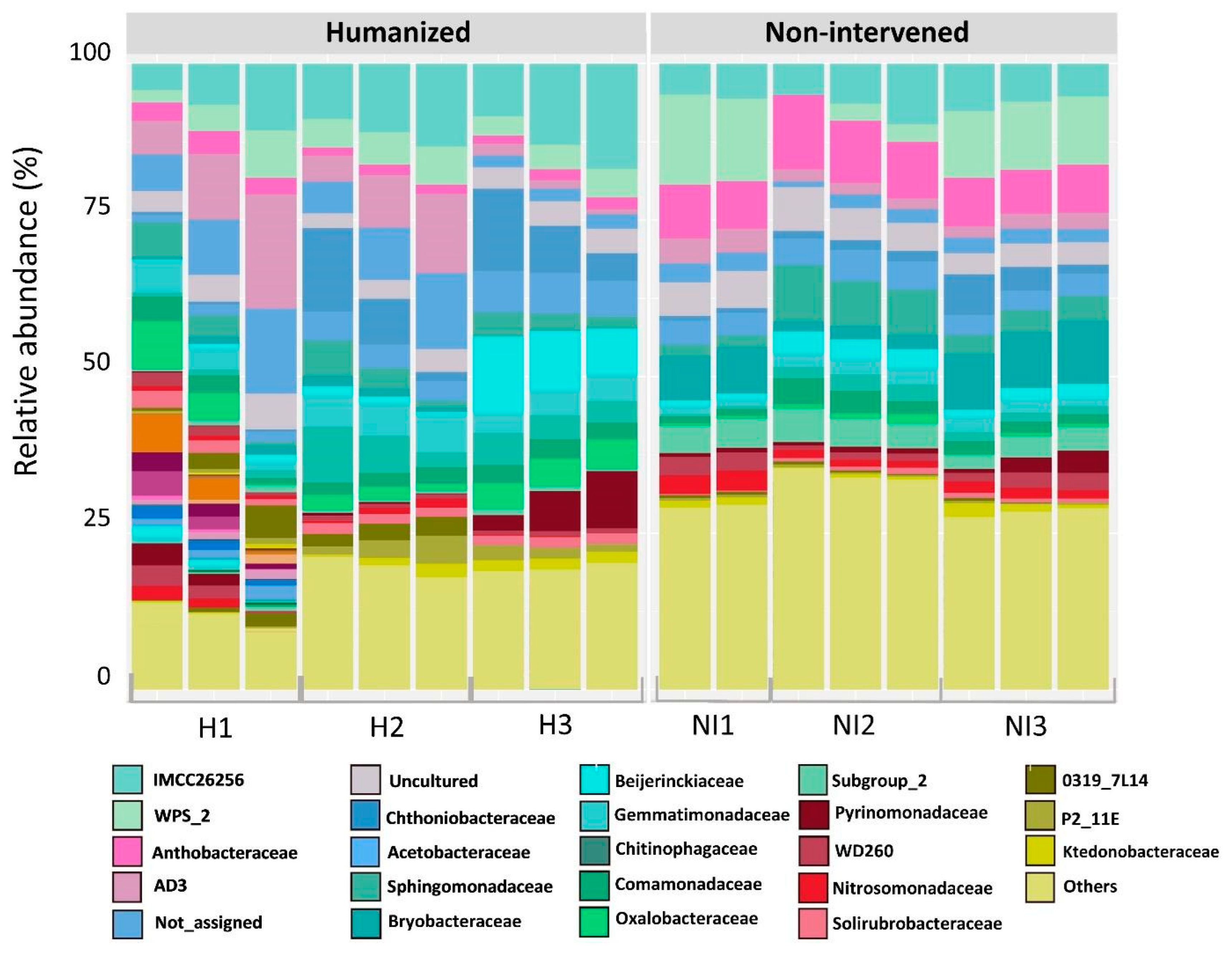
3.2. Soil Bacterial Community Composition
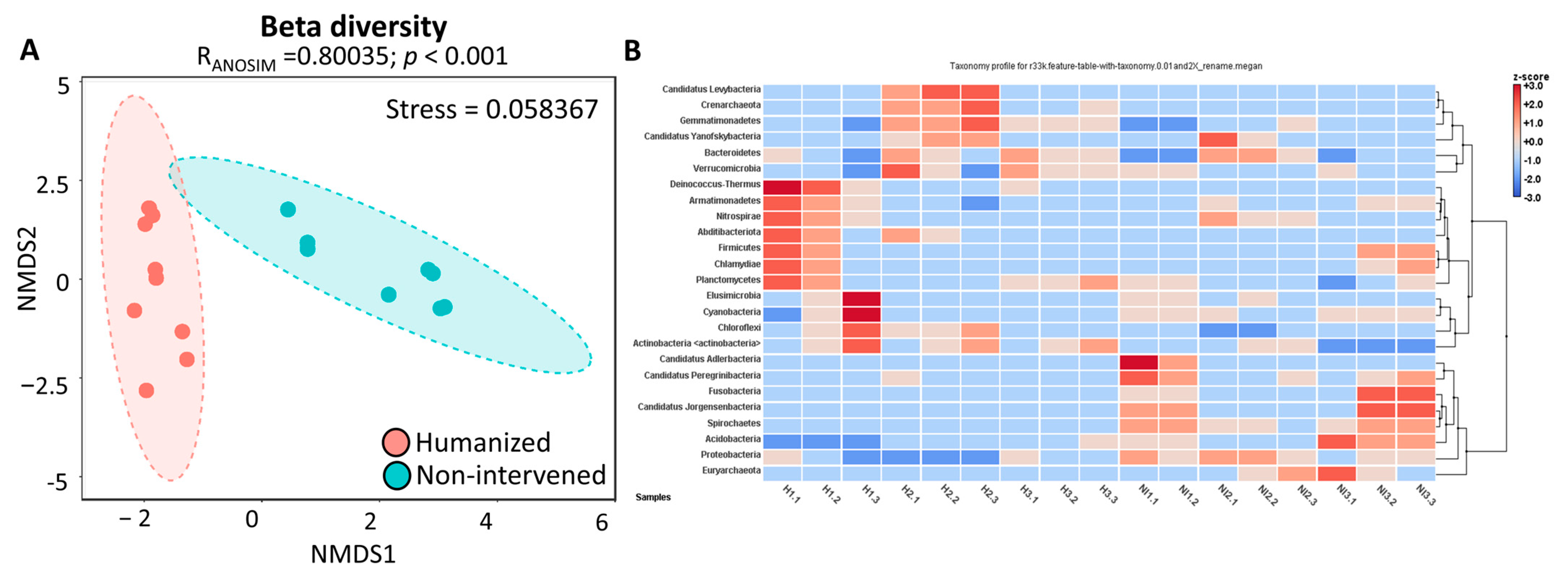
3.3. Alpha and Beta Diversity
3.4. Heatmap and Cluster Analysis
3.5. Differential Abundance Analysis
3.6. Potential Functional Diversity and Distribution Patterns of Functional Microbes
3.7. Relative Contribution of Physicochemical Soil Characteristics to Bacterial Community Structure
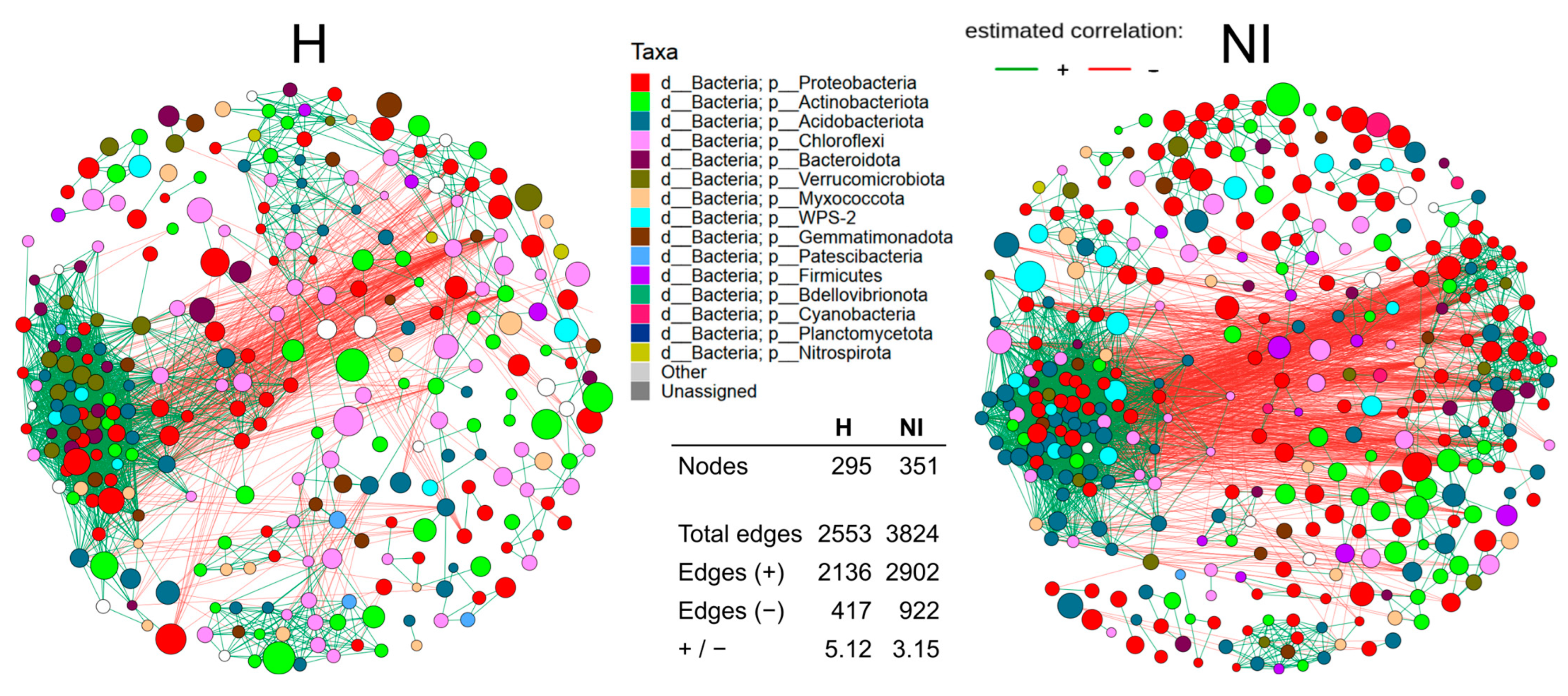
3.8. Co-Occurrence Network Structure and Modularity in Volcanic Soils
3.8.1. Overall Network Properties and Differential Connectivity of ASVs
3.8.2. Modular Structure and Taxonomic Composition of Clusters
3.8.3. Potentially Keystone Taxa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Titeux, N.; Brotons, L.; Settele, J. IPBES Promotes Integration of Multiple Threats to Biodiversity. Trends Ecol. Evol. 2019, 34, 969–970. [Google Scholar] [CrossRef] [PubMed]
- Hudson, L.N.; Newbold, T.; Contu, S.; Hill, S.L.L.; Lysenko, I.; De Palma, A.; Phillips, H.R.P.; Alhusseini, T.I.; Bedford, F.E.; Bennett, D.J.; et al. The database of the PREDICTS (Projecting Responses of Ecological Diversity in Changing Terrestrial Systems) project. Ecol. Evol. 2017, 7, 145–188. [Google Scholar] [CrossRef]
- Van Bruggen, A.H.C.; Goss, E.M.; Havelaar, A.; van Diepeningen, A.D.; Finckh, M.R.; Morris, J.G. One Health-Cycling of Diverse Microbial Communities as a Connecting Force for Soil, Plant, Animal, Human and Ecosystem Health. Sci. Total Environ. 2019, 664, 927–937. [Google Scholar] [CrossRef]
- Guerra, C.A.; Heintz-Buschart, A.; Sikorski, J.; Chatzinotas, A.; Guerrero-Ramírez, N.; Cesarz, S.; Beaumelle, L.; Rillig, M.C.; Maestre, F.T.; Delgado-Baquerizo, M.; et al. Blind spots in global soil biodiversity and ecosystem function research. Nat. Commun. 2020, 11, 3870. [Google Scholar] [CrossRef]
- Nizamani, M.M.; Hughes, A.C.; Qureshi, S.; Zhang, Q.; Tarafder, E.; Das, D.; Acharya, K.; Wang, Y.; Zhang, Z.-G. Microbial biodiversity and plant functional trait interactions in multifunctional ecosystems. Appl. Soil Ecol. 2024, 201, 105515. [Google Scholar] [CrossRef]
- Hernández, M.; Calabi, M.; Conrad, R.; Dumont, M.G. Analysis of the microbial communities in soils of different ages following volcanic eruptions. Pedosphere 2020, 30, 126–134. [Google Scholar] [CrossRef]
- Anderson, D.; Song, Y.P.; Wu, Y.T. Environmental variables including heavy metals significantly shape the soil bacterial community structure in the Tatun volcano group, Northern Taiwan. Microbes Environ. 2022, 37, ME22005. [Google Scholar] [CrossRef]
- Hadland, N.; Hamilton, C.W.; Duhamel, S. Young volcanic terrains are windows into early microbial colonization. Commun. Earth Environ. 2024, 5, 114. [Google Scholar] [CrossRef]
- Garcia-López, E.; Ruiz-Blas, F.; Sanchez-Casanova, S.; Peña Pérez, S.; Martin-Cerezo, M.L.; Cid, C. Microbial communities in volcanic glacier ecosystems. Front. Microbiol. 2022, 13, 825632. [Google Scholar] [CrossRef] [PubMed]
- Hernández, M.; Vera-Gargallo, B.; Calabi-Floody, M.; King, G.M.; Conrad, R.; Tebbe, C.C. Reconstructing genomes of carbon monoxide oxidisers in volcanic deposits including members of the class Ktedonobacteria. Microorganisms 2020, 8, 1880. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Fujimura, R.; Sato, Y.; Suda, W.; Kim, S.W.; Oshima, K.; Hattori, M.; Kamijo, T.; Narisawa, K.; Ohta, H. Characterization of early microbial communities on volcanic deposits along a vegetation gradient on the island of Miyake, Japan. Microbes Environ. 2014, 29, 38–49. [Google Scholar] [CrossRef]
- Yang, C.H.; Crowley, D.E. Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 2000, 66, 345–351. [Google Scholar] [CrossRef]
- Ciccazzo, S.; Esposito, A.; Rolli, E.; Zerbe, S.; Daffonchio, D.; Brusetti, L. Different pioneer plant species select specific rhizosphere bacterial communities in a high mountain environment. SpringerPlus 2014, 3, 391. [Google Scholar] [CrossRef]
- Lathifah, A.N.; Guo, Y.; Sakagami, N.; Suda, W.; Higuchi, M.; Nishizawa, T.; Prijambada, I.D.; Ohta, H. Comparative Characterization of Bacterial Communities in Moss-Covered and Unvegetated Volcanic Deposits of Mount Merapi, Indonesia. Microbes Environ. 2019, 34, 268–277. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yabe, S.; Sakai, Y.; Abe, K.; Yokota, A. Diversity of Ktedonobacteria with actinomycetes-like morphology in terrestrial environments. Microbes Environ. 2017, 32, 61–70. [Google Scholar] [CrossRef]
- Fujimura, R.; Sato, Y.; Nishizawa, T.; Nanba, K.; Oshima, K.; Hattori, M.; Kamijo, T.; Ohta, H. Analysis of Early Bacterial Communities on Volcanic Deposits on the Island of Miyake (Miyake-jima), Japan: A 6-year Study at a Fixed Site. Microbes Environ. 2012, 27, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Dawson, R.A.; Fantom, N.; Martin-Pozas, T.; Aguila, P.; King, G.; Hernández, M. Carbon monoxide-oxidising Pseudomonadota on volcanic deposits. Environ. Microbiome 2025, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Byloos, B.; Monsieurs, P.; Mysara, M.; Leys, N.; Boon, N.; Van Houdt, R. Characterization of the bacterial communities on recent Icelandic volcanic deposits of different ages. BMC Microbiol. 2018, 18, 122. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, Y.; Guo, Y.; Li, F.; Xu, D.; Chao, L.; Qu, H.; Wang, B.; Ma, X.; Wang, S.; et al. Differences in microbial communities from Quaternary volcanic soils at different stages of development: Evidence from Late Pleistocene and Holocene volcanoes. CATENA 2021, 201, 10521. [Google Scholar] [CrossRef]
- Stern, C.R.; Moreno, H.; López-Escobar, L.; Clavero, J.E.; Lara, L.E.; Naranjo, J.A.; Parada, M.A.; Skewes, M.A. Chilean volcanoes. In The Geology of Chile; Moreno, T., Gibbons, W., Eds.; The Geological Society of London: London, UK, 2007; pp. 149–180. [Google Scholar] [CrossRef]
- Moreno, H.; Lara, L.E.; Orozco, G. Geología del Volcán Osorno, Región de Los Lagos; Serie Geología Básica n.126; Servicio Nacional de Geología y Minería: Santiago, Chile, 2010; p. 31. [Google Scholar]
- Díaz, D.; Zúñiga, F.; Castruccio, A. The interaction between active crustal faults and volcanism: A case study of the Liquiñe-Ofqui Fault Zone and Osorno volcano, southern Andes, using magnetotellurics. J. Volcanol. Geotherm. Res. 2020, 393, 106806. [Google Scholar] [CrossRef]
- Fang, X.; Wang, H.; Zhao, L.; Wang, M.; Sun, M. Diversity and structure of the rhizosphere microbial communities of wild and cultivated ginseng. BMC Microbiol. 2022, 22, 2. [Google Scholar] [CrossRef]
- Hua, H.; Sui, X.; Liu, Y.; Liu, X.; Chang, Q.; Xu, R.; Li, M.; Mu, L. Effects of Land Use Type Transformation on the Structure and Diversity of Soil Bacterial Communities. Life 2024, 14, 252. [Google Scholar] [CrossRef]
- Corporación Nacional Forestal, CONAF. Parque Nacional Vicente Pérez Rosales. 2020. Available online: https://www.conaf.cl/parque_nacionales/parque-nacional-vicente-perez-rosales/ (accessed on 2 January 2022).
- Huang, Q.; Yang, F.; Cao, H.; Cheng, J.; Jiang, M.; Li, M.; Ni, H.; Xie, L. Comparison of Microbial Diversity of Two Typical Volcanic Soils in Wudalianchi, China. Microorganisms 2024, 12, 656. [Google Scholar] [CrossRef] [PubMed]
- Sadzawka, R.A.; Carrasco, R.M.A.; Grez, Z.R.; de la Luz Mora, M.G.; Flores, P.H.; Neaman, A. Métodos de Análisis Recomendados para los Suelos de Chile; Serie Actas INIA 163 (2006); Instituto de Investigaciones Agropecuarias: Santiago, Chile, 2006; Revision 2006; Available online: https://www.schcs.cl/wp-content/uploads/2018/11/Analisis-de-suelos.pdf (accessed on 2 January 2022).
- Mandakovic, D.; Maldonado, J.; Pulgar, R.; Cabrera, P.; Gaete, A.; Urtuvia, V.; Seeger, M.; Cambiazo, V.; González, M. Microbiome analysis and bacterial isolation from Lejía Lake soil in Atacama Desert. Extremophiles 2018, 22, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Handl, S.; Dowd, S.E.; Garcia-Mazcorro, J.F.; Steiner, J.M.; Suchodolski, J.S. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. 2011, 76, 301–310. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. SCIKIT-LEARN: Machine learning in PYTHON. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef]
- Smyth, G.K. limma: Linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V., Huber, W., Irizarry, R., Dudoit, S., Eds.; Springer: New York, NY, USA, 2005; pp. 397–420. [Google Scholar] [CrossRef]
- Oliveros, J.C. (2007–2015) Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 2 January 2022).
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Maldonado, J.E.; Gaete, A.; Mandakovic, D.; Aguado-Norese, C.; Aguilar, M.; Gutiérrez, R.A.; González, M. Partners to survive: Hoffmannseggia doellii root-associated microbiome at the Atacama Desert. New Phytol. 2022, 234, 2126–2139. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Gorska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.J.; Tappu, R. MEGAN Community Edition–interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Aguila-Torres, P.; González, M.; Maldonado, J.E.; Miranda, R.; Zhang, L.; González-Stegmaier, R.; Rojas, L.A.; Gaete, A. Associations between bacterial communities and microplastics from surface seawater of the Northern Patagonian area of Chile. Environ. Pollut. 2022, 306, 119313. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1. [Google Scholar]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, G.; Ewald, J.; Pang, Z.; Shiri, T.; Xia, J. MicrobiomeAnalyst 2.0: Comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Res. 2023, 51, W310–W318. [Google Scholar] [CrossRef] [PubMed]
- Calle, M.L. Statistical Analysis of Metagenomics Data. Genom. Inform. 2019, 17, e6. [Google Scholar] [CrossRef]
- Peschel, S.; Müller, C.L.; von Mutius, E.; Boulesteix, A.-L.; Depner, M. NetCoMi: Network construction and comparison for microbiome data in R. Brief. Bioinform. 2021, 22, bbaa290. [Google Scholar] [CrossRef]
- Menzel, U. RMThreshold: Signal-Noise Separation in Random Matrices by Using Eigenvalue Spectrum Analysis, Version 1.1; CRAN: Online. Available online: https://cran.r-project.org/web/packages/RMThreshold/index.html (accessed on 2 January 2022).
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Han, L.; Zhang, J.; Zhang, Y.; Lang, Q.; Li, F.; Han, A.; Bao, Y.; Li, K.; Alu, S. Environmental Risk Assessment of Metals in the Volcanic Soil of Changbai Mountain. Int. J. Environ. Res. Public Health 2019, 16, 2047. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I.; Head, I.M.; Stein, L.Y. The Family Nitrosomonadaceae. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014. [Google Scholar] [CrossRef]
- Flynn, D.F.; Mirotchnick, N.; Jain, M.; Palmer, M.I.; Naeem, S. Functional and phylogenetic diversity as predictors of biodiversity—ecosystem-function relationships. Ecology 2011, 92, 1573–1581. [Google Scholar] [CrossRef]
- Matus, F.; Salazar, O.; Aburto, F.; Zamorano, D.; Nájera, F.; Jovanović, R.; Guerra, C.; Reyes-Rojas, L.; Seguel, O.; Pfeiffer, M.; et al. Perspective of soil carbon sequestration in Chilean volcanic soils. npj Mater. Sustain. 2024, 2, 32. [Google Scholar] [CrossRef]
- Natasha, N.; Shahid, M.; Bibi, I.; Iqbal, J.; Khalid, S.; Murtaza, B.; Bakhat, H.F.; Umer, A.B.; Amjad, M.; Hammad, H.M.; et al. Zinc in soil-plant-human system: A data-analysis review. Sci. Total Environ. 2022, 808, 152024. [Google Scholar] [CrossRef]
- Vargas, R.A.; Valdés, N.; Balic, I.; Contreras, E.I.; Venegas, C.; Aranda, C.P.; Tello, M.; González, A.R. Amplicon of 16S rRNA Gene Sequencing of Fertilized Volcanic Soils from Southern Chile. Microbiol. Res. Announ. 2021, 10, e00590-20. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Alvarez, V.; King, G.M.; Nusslein, K. Comparative bacterial diversity in recent Hawaiian volcanic deposits of different ages. FEMS Microbiol. Ecol. 2007, 60, 60–73. [Google Scholar] [CrossRef]
- King, C.E.; King, G.M. Description of Thermogemmatispora carboxidivorans sp. nov., a carbon-monoxide-oxidizing member of the class Ktedonobacteria isolated from a geothermally heated biofilm, and analysis of carbon monoxide oxidation by members of the class Ktedonobacteria. Int. J. Syst. Evol. Microbiol. 2014, 64 Pt 4, 1244–1251. [Google Scholar] [CrossRef]
- Faust, K.; Raes, J. Microbial interactions: From networks to models. Nat. Rev. Microbiol. 2012, 10, 538–550. [Google Scholar] [CrossRef]
- Berry, D.; Widder, S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front. Microbiol. 2014, 5, 219. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Microbiol. 2019, 4, 2329–2345. [Google Scholar] [CrossRef]
- Maurice, K.; Bourceret, A.; Youssef, S.; Boivin, S.; Laurent-Webb, L.; Damasio, C.; Boukcim, H.; Selosse, M.A.; Ducousso, M. Anthropic disturbances impact the soil microbial network structure and stability to a greater extent than natural disturbances in an arid ecosystem. Sci. Total Environ. 2024, 907, 167969. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, H.; Li, P. Urbanization effects on soil microbial co-occurrence networks in Shanghai. Sci. Total Environ. 2023, 874, 162420. [Google Scholar] [CrossRef]
- Cornell, H.V.; Adler, P.B.; Levine, J.M. Disturbance and the paradox of complexity in ecological networks. Ecol. Lett. 2023, 26, 233–247. [Google Scholar] [CrossRef]
- Sun, Z.; Zhang, T.; Liu, J.; Wang,, X.; Feng, J.; Li, S.; Tang, S.; Jin, K. Soil microbial community variation among different land use types in the agro-pastoral ecotone of northern China is likely to be caused by anthropogenic activities. Front. Microbiol. 2024, 15, 1390286. [Google Scholar] [CrossRef]
- Fantom, N.; Dawson, R.A.; Prondvai, E.; Constant, P.; King, G.M.; Schäfer, H.; Hernández, M. Metabolism of CO and H2 by pioneer bacteria in volcanic soils and the phyllosphere. ISME J. 2025, 19, wraf053. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- King, G.M.; Weber, C.F.; Nanba, K.; Sato, Y.; Ohta, H. Atmospheric CO and hydrogen uptake and CO oxidizer phylogeny for Miyake-jima, Japan volcanic deposits. Microbes Environ. 2008, 23, 299–305. [Google Scholar] [CrossRef]
- Pal, C.; Bengtsson-Palme, J.; Kristiansson, E.; Larsson, D.J. Co-occurrence of resistance genes to antibiotics, biocides and metals reveals novel insights into their co-selection potential. BMC Genom. 2015, 16, 964. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.; Zhang, H.; Shi, W.; Liu, Y. Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front. Microbiol. 2019, 10, 1916. [Google Scholar] [CrossRef]
- Bazzi, W.; Fayad, A.G.A.; Nasser, A.; Haraoui, L.P.; Dewachi, O.; Abou-Sitta, G.; Nguyen, V.-K.; Abara, A.; Karah, N.; Landecker, H.; et al. Heavy metal toxicity in armed conflicts potentiates AMR in A. baumannii by selecting for antibiotic and heavy metal co-resistance mechanisms. Front. Microbiol. 2020, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa among bacterial and fungal communities in agricultural soils. Nat. Microbiol. 2018, 3, 890–899. [Google Scholar] [CrossRef]
- Aguado-Norese, C.; Maldonado, J.E.; Hodar, C.; Galvez, G.; Palma, D.E.; Cambiazo, V.; González, M. Ironing out the conflicts: Iron supplementation reduces negatives bacterial interactions in the rhizosphere of an Atacama-endemic perennial grass. Environ. Microbiome 2025, 20, 29. [Google Scholar] [CrossRef]
- Chen, J.; Xiao, Q.; Xu, D.; Li, Z.; Chao, L.; Li, X.; Liu, H.; Wang, P.; Zheng, Y.; Liu, X.; et al. Soil microbial community composition and co-occurrence network responses to mild and severe disturbances in volcanic areas. Sci. Total Environ. 2023, 901, 165889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tang, J.; Wang, L.; He, J. Human-impacted land uses enhance network complexity but impair stability of soil microbial communities. J. Clean. Prod. 2025, 512, 145665. [Google Scholar] [CrossRef]

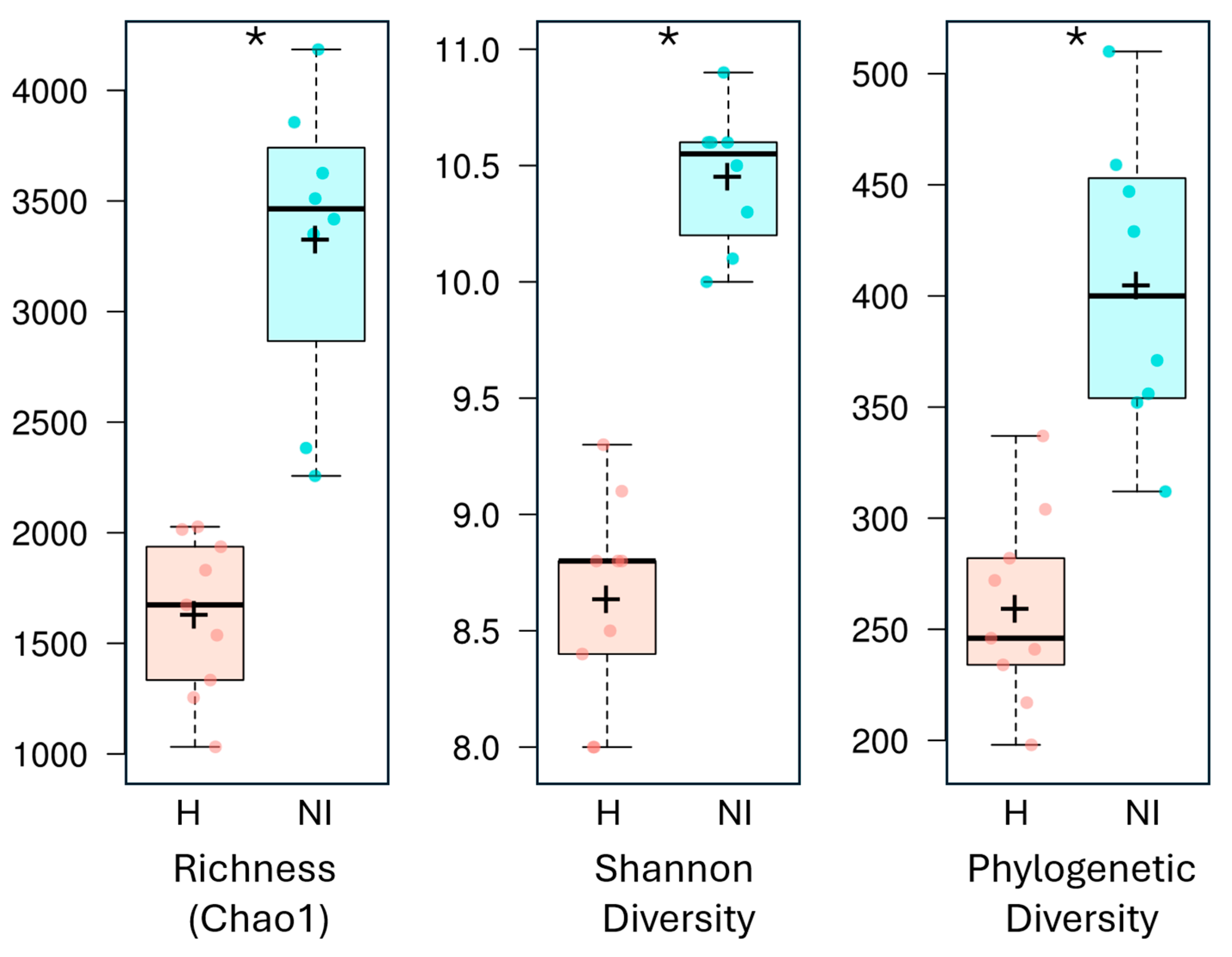

| Sample Internal Code | Type of Area | Assigned Sequences | Total ASVs (Richness) | Chao1 Index | Shannon Index | Phylogenetic Diversity (PD) | Good’s Coverage | NCBI Accession |
|---|---|---|---|---|---|---|---|---|
| H1.1 | Humanized | 68,525 | 947 | 1,032 | 8 | 198 | 99.5 | SAMN35159685 |
| H1.2 | Humanized | 63,869 | 1,193 | 1,255 | 8.8 | 217 | 99.6 | SAMN35159686 |
| H1.3 | Humanized | 63,284 | 1,841 | 2,015 | 8.8 | 304 | 99 | SAMN35159687 |
| H2.1 | Humanized | 55,326 | 1,836 | 1,937 | 9.3 | 272 | 99.3 | SAMN35159688 |
| H2.2 | Humanized | 59,035 | 1,711 | 1,831 | 9.1 | 234 | 99.2 | SAMN35159689 |
| H2.3 | Humanized | 59,328 | 1,244 | 1,334 | 8 | 246 | 99.4 | SAMN35159690 |
| H3.1 | Humanized | 58,607 | 1,887 | 2,027 | 8.8 | 337 | 99.1 | SAMN35159691 |
| H3.2 | Humanized | 58,755 | 1,470 | 1,537 | 8.5 | 282 | 99.5 | SAMN35159692 |
| H3.3 | Humanized | 60,309 | 1,561 | 1,674 | 8.4 | 241 | 99.3 | SAMN35159693 |
| NI1.1 | Non-intervened | 34,563 | 2,256 | 2,257 | 10 | 356 | 99.9 | SAMN35159694 |
| NI1.2 | Non-intervened | 33,802 | 2,383 | 2,383 | 10.1 | 312 | 99.9 | SAMN35159695 |
| NI2.1 | Non-intervened | 53,979 | 3,188 | 3,350 | 10.5 | 371 | 98.7 | SAMN35159696 |
| NI2.2 | Non-intervened | 54,642 | 3,232 | 3,418 | 10.6 | 352 | 98.6 | SAMN35159697 |
| NI2.3 | Non-intervened | 58,711 | 3,862 | 4,185 | 10.9 | 510 | 97.9 | SAMN35159698 |
| NI3.1 | Non-intervened | 53,866 | 3,308 | 3,511 | 10.3 | 459 | 98.4 | SAMN35159699 |
| NI3.2 | Non-intervened | 57,031 | 3,607 | 3,856 | 10.6 | 429 | 98.2 | SAMN35159700 |
| NI3.3 | Non-intervened | 59,409 | 3,436 | 3,626 | 10.6 | 447 | 98.6 | SAMN35159701 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguila-Torres, P.; González, M.; Hernández, M.; Aguado-Norese, C.; Maldonado, J.E.; Miranda, R.M.; González-Stegmaier, R.; Palma, D.E.; Rojas, L.A.; Mellado, M. Characterization of Bacterial Communities in Volcanic Soil from Northern Patagonian Area of Chile. Microorganisms 2025, 13, 2519. https://doi.org/10.3390/microorganisms13112519
Aguila-Torres P, González M, Hernández M, Aguado-Norese C, Maldonado JE, Miranda RM, González-Stegmaier R, Palma DE, Rojas LA, Mellado M. Characterization of Bacterial Communities in Volcanic Soil from Northern Patagonian Area of Chile. Microorganisms. 2025; 13(11):2519. https://doi.org/10.3390/microorganisms13112519
Chicago/Turabian StyleAguila-Torres, Patricia, Mauricio González, Marcela Hernández, Constanza Aguado-Norese, Jonathan E. Maldonado, Richard M. Miranda, Roxana González-Stegmaier, Daniel E. Palma, Luis A. Rojas, and Macarena Mellado. 2025. "Characterization of Bacterial Communities in Volcanic Soil from Northern Patagonian Area of Chile" Microorganisms 13, no. 11: 2519. https://doi.org/10.3390/microorganisms13112519
APA StyleAguila-Torres, P., González, M., Hernández, M., Aguado-Norese, C., Maldonado, J. E., Miranda, R. M., González-Stegmaier, R., Palma, D. E., Rojas, L. A., & Mellado, M. (2025). Characterization of Bacterial Communities in Volcanic Soil from Northern Patagonian Area of Chile. Microorganisms, 13(11), 2519. https://doi.org/10.3390/microorganisms13112519





