Clinical and Genomic Insights into Antifungal Resistance in Aspergillus Isolates from Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Strains, Media, and Conditions
2.2. Broth Microdilution Assay
2.3. Sequencing Analysis
2.4. Profiling the cyp51A Gene for Phylogenetic Analysis
2.5. Statistical Analysis
2.6. Ethics Statement
3. Results
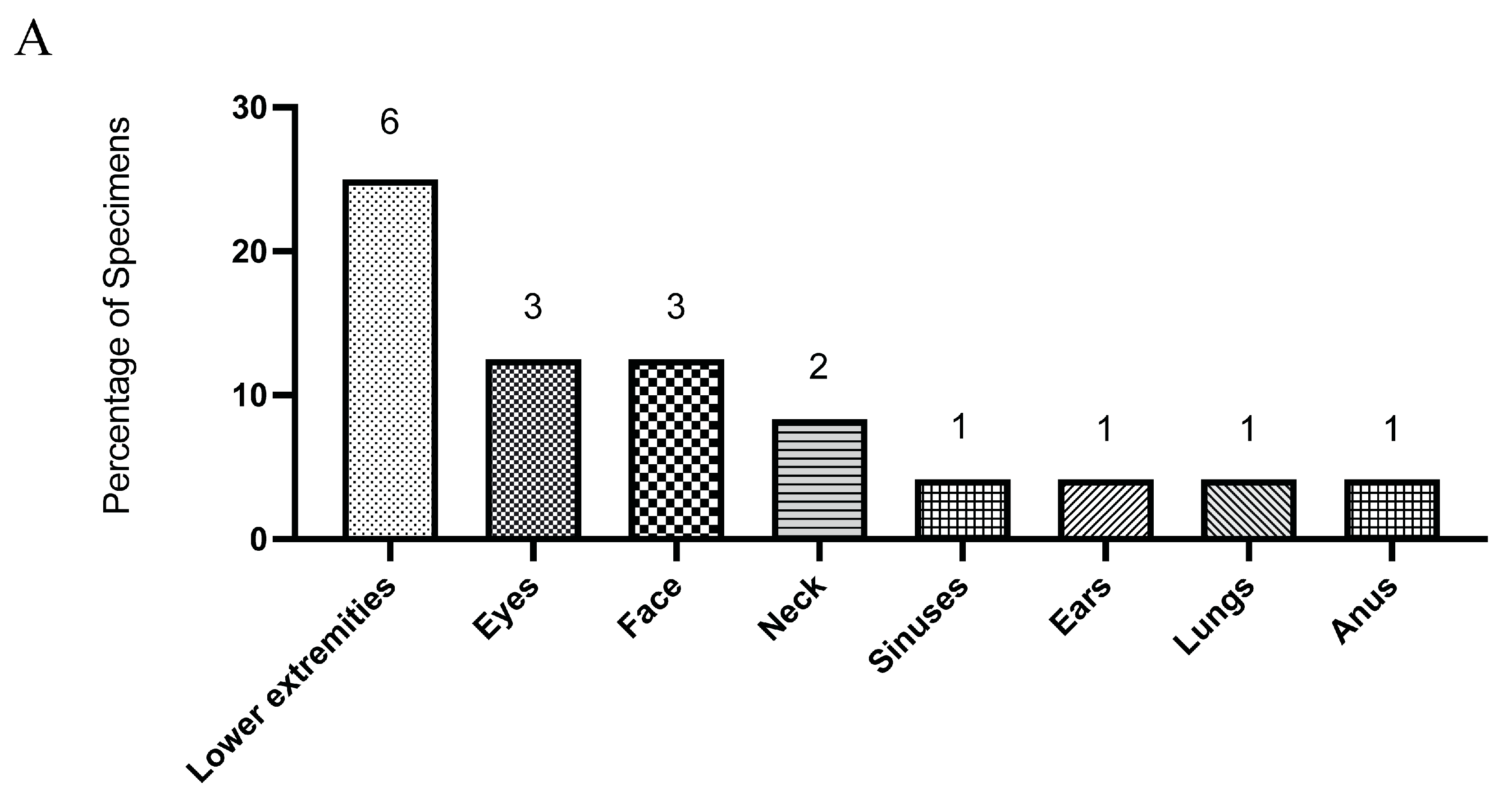
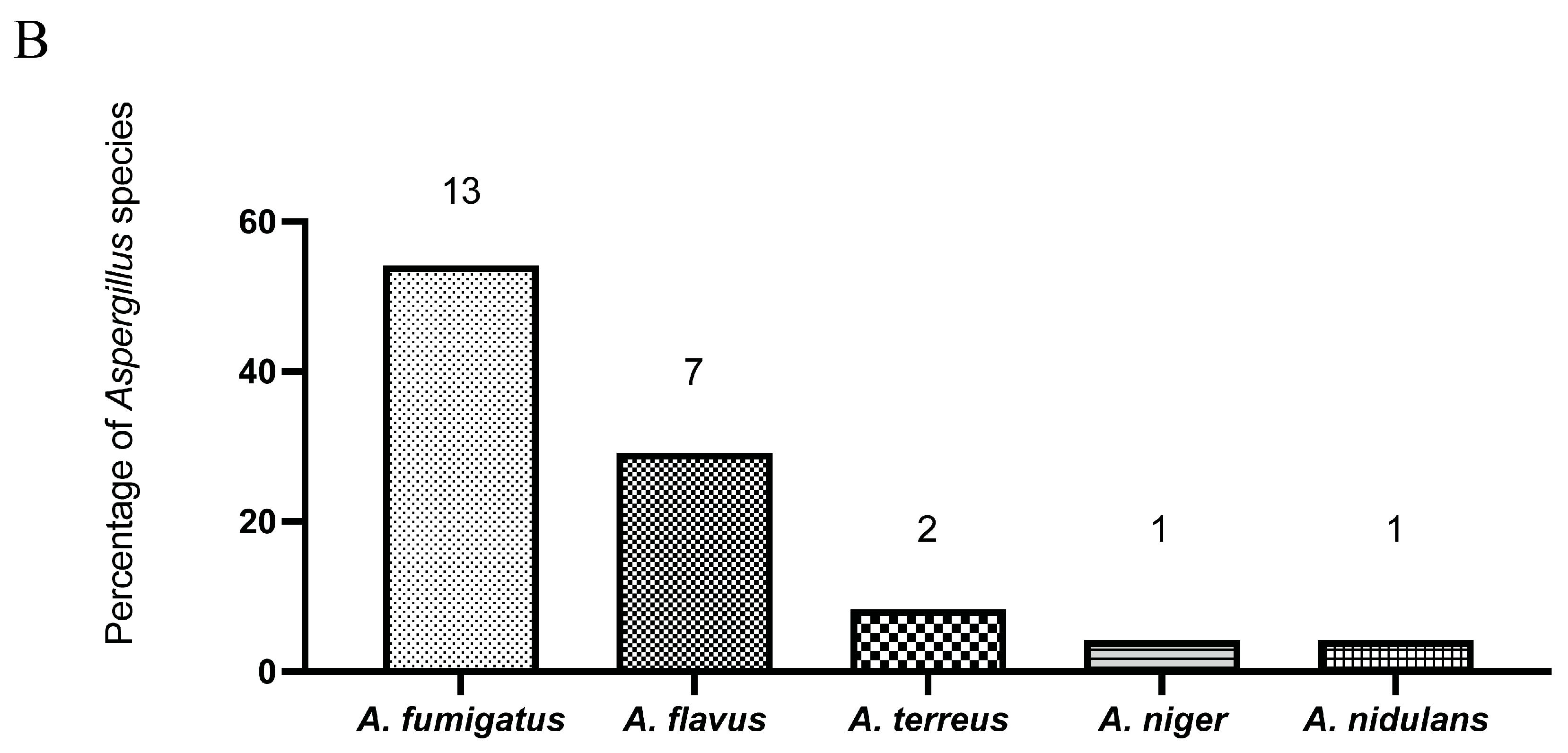
3.1. Aspergillus fumigatus Is the Most Common Isolate from Sterile Clinical Specimens
3.2. The Minimum Inhibitory Concentrations (MICs) of Voriconazole Remain Low for Most Aspergillus Isolates
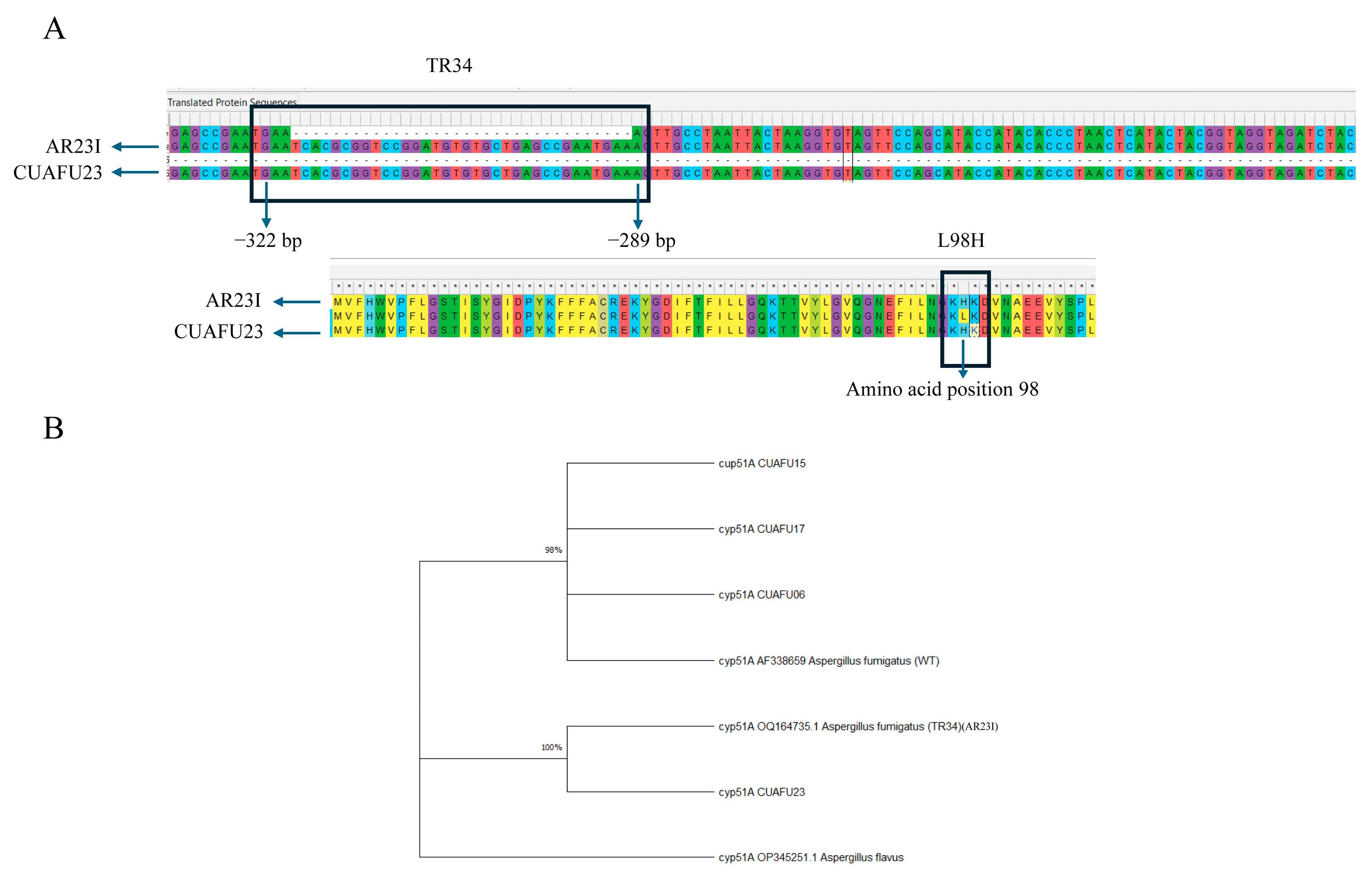
3.3. The cyp51A Gene of Strain CUAFU23 Carried the TR34/L98H Mutation
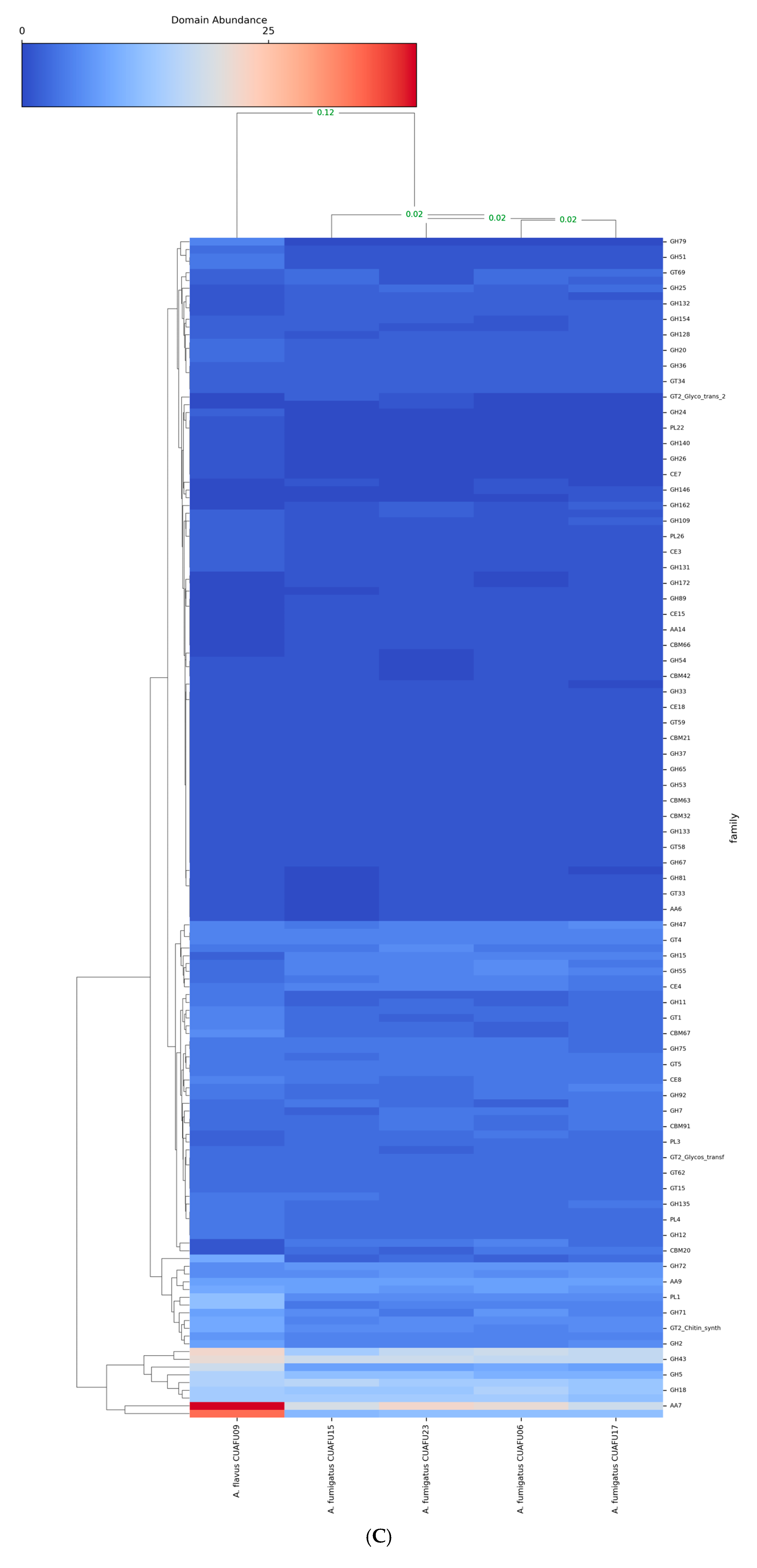
3.4. The cyp51A-Mutated Strain Exhibited Distinct Genomic Domain Profiles
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lass-Florl, C. The changing face of epidemiology of invasive fungal disease in Europe. Mycoses 2009, 52, 197–205. [Google Scholar] [CrossRef]
- Lee, M.J.; Liu, H.; Barker, B.M.; Snarr, B.D.; Gravelat, F.N.; Al Abdallah, Q.; Gavino, C.; Baistrocchi, S.R.; Ostapska, H.; Xiao, T.; et al. The Fungal Exopolysaccharide Galactosaminogalactan Mediates Virulence by Enhancing Resistance to Neutrophil Extracellular Traps. PLoS Pathog. 2015, 11, e1005187. [Google Scholar] [CrossRef]
- Bottery, M.J.; van Rhijn, N.; Chown, H.; Rhodes, J.L.; Celia-Sanchez, B.N.; Brewer, M.T.; Momany, M.; Fisher, M.C.; Knight, C.G.; Bromley, M.J. Elevated mutation rates in multi-azole resistant Aspergillus fumigatus drive rapid evolution of antifungal resistance. Nat. Commun. 2024, 15, 10654. [Google Scholar] [CrossRef]
- Patterson, T.F.; Thompson, G.R., 3rd; Denning, D.W.; Fishman, J.A.; Hadley, S.; Herbrecht, R.; Kontoyiannis, D.P.; Marr, K.A.; Morrison, V.A.; Nguyen, M.H.; et al. Practice Guidelines for the Diagnosis and Management of Aspergillosis: 2016 Update by the Infectious Diseases Society of America. Clin. Infect. Dis. 2016, 63, e1–e60. [Google Scholar] [CrossRef]
- Gonzalez-Jimenez, I.; Lucio, J.; Amich, J.; Cuesta, I.; Sanchez Arroyo, R.; Alcazar-Fuoli, L.; Mellado, E. A Cyp51B Mutation Contributes to Azole Resistance in Aspergillus fumigatus. J. Fungi 2020, 6, 315. [Google Scholar] [CrossRef]
- He, X.; Kusuya, Y.; Hagiwara, D.; Toyotome, T.; Arai, T.; Bian, C.; Nagayama, M.; Shibata, S.; Watanabe, A.; Takahashi, H. Genomic diversity of the pathogenic fungus Aspergillus fumigatus in Japan reveals the complex genomic basis of azole resistance. Commun. Biol. 2024, 7, 274. [Google Scholar] [CrossRef]
- De Pauw, B.; Walsh, T.J.; Donnelly, J.P.; Stevens, D.A.; Edwards, J.E.; Calandra, T.; Pappas, P.G.; Maertens, J.; Lortholary, O.; Kauffman, C.A.; et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 2008, 46, 1813–1821. [Google Scholar] [PubMed]
- Clinical and Laboratory Standards Institute. Approved Standard. In Reference Method for Broth Dilution Antifungal Susceptibility Testing of Filamentous Fungi, 3rd ed.; CLSI Document M38-A3: Wayne, PA, USA, 2017. [Google Scholar]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33 (Suppl. 1), D501–D504. [Google Scholar] [CrossRef]
- Tarailo-Graovac, M.; Chen, N. Using RepeatMasker to Identify Repetitive Elements in Genomic Sequences. Curr. Protoc. Bioinform. 2009, 25, 4–10. [Google Scholar] [CrossRef]
- Jonathan, M.; Palmer, J.S. Funannotate, v1.8.1; Eukaryotic Genome Annotation; Zenodo: Genève, Switzerland, 2020.
- Stanke, M.; Diekhans, M.; Baertsch, R.; Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 2008, 24, 637–644. [Google Scholar] [CrossRef]
- Johnson, A.D.; Handsaker, R.E.; Pulit, S.L.; Nizzari, M.M.; O’Donnell, C.J.; de Bakker, P.I.W. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008, 24, 2938–2939. [Google Scholar] [CrossRef]
- Majoros, W.H.; Pertea, M.; Salzberg, S.L. TigrScan and GlimmerHMM: Two open source ab initio eukaryotic gene-finders. Bioinformatics 2004, 20, 2878–2879. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Haas, B.J.; Salzberg, S.L.; Zhu, W.; Pertea, M.; Allen, J.E.; Orvis, J.; White, O.; Buell, C.R.; Wortman, J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008, 9, R7. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, G.R., 3rd; Chen, S.C.; Alfouzan, W.A.; Izumikawa, K.; Colombo, A.L.; Maertens, J. A global perspective of the changing epidemiology of invasive fungal disease and real-world experience with the use of isavuconazole. Med. Mycol. 2024, 62, myae083. [Google Scholar] [CrossRef]
- Thammahong, A.; Thayidathara, P.; Suksawat, K.; Chindamporn, A. Invasive Aspergillus Infections in a Thai Tertiary-Care Hospital during 2006–2011. Adv. Microbiol. 2015, 05, 298–306. [Google Scholar] [CrossRef][Green Version]
- Rudramurthy, S.M.; Paul, R.A.; Chakrabarti, A.; Mouton, J.W.; Meis, J.F. Invasive Aspergillosis by Aspergillus flavus: Epidemiology, Diagnosis, Antifungal Resistance, and Management. J. Fungi 2019, 5, 55. [Google Scholar] [CrossRef] [PubMed]
- Tangwattanachuleeporn, M.; Minarin, N.; Saichan, S.; Sermsri, P.; Mitkornburee, R.; Gross, U.; Chindamporn, A.; Bader, O. Prevalence of azole-resistant Aspergillus fumigatus in the environment of Thailand. Med. Mycol. 2017, 55, 429–435. [Google Scholar]
- Daloh, M.; Wisessombat, S.; Pinchai, N.; Santajit, S.; Bhoopong, P.; Soaart, A.; Chueajeen, K.; Jitlang, A.; Sama-Ae, I. High prevalence and genetic diversity of a single ancestral origin azole-resistant Aspergillus fumigatus in indoor environments at Walailak University, Southern Thailand. Environ. Microbiol. 2022, 24, 4641–4651. [Google Scholar] [CrossRef]
- Chowdhary, A.; Kathuria, S.; Xu, J.; Meis, J.F. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013, 9, e1003633, Erratum in PLoS Pathog. 2013, 9. [Google Scholar] [CrossRef]
- Rocchi, S.; Daguindau, E.; Grenouillet, F.; Deconinck, E.; Bellanger, A.-P.; Garcia-Hermoso, D.; Bretagne, S.; Reboux, G.; Millon, L. Azole-resistant Aspergillus fumigatus isolate with the TR34/L98H mutation in both a fungicide-sprayed field and the lung of a hematopoietic stem cell transplant recipient with invasive aspergillosis. J. Clin. Microbiol. 2014, 52, 1724–1726. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Huang, J.; Liu, Y.; Zhang, Y.; Gao, Y. Unveiling environmental transmission risks: Comparative analysis of azole resistance in Aspergillus fumigatus clinical and environmental isolates from Yunnan, China. Microbiol. Spectr. 2024, 12, e0159424. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.; Ajayababu, A.; Chowdhury, S.; Singh, G.; Ray, A. Epidemiology of Triazole Resistant Aspergillus fumigatus in Asia: A Systematic Review and Meta-Analysis. Mycoses 2025, 68, e70099. [Google Scholar] [CrossRef]
- Hokken, M.W.J.; Coolen, J.P.M.; Steenbreker, H.; Zoll, J.; Baltussen, T.J.H.; Verweij, P.E.; Melchers, W.J.G. The Transcriptome Response to Azole Compounds in Aspergillus fumigatus Shows Differential Gene Expression across Pathways Essential for Azole Resistance and Cell Survival. J. Fungi 2023, 9, 807. [Google Scholar] [CrossRef]
- Bultman, K.M.; Kowalski, C.H.; Cramer, R.A. Aspergillus fumigatus virulence through the lens of transcription factors. Med. Mycol. 2017, 55, 24–38. [Google Scholar] [CrossRef][Green Version]


| Fungal Name (Total Number)/Antifungal | MIC (µg/mL) | MIC Range (µg/mL) | ECV (µg/mL) (CLSI M57S, 2022) | Breakpoint (µg/mL) | WT (%)(N) | Non-W (%)(N) |
|---|---|---|---|---|---|---|
| Aspergillus fumigatus (13) | ||||||
| Amphotericin B | 3.15 | 1–8 | 2 | - | 23.08% (3) | 76.92% (10) |
| Voriconazole | 0.69 | 0.0625–4 | 0.5;1;2 (2024) | 85% (11) | 15% (2) | |
| Aspergillus flavus (7) | ||||||
| Amphotericin B | 4.86 | 2–8 | 4 | - | 14.29% (1) | 85.71% (6) |
| Voriconazole | 0.19 | 0.0625–0.25 | 2 | - | 100% (7) | 0% (0) |
| Aspergillus terreus (2) | ||||||
| Amphotericin B | 5 | 2–8 | 4 | - | 50% (1) | 50% (1) |
| Voriconazole | 0.125 | 0.125 | 2 | - | 100% (2) | 0% (0) |
| Aspergillus niger (1) | ||||||
| Amphotericin B | 16 | 16 | 2 | - | 0% (0) | 100% (1) |
| Voriconazole | 0.25 | 0.25 | 2 | - | 100% (1) | 0% (0) |
| Aspergillus nidulans (1) | ||||||
| Amphotericin B | 4 | 4 | - | - | - | - |
| Voriconazole | 0.125 | 0.125 | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Onchan, T.; Langsiri, N.; Thammahong, A. Clinical and Genomic Insights into Antifungal Resistance in Aspergillus Isolates from Thailand. Microorganisms 2025, 13, 2495. https://doi.org/10.3390/microorganisms13112495
Onchan T, Langsiri N, Thammahong A. Clinical and Genomic Insights into Antifungal Resistance in Aspergillus Isolates from Thailand. Microorganisms. 2025; 13(11):2495. https://doi.org/10.3390/microorganisms13112495
Chicago/Turabian StyleOnchan, Thanyarat, Nattapong Langsiri, and Arsa Thammahong. 2025. "Clinical and Genomic Insights into Antifungal Resistance in Aspergillus Isolates from Thailand" Microorganisms 13, no. 11: 2495. https://doi.org/10.3390/microorganisms13112495
APA StyleOnchan, T., Langsiri, N., & Thammahong, A. (2025). Clinical and Genomic Insights into Antifungal Resistance in Aspergillus Isolates from Thailand. Microorganisms, 13(11), 2495. https://doi.org/10.3390/microorganisms13112495







