Case Study on Shifts in Human Skin Microbiome During Antarctica Expeditions
Abstract
1. Introduction
2. Materials and Methods
2.1. Expedition Routes and Skin Swap Sample Collection
2.2. DNA Extraction and Amplicon Sequencing
2.3. Metadata Preprocessing
2.4. Skin Microbiome Analysis
2.5. Removal of Batch Effects
2.6. Statistical Analysis
3. Results
3.1. Environmental and Lifestyle Characteristics of Hosts During Antarctic Expedition
3.2. Skin Microbial Communities in Expedition Hosts During Trips to Antarctica
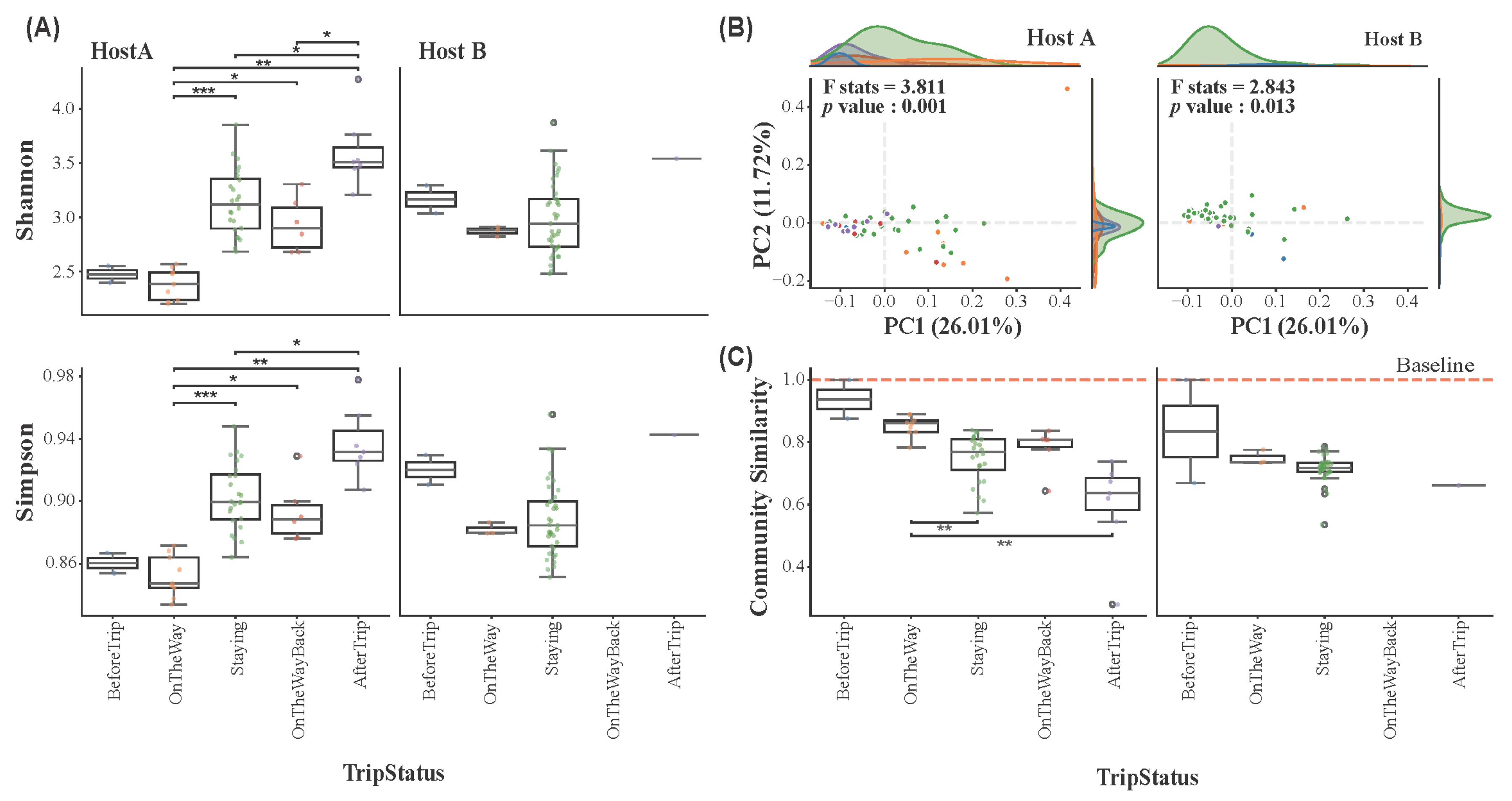
3.3. Skin Microbiome Diversity by Trip Status
3.4. Host-Specific Skin Microbiome Resilience in Response to Environmental Stress
3.5. Correlation of Environmental and Lifestyle Factors and Skin Microbiome Changes
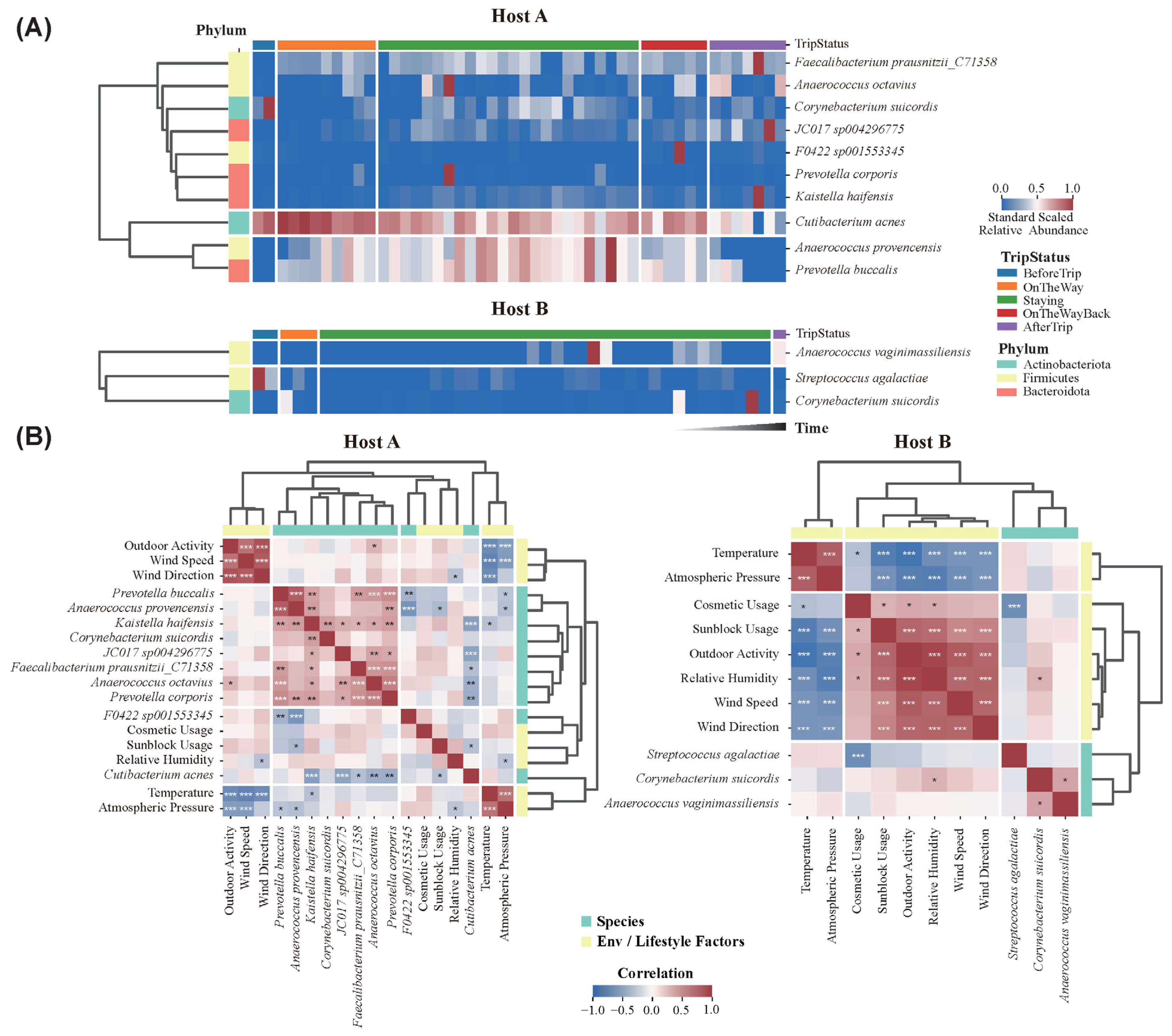
3.6. Temporal Shifts and Environmental Correlations of Key Skin Microbes
3.7. Representative Skin Microbes Are Influenced by Environmental and Lifestyle Factors During Antarctic Expedition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
References
- Grice, E.A.; Segre, J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The Human Skin Microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Han, J.H.; Kim, H.S. Skin Deep: The Potential of Microbiome Cosmetics. J. Microbiol. 2024, 62, 181–199. [Google Scholar] [CrossRef] [PubMed]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human Skin Microbiome: Impact of Intrinsic and Extrinsic Factors on Skin Microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef]
- Dimitriu, P.A.; Iker, B.; Malik, K.; Leung, H.; Mohn, W.W.; Hillebrand, G.G. New Insights into the Intrinsic and Extrinsic Factors That Shape the Human Skin Microbiome. mBio 2019, 10, e00839-19. [Google Scholar] [CrossRef]
- Szabó, K.; Erdei, L.; Bolla, B.S.; Tax, G.; Bíró, T.; Kemény, L. Factors Shaping the Composition of the Cutaneous Microbiota. Br. J. Dermatol. 2017, 176, 344–351. [Google Scholar] [CrossRef]
- Schommer, N.N.; Gallo, R.L. Structure and Function of the Human Skin Microbiome. Trends Microbiol. 2013, 21, 660–668. [Google Scholar] [CrossRef]
- Leo, C.; Nardi, F.; Cucini, C.; Frati, F.; Convey, P.; Weedon, J.T.; Roelofs, D.; Carapelli, A. Evidence for Strong Environmental Control on Bacterial Microbiomes of Antarctic Springtails. Sci. Rep. 2021, 11, 2973. [Google Scholar] [CrossRef]
- Santos, A.; Gómez-Espinoza, O.; Núñez-Montero, K.; Zárate, A.; Andreote, F.D.; Pylro, V.S.; Bravo, L.; Barrientos, L. Measuring the Effect of Climate Change in Antarctic Microbial Communities: Toward Novel Experimental Approaches. Curr. Opin. Biotechnol. 2023, 81, 102918. [Google Scholar] [CrossRef]
- Cannone, N.; Malfasi, F.; Favero-Longo, S.E.; Convey, P.; Guglielmin, M. Acceleration of Climate Warming and Plant Dynamics in Antarctica. Curr. Biol. 2022, 32, 1599–1606.e2. [Google Scholar] [CrossRef]
- Vincent, W.F. Microbial Ecosystem Responses to Rapid Climate Change in the Arctic. ISME J. 2010, 4, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Koo, J.-H.; Choi, T.; Cho, Y.; Lee, H.; Kim, J.; Ahn, D.H.; Kim, J.; Lee, Y.G. The Variation in Aerosol Optical Depth over the Polar Stations of Korea. Aerosol Air Qual. Res. 2018, 18, 3020–3210. [Google Scholar] [CrossRef]
- Park, B.-K.; Chang, S.-K.; Yoon, H.I.; Chung, H. Recent Retreat of Ice Cliffs, King George Island, South Shetland Islands, Antarctic Peninsula. Ann. Glaciol. 1998, 27, 633–635. [Google Scholar] [CrossRef]
- Hur, S.D.; Chung, J.; Namgerel, Y.; Lee, J. Seasonality of Isotopic and Chemical Composition of Snowpack in the Vicinity of Jang Bogo Station, East Antarctica. Pol. Polar Res. 2021, 42, 221–236. [Google Scholar] [CrossRef]
- Kanao, M.; Murayama, T.; Park, Y. Infrasound Signals in Coastal Environment at Jang Bogo Station, Terra Nova Bay, Antarctica. Int. J. Geosci. 2022, 13, 1103–1114. [Google Scholar] [CrossRef]
- Yoo, J.-H.; Choi, T.; Chun, H.-Y.; Kim, Y.-H.; Song, I.-S.; Song, B.-G. Inertia-Gravity Waves Revealed in Radiosonde Data at Jang Bogo Station, Antarctica (74°37′ S, 164°13′ E): 1. Characteristics, Energy, and Momentum Flux. JGR Atmos. 2018, 123, 13305–13331. [Google Scholar] [CrossRef]
- Turroni, S.; Rampelli, S.; Biagi, E.; Consolandi, C.; Severgnini, M.; Peano, C.; Quercia, S.; Soverini, M.; Carbonero, F.G.; Bianconi, G.; et al. Temporal Dynamics of the Gut Microbiota in People Sharing a Confined Environment, a 520-Day Ground-Based Space Simulation, MARS500. Microbiome 2017, 5, 39. [Google Scholar] [CrossRef]
- Garrett-Bakelman, F.E.; Darshi, M.; Green, S.J.; Gur, R.C.; Lin, L.; Macias, B.R.; McKenna, M.J.; Meydan, C.; Mishra, T.; Nasrini, J.; et al. The NASA Twins Study: A Multidimensional Analysis of a Year-Long Human Spaceflight. Science 2019, 364, eaau8650. [Google Scholar] [CrossRef]
- Li, K.; Dan, Z.; Gesang, L.; Wang, H.; Zhou, Y.; Du, Y.; Ren, Y.; Shi, Y.; Nie, Y. Comparative Analysis of Gut Microbiota of Native Tibetan and Han Populations Living at Different Altitudes. PLoS ONE 2016, 11, e0155863. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Bhushan, B.; Eslavath, M.R.; Srivastava, A.K.; Meena, R.C.; Varshney, R.; Ganju, L. Impact of High Altitude on Composition and Functional Profiling of Oral Microbiome in Indian Male Population. Sci. Rep. 2023, 13, 4038. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Rohil, V.; Bhushan, B.; Eslavath, M.R.; Gupta, H.; Chanda, S.; Kumar, B.; Varshney, R.; Ganju, L. Probiotics Maintain the Gut Microbiome Homeostasis during Indian Antarctic Expedition by Ship. Sci. Rep. 2021, 11, 18793. [Google Scholar] [CrossRef] [PubMed]
- Grant, G.J.; Kohli, I.; Mohammad, T.F. A Narrative Review of the Impact of Ultraviolet Radiation and Sunscreen on the Skin Microbiome. Photodermatol. Photoimmunol. Photomed. 2024, 40, e12943. [Google Scholar] [CrossRef] [PubMed]
- NIST IR 8250; Simulation Interoperability Standards Organization. Recommended Practices for Modeling and Simulation (M&S) Ecosystem Framework. National Institute of Standards and Technology: Gaithersburg, MD, USA, 2019. [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Duchesnay, É.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Correction in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McDonald, D.; Jiang, Y.; Balaban, M.; Cantrell, K.; Zhu, Q.; Gonzalez, A.; Morton, J.T.; Nicolaou, G.; Parks, D.H.; Karst, S.M.; et al. Greengenes2 Unifies Microbial Data in a Single Reference Tree. Nat. Biotechnol. 2024, 42, 715–718, Correction in Nat. Biotechnol. 2024, 42, 813. [Google Scholar] [CrossRef]
- Katoh, K. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Rideout, J.R.; Caporaso, G.; Bolyen, E.; McDonald, D.; Baeza, Y.V.; Alastuey, J.C.; Pitman, A.; Morton, J.; Navas, J.; Kestrel, G.; et al. Biocore/Scikit-Bio, Scikit-Bio 0.5.9. Maintenance Release 2023. Zenodo: Geneva, Switzerland, 2023. [CrossRef]
- Kim, B.-R.; Shin, J.; Guevarra, R.B.; Lee, J.H.; Kim, D.W.; Seol, K.-H.; Lee, J.-H.; Kim, H.B.; Isaacson, R.E. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Kemp, P.F.; Aller, J.Y. Bacterial Diversity in Aquatic and Other Environments: What 16S rDNA Libraries Can Tell Us. FEMS Microbiol. Ecol. 2004, 47, 161–177. [Google Scholar] [CrossRef]
- Behdenna, A.; Colange, M.; Haziza, J.; Gema, A.; Appé, G.; Azencott, C.-A.; Nordor, A. pyComBat, a Python Tool for Batch Effects Correction in High-Throughput Molecular Data Using Empirical Bayes Methods. BMC Bioinform. 2023, 24, 459. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. Umap: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; Kenett, R.S., Longford, N.T., Piegorsch, W.W., Ruggeri, F., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 1–15. ISBN 978-1-118-44511-2. [Google Scholar]
- Shade, A.; Peter, H.; Allison, S.D.; Baho, D.L.; Berga, M.; Bürgmann, H.; Huber, D.H.; Langenheder, S.; Lennon, J.T.; Martiny, J.B.H.; et al. Fundamentals of Microbial Community Resistance and Resilience. Front. Microbiol. 2012, 3, 417. [Google Scholar] [CrossRef] [PubMed]
- Mallick, H.; Rahnavard, A.; McIver, L.J.; Ma, S.; Zhang, Y.; Nguyen, L.H.; Tickle, T.L.; Weingart, G.; Ren, B.; Schwager, E.H.; et al. Multivariable Association Discovery in Population-Scale Meta-Omics Studies. PLoS Comput. Biol. 2021, 17, e1009442. [Google Scholar] [CrossRef] [PubMed]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Hunter, J.D. Matplotlib: A 2D Graphics Environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Waskom, M. Seaborn: Statistical Data Visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]
- Flowers, L.; Grice, E.A. The Skin Microbiota: Balancing Risk and Reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef]
- Harris-Tryon, T.A.; Grice, E.A. Microbiota and Maintenance of Skin Barrier Function. Science 2022, 376, 940–945. [Google Scholar] [CrossRef]
- Dekio, I.; Asahina, A.; Shah, H.N. Unravelling the Eco-Specificity and Pathophysiological Properties of Cutibacterium Species in the Light of Recent Taxonomic Changes. Anaerobe 2021, 71, 102411. [Google Scholar] [CrossRef]
- Tsankov, N.; Mateev, D.; Darlenski, R. Skin Hydration, Microrelief and Greasiness of Normal Skin in Antarctica. Acad. Dermatol. Venereol. 2018, 32, 482–485. [Google Scholar] [CrossRef]
- Monsalves, M.T.; Ollivet-Besson, G.P.; Amenabar, M.J.; Blamey, J.M. Isolation of a Psychrotolerant and UV-C-Resistant Bacterium from Elephant Island, Antarctica with a Highly Thermoactive and Thermostable Catalase. Microorganisms 2020, 8, 95. [Google Scholar] [CrossRef]
- Sugita, T.; Yamazaki, T.; Yamada, S.; Takeoka, H.; Cho, O.; Tanaka, T.; Ohno, G.; Watanabe, K.; Makimura, K.; Ohshima, H.; et al. Temporal Changes in the Skin Malassezia Microbiota of Members of the Japanese Antarctic Research Expedition (JARE): A Case Study in Antarctica as a Pseudo-Space Environment. Med. Mycol. 2015, 53, 717–724. [Google Scholar] [CrossRef]
- Tozzo, P.; Delicati, A.; Caenazzo, L. Skin Microbial Changes during Space Flights: A Systematic Review. Life 2022, 12, 1498. [Google Scholar] [CrossRef]
- Troitsky, T.S.; Laine, V.N.; Lilley, T.M. When the Host’s Away, the Pathogen Will Play: The Protective Role of the Skin Microbiome during Hibernation. Anim. Microbiome 2023, 5, 66. [Google Scholar] [CrossRef]
- Castro-Severyn, J.; Pardo-Esté, C.; Mendez, K.N.; Fortt, J.; Marquez, S.; Molina, F.; Castro-Nallar, E.; Remonsellez, F.; Saavedra, C.P. Living to the High Extreme: Unraveling the Composition, Structure, and Functional Insights of Bacterial Communities Thriving in the Arsenic-Rich Salar de Huasco Altiplanic Ecosystem. Microbiol. Spectr. 2021, 9, e00444-21. [Google Scholar] [CrossRef] [PubMed]
- Coppola, D.; Lauritano, C.; Zazo, G.; Nuzzo, G.; Fontana, A.; Ianora, A.; Costantini, M.; Verde, C.; Giordano, D. Biodiversity of UV-Resistant Bacteria in Antarctic Aquatic Environments. JMSE 2023, 11, 968. [Google Scholar] [CrossRef]
- Armstrong, A.; Valverde, A.; Ramond, J.-B.; Makhalanyane, T.P.; Jansson, J.K.; Hopkins, D.W.; Aspray, T.J.; Seely, M.; Trindade, M.I.; Cowan, D.A. Temporal Dynamics of Hot Desert Microbial Communities Reveal Structural and Functional Responses to Water Input. Sci. Rep. 2016, 6, 34434. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Löber, U.; Bartolomaeus, H.; Maier, L.; Müller, D.N.; Wilck, N.; Jarquín-Díaz, V.H.; Forslund-Startceva, S.K. Baseline Microbiome Composition Impacts Resilience to and Recovery Following Antibiotics. bioRxiv 2024. [Google Scholar] [CrossRef]
- Tierney, B.T.; Kim, J.; Overbey, E.G.; Ryon, K.A.; Foox, J.; Sierra, M.A.; Bhattacharya, C.; Damle, N.; Najjar, D.; Park, J.; et al. Longitudinal Multi-Omics Analysis of Host Microbiome Architecture and Immune Responses during Short-Term Spaceflight. Nat. Microbiol. 2024, 9, 1661–1675. [Google Scholar] [CrossRef]
- Caswell, G.; Eshelby, B. Skin Microbiome Considerations for Long Haul Space Flights. Front. Cell Dev. Biol. 2022, 10, 956432. [Google Scholar] [CrossRef]
- Schwendner, P.; Mahnert, A.; Koskinen, K.; Moissl-Eichinger, C.; Barczyk, S.; Wirth, R.; Berg, G.; Rettberg, P. Preparing for the Crewed Mars Journey: Microbiota Dynamics in the Confined Mars500 Habitat during Simulated Mars Flight and Landing. Microbiome 2017, 5, 129. [Google Scholar] [CrossRef] [PubMed]
- Tesei, D.; Jewczynko, A.; Lynch, A.; Urbaniak, C. Understanding the Complexities and Changes of the Astronaut Microbiome for Successful Long-Duration Space Missions. Life 2022, 12, 495. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, S.M.J.; Said-Salman, I.; Mortazavi, A.R.; El Khatib, S.; Sihver, L. How the Adaptation of the Human Microbiome to Harsh Space Environment Can Determine the Chances of Success for a Space Mission to Mars and Beyond. Front. Microbiol. 2024, 14, 1237564. [Google Scholar] [CrossRef] [PubMed]
- Voorhies, A.A.; Mark Ott, C.; Mehta, S.; Pierson, D.L.; Crucian, B.E.; Feiveson, A.; Oubre, C.M.; Torralba, M.; Moncera, K.; Zhang, Y.; et al. Study of the Impact of Long-Duration Space Missions at the International Space Station on the Astronaut Microbiome. Sci. Rep. 2019, 9, 9911. [Google Scholar] [CrossRef]
- Francioli, D.; Cid, G.; Kanukollu, S.; Ulrich, A.; Hajirezaei, M.-R.; Kolb, S. Flooding Causes Dramatic Compositional Shifts and Depletion of Putative Beneficial Bacteria on the Spring Wheat Microbiota. Front. Microbiol. 2021, 12, 773116. [Google Scholar] [CrossRef]
- Boxberger, M.; Cenizo, V.; Cassir, N.; La Scola, B. Challenges in Exploring and Manipulating the Human Skin Microbiome. Microbiome 2021, 9, 125. [Google Scholar] [CrossRef]
- Naik, S.; Bouladoux, N.; Wilhelm, C.; Molloy, M.J.; Salcedo, R.; Kastenmuller, W.; Deming, C.; Quinones, M.; Koo, L.; Conlan, S.; et al. Compartmentalized Control of Skin Immunity by Resident Commensals. Science 2012, 337, 1115–1119. [Google Scholar] [CrossRef]
- Meisel, J.S.; Sfyroera, G.; Bartow-McKenney, C.; Gimblet, C.; Bugayev, J.; Horwinski, J.; Kim, B.; Brestoff, J.R.; Tyldsley, A.S.; Zheng, Q.; et al. Commensal Microbiota Modulate Gene Expression in the Skin. Microbiome 2018, 6, 20. [Google Scholar] [CrossRef]
- Kono, H.; Rock, K.L. How Dying Cells Alert the Immune System to Danger. Nat. Rev. Immunol. 2008, 8, 279–289. [Google Scholar] [CrossRef]
- Aguilera, Á.; De Diego-Castilla, G.; Osuna, S.; Bardera, R.; Sor Mendi, S.; Blanco, Y.; González-Toril, E. Microbial Ecology in the Atmosphere: The Last Extreme Environment. In Extremophilic Microbes and Metabolites—Diversity, Bioprospecting and Biotechnological Applications; Najjari, A., Cherif, A., Sghaier, H., Imene Ouzari, H., Eds.; IntechOpen: London, UK, 2021; ISBN 978-1-83969-038-9. [Google Scholar]
- Van Houdt, R.; De Boever, P.; Coninx, I.; Le Calvez, C.; Dicasillati, R.; Mahillon, J.; Mergeay, M.; Leys, N. Evaluation of the Airborne Bacterial Population in the Periodically Confined Antarctic Base Concordia. Microb. Ecol. 2009, 57, 640–648. [Google Scholar] [CrossRef]
- Parro, V.; Lezcano, M.Á.; Moreno-Paz, M.; Davila, A.F.; Azua-Bustos, A.; García-Villadangos, M.; Wierzchos, J.; Fernández-Martínez, M.Á.; Larramendi, R.; Moreno, H.; et al. Microbial Biogeography along a 2578 Km Transect on the East Antarctic Plateau. Nat. Commun. 2025, 16, 775. [Google Scholar] [CrossRef]
- Després, V.R.; Huffman, J.A.; Burrows, S.M.; Hoose, C.; Safatov, A.S.; Buryak, G.; Fröhlich-Nowoisky, J.; Elbert, W.; Andreae, M.O.; Pöschl, U.; et al. Primary Biological Aerosol Particles in the Atmosphere: A Review. Tellus B Chem. Phys. Meteorol. 2012, 64, 15598. [Google Scholar] [CrossRef]
- Alekseev, I.; Zverev, A.; Abakumov, E. Microbial Communities in Permafrost Soils of Larsemann Hills, Eastern Antarctica: Environmental Controls and Effect of Human Impact. Microorganisms 2020, 8, 1202. [Google Scholar] [CrossRef]
- Varliero, G.; Lebre, P.H.; Adams, B.; Chown, S.L.; Convey, P.; Dennis, P.G.; Fan, D.; Ferrari, B.; Frey, B.; Hogg, I.D.; et al. Biogeographic Survey of Soil Bacterial Communities across Antarctica. Microbiome 2024, 12, 9. [Google Scholar] [CrossRef]
- Smith, M.L.; O’Neill, C.A.; Dickinson, M.R.; Chavan, B.; McBain, A.J. Exploring Associations between Skin, the Dermal Microbiome, and Ultraviolet Radiation: Advancing Possibilities for next-Generation Sunscreens. Front. Microbiomes 2023, 2, 1102315. [Google Scholar] [CrossRef]
- Das, S.; Yadav, A.; Debnath, N. Entomotoxic Efficacy of Aluminium Oxide, Titanium Dioxide and Zinc Oxide Nanoparticles against Sitophilus Oryzae (L.): A Comparative Analysis. J. Stored Prod. Res. 2019, 83, 92–96. [Google Scholar] [CrossRef]
- Pistone, D.; Meroni, G.; Panelli, S.; D’Auria, E.; Acunzo, M.; Pasala, A.R.; Zuccotti, G.V.; Bandi, C.; Drago, L. A Journey on the Skin Microbiome: Pitfalls and Opportunities. Int. J. Mol. Sci. 2021, 22, 9846. [Google Scholar] [CrossRef]
- Wallen-Russell, C.; Wallen-Russell, S. New Link between Skin Microbiota Diversity and Skin Health: Proposal to Use This as a Mechanism to Determine Whether Cosmetic Products Damage the Skin and Are a Cause of the Skin Allergy Epidemic. Cosmetics 2017, 4, 14. [Google Scholar] [CrossRef]
- Lee, H.J.; Jeong, S.E.; Lee, S.; Kim, S.; Han, H.; Jeon, C.O. Effects of Cosmetics on the Skin Microbiome of Facial Cheeks with Different Hydration Levels. Microbiologyopen 2018, 7, e00557. [Google Scholar] [CrossRef] [PubMed]
- Corsini, E.; Galbiati, V. Contact Allergy to Fragrances: In Vitro Opportunities for Safety Assessment. Cosmetics 2019, 6, 3. [Google Scholar] [CrossRef]
- Rajkumar, J.; Chandan, N.; Lio, P.; Shi, V. The Skin Barrier and Moisturization: Function, Disruption, and Mechanisms of Repair. Skin. Pharmacol. Physiol. 2023, 36, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zeng, D.-N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.-X. The Influence of Age and Gender on Skin-Associated Microbial Communities in Urban and Rural Human Populations. PLoS ONE 2015, 10, e0141842. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Guichard, A.; Cheng, Y.; Li, J.; Qin, O.; Wang, X.; Liu, W.; Tan, Y. Sensitive Scalp Is Associated with Excessive Sebum and Perturbed Microbiome. J. Cosmet. Dermatol. 2019, 18, 922–928. [Google Scholar] [CrossRef] [PubMed]
- Giacomoni, P.U.; Mammone, T.; Teri, M. Gender-Linked Differences in Human Skin. J. Dermatol. Sci. 2009, 55, 144–149. [Google Scholar] [CrossRef]
- Maarouf, M.; Maarouf, C.L.; Yosipovitch, G.; Shi, V.Y. The Impact of Stress on Epidermal Barrier Function: An Evidence-based Review. Br. J. Dermatol. 2019, 181, 1129–1137. [Google Scholar] [CrossRef]
- Kortekaas Krohn, I.; Callewaert, C.; Belasri, H.; De Pessemier, B.; Diez Lopez, C.; Mortz, C.G.; O’Mahony, L.; Pérez-Gordo, M.; Sokolowska, M.; Unger, Z.; et al. The Influence of Lifestyle and Environmental Factors on Host Resilience through a Homeostatic Skin Microbiota: An EAACI Task Force Report. Allergy 2024, 79, 3269–3284. [Google Scholar] [CrossRef]
- Bouslimani, A.; Da Silva, R.; Kosciolek, T.; Janssen, S.; Callewaert, C.; Amir, A.; Dorrestein, K.; Melnik, A.V.; Zaramela, L.S.; Kim, J.-N.; et al. The Impact of Skin Care Products on Skin Chemistry and Microbiome Dynamics. BMC Biol. 2019, 17, 47. [Google Scholar] [CrossRef]
- Jugé, R.; Rouaud-Tinguely, P.; Breugnot, J.; Servaes, K.; Grimaldi, C.; Roth, M.-P.; Coppin, H.; Closs, B. Shift in Skin Microbiota of Western European Women across Aging. J. Appl. Microbiol. 2018, 125, 907–916. [Google Scholar] [CrossRef]
- Swaney, M.H.; Kalan, L.R. Living in Your Skin: Microbes, Molecules, and Mechanisms. Infect. Immun. 2021, 89, e00695-20. [Google Scholar] [CrossRef]
- Han, S.H.; Park, J.W. Diabetic and Sympathetic Influences on the Water Permeability Barrier Function of Human Skin as Measured Using Transepidermal Water Loss: A Case-Control Study. Medicine 2017, 96, e8611. [Google Scholar] [CrossRef]
- Mohammed, D.; Hirata, K.; Hadgraft, J.; Lane, M.E. Influence of Skin Penetration Enhancers on Skin Barrier Function and Skin Protease Activity. Eur. J. Pharm. Sci. 2014, 51, 118–122. [Google Scholar] [CrossRef]
- Wang, D.-Q.; Li, X.; Zhang, R.-Y.; Yuan, C.; Yan, B.; Humbert, P.; Quan, Z.-X. Effects of Investigational Moisturizers on the Skin Barrier and Microbiome Following Exposure to Environmental Aggressors: A Randomized Clinical Trial and Ex Vivo Analysis. J. Clin. Med. 2023, 12, 6078. [Google Scholar] [CrossRef]
- Li, M.; Budding, A.E.; Van Der Lugt-Degen, M.; Du-Thumm, L.; Vandeven, M.; Fan, A. The Influence of Age, Gender and Race/Ethnicity on the Composition of the Human Axillary Microbiome. Intern. J. Cosmet. Sci. 2019, 41, 371–377. [Google Scholar] [CrossRef]
- Ji, X.; Wu, Z.; Sung, S.; Lee, P.-H. Metagenomics and Metatranscriptomics Analyses Reveal Oxygen Detoxification and Mixotrophic Potentials of an Enriched Anammox Culture in a Continuous Stirred-Tank Reactor. Water Res. 2019, 166, 115039. [Google Scholar] [CrossRef]



| Phylum | Host A | Host B | |||||||
|---|---|---|---|---|---|---|---|---|---|
| BeforeTrip | OnTheWay | Staying | OnTheWayBack | AfterTrip | BeforeTrip | OnTheWay | Staying | AfterTrip | |
| p__Actinomycetota | 91.97 | 91.70 | 76.12 | 80.52 | 61.68 | 74.58 | 83.93 | 80.51 | 68.66 |
| p__Pseudomonadota | 2.65 | 2.50 | 10.37 | 8.57 | 11.90 | 9.85 | 5.44 | 8.67 | 14.41 |
| p__Bacillota_D | 3.27 | 3.48 | 8.25 | 7.61 | 17.54 | 12.90 | 6.79 | 5.98 | 7.34 |
| p__Bacteroidota | 1.06 | 0.71 | 2.86 | 1.83 | 5.51 | 0.87 | 1.58 | 2.46 | 4.96 |
| p__Bacillota_A | 0.44 | 0.85 | 0.89 | 0.49 | 2.39 | 1.29 | 1.14 | 0.91 | 4.39 |
| p__Bacillota_C | 0.11 | 0.29 | 0.62 | 0.30 | 0.44 | 0.19 | 0.43 | 0.39 | 0.24 |
| p__Cyanobacteria | 0.02 | 0.02 | 0.29 | 0.01 | 0.02 | 0.00 | 0.00 | 0.44 | 0.00 |
| p__Deinococcota | 0.00 | 0.00 | 0.18 | 0.17 | 0.13 | 0.04 | 0.03 | 0.23 | 0.00 |
| p__Fusobacteriota | 0.01 | 0.01 | 0.06 | 0.05 | 0.12 | 0.00 | 0.20 | 0.06 | 0.00 |
| p__Planctomycetota | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.05 | 0.00 |
| p__Eremiobacterota | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 |
| p__Gemmatimonadota | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.02 | 0.00 |
| Unclassified | 0.47 | 0.43 | 0.36 | 0.45 | 0.28 | 0.29 | 0.45 | 0.28 | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.-C.; Lee, H.; Kim, O.-S.; Sul, W.J.; Lee, H.; Kim, H.-J. Case Study on Shifts in Human Skin Microbiome During Antarctica Expeditions. Microorganisms 2025, 13, 2491. https://doi.org/10.3390/microorganisms13112491
Lee K-C, Lee H, Kim O-S, Sul WJ, Lee H, Kim H-J. Case Study on Shifts in Human Skin Microbiome During Antarctica Expeditions. Microorganisms. 2025; 13(11):2491. https://doi.org/10.3390/microorganisms13112491
Chicago/Turabian StyleLee, Kyu-Chan, Hanbyul Lee, Ok-Sun Kim, Woo Jun Sul, Hyeonah Lee, and Hye-Jin Kim. 2025. "Case Study on Shifts in Human Skin Microbiome During Antarctica Expeditions" Microorganisms 13, no. 11: 2491. https://doi.org/10.3390/microorganisms13112491
APA StyleLee, K.-C., Lee, H., Kim, O.-S., Sul, W. J., Lee, H., & Kim, H.-J. (2025). Case Study on Shifts in Human Skin Microbiome During Antarctica Expeditions. Microorganisms, 13(11), 2491. https://doi.org/10.3390/microorganisms13112491


_Di_Marco.png)



