Potential Bacterial Biomarkers Associated with Penaeus stylirostris Shrimp Larvae to Infer Holobiont Health and Dysbiosis Across Larvae Stages
Abstract
1. Introduction
2. Materials and Methods
2.1. Rearing Experiment
2.2. Sample Collection, Total RNA Extractions, Reverse Transcription, and Sequencing
2.3. Bioinformatics and Downstream Analysis
2.4. Downstream Data Analysis
3. Results
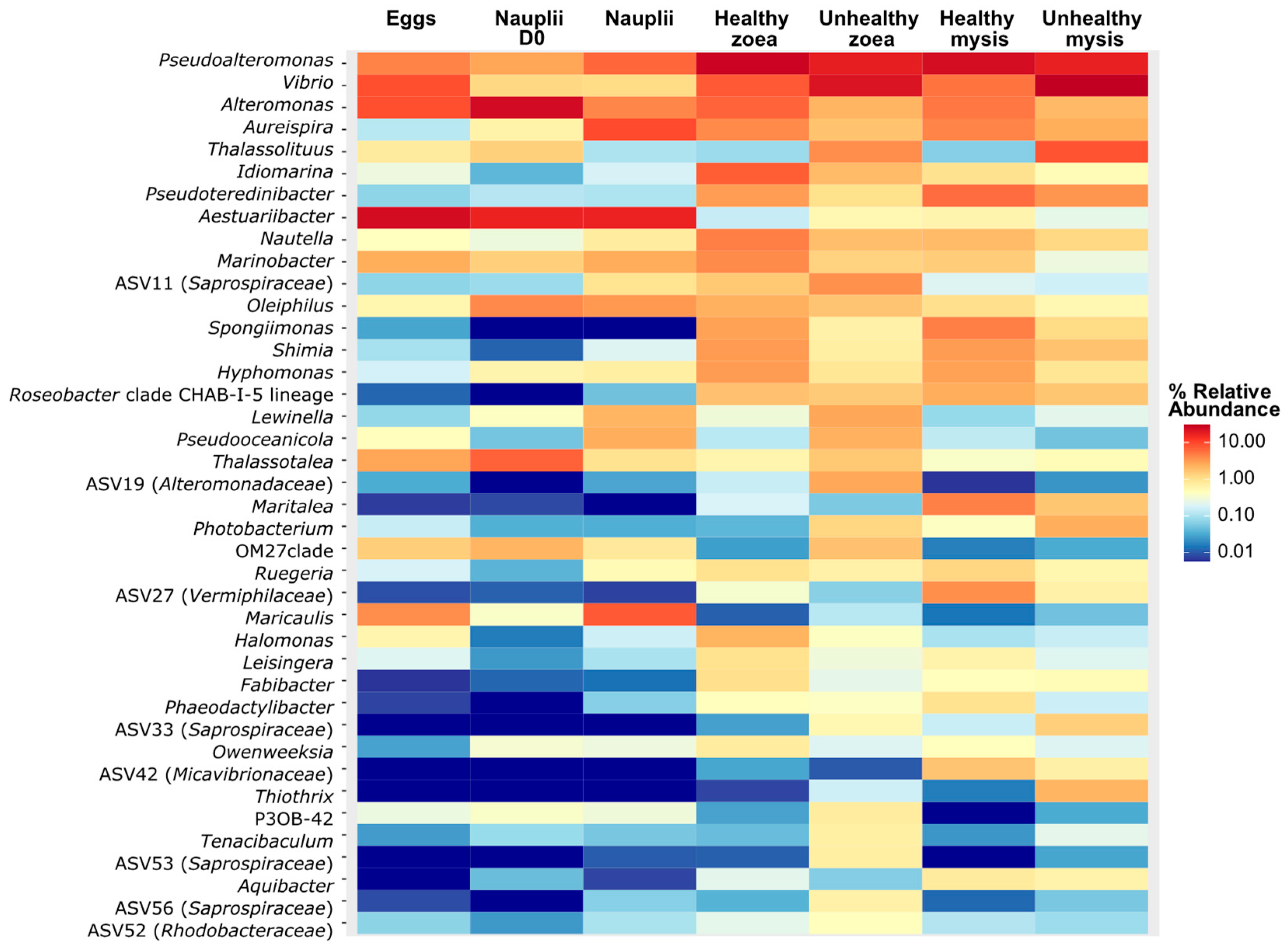
3.1. Main Genera Related to the Larval Stage and Health
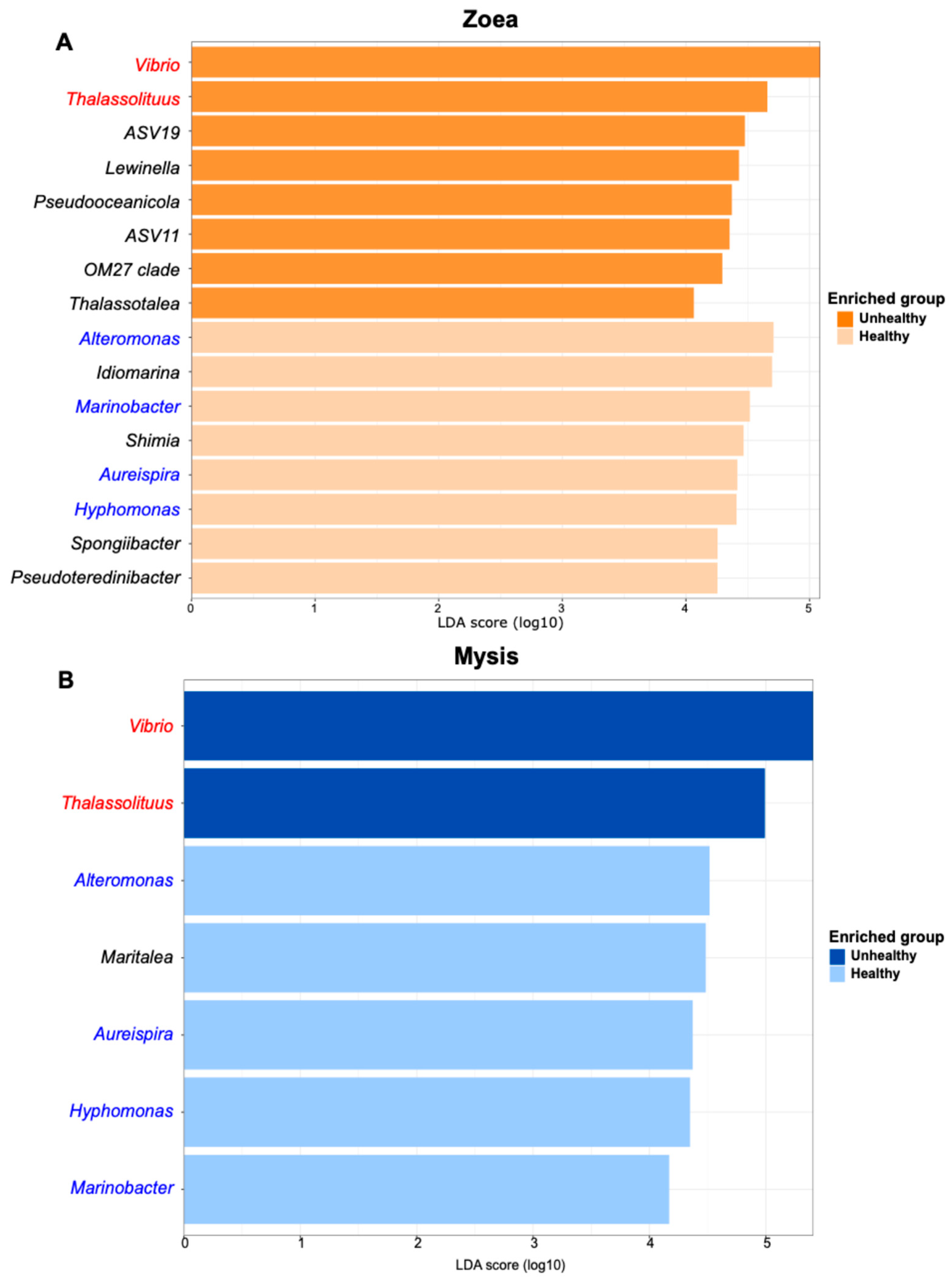
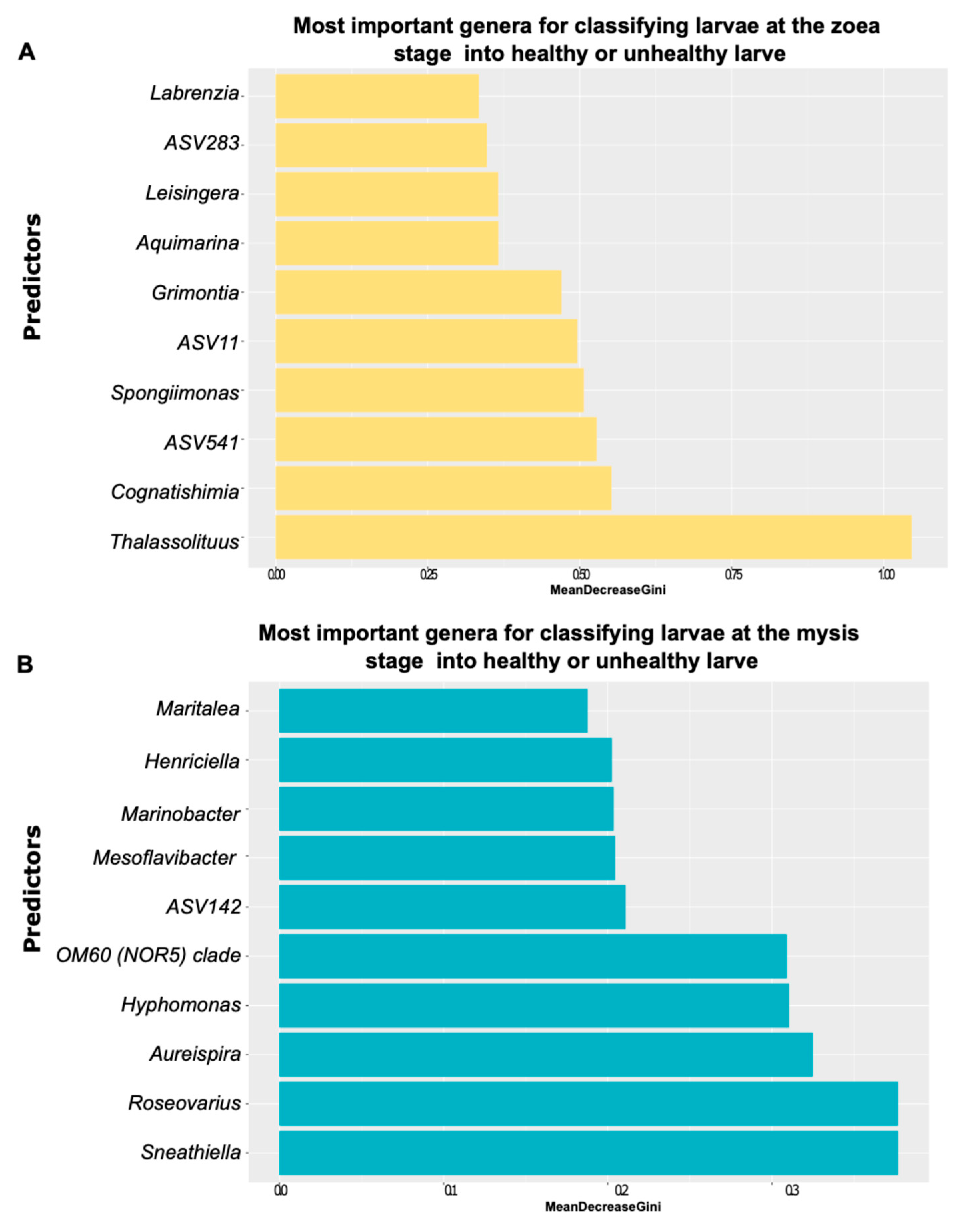
3.2. Investigation of Biomarkers Related to the Larval Stage and Health as Indicators of Larval Health
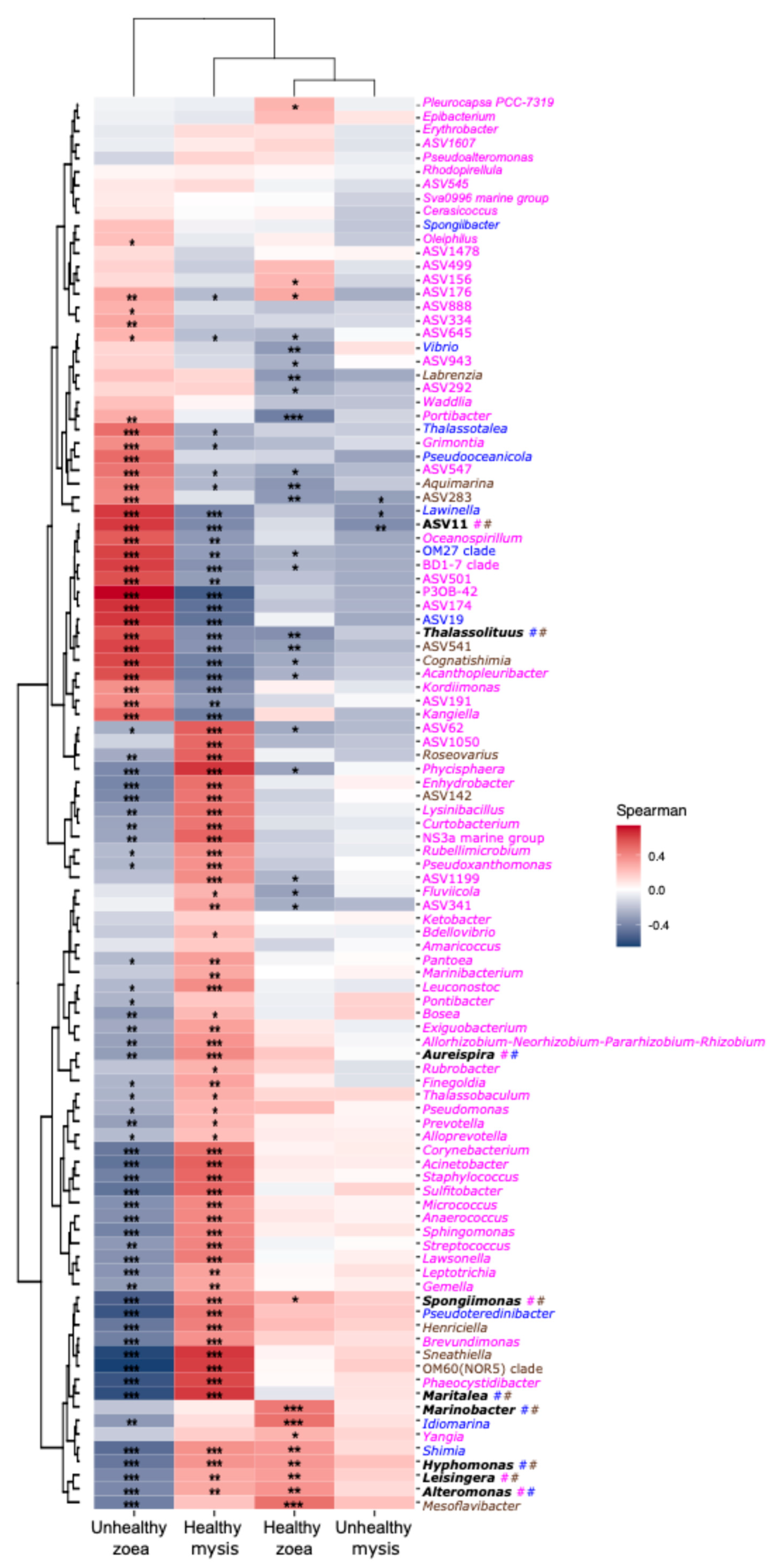
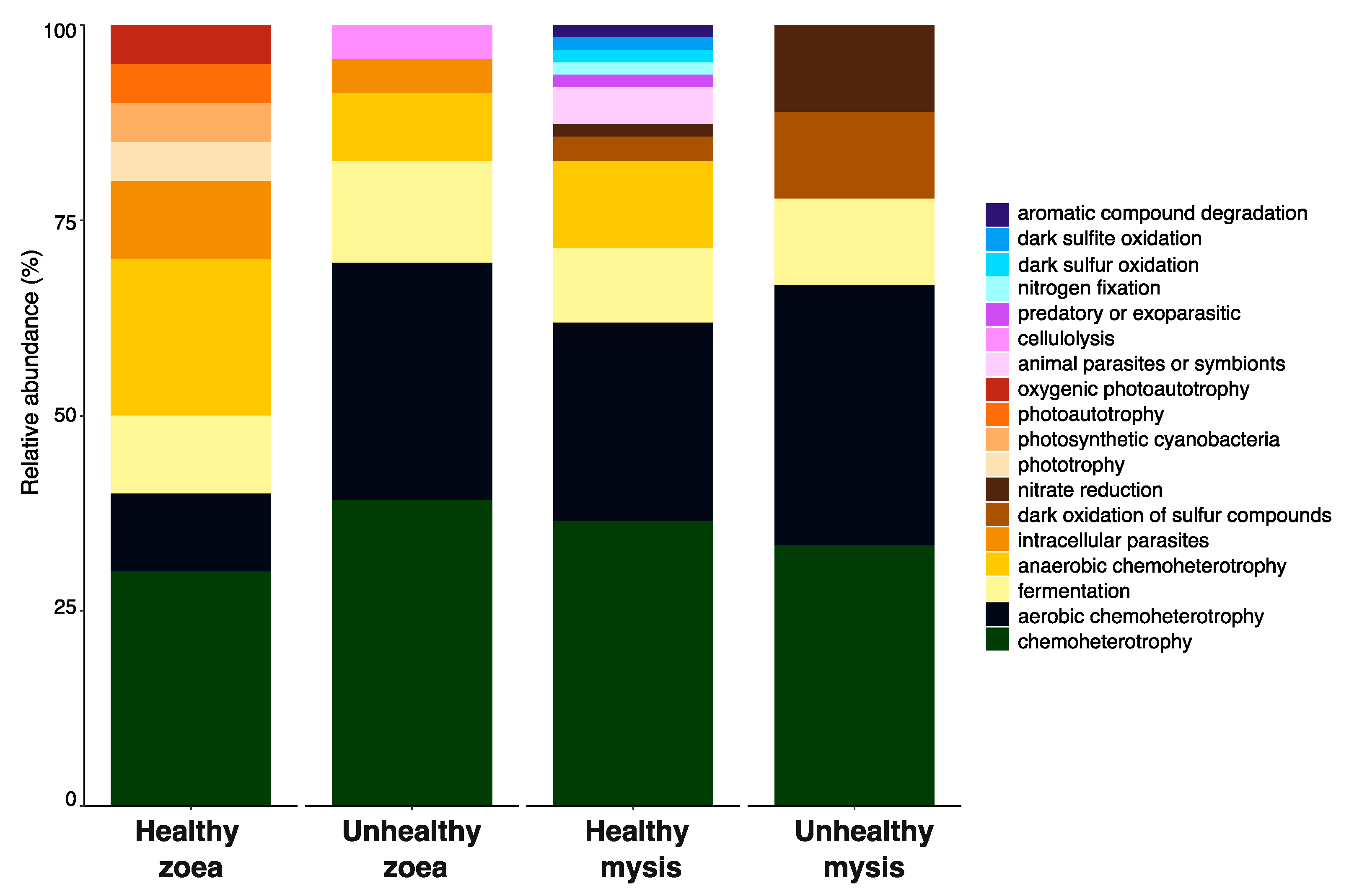
3.3. Biomarkers and Putative Functions Associated with Larval Stage and Health
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ganesan, R.; Wierz, J.C.; Kaltenpoth, M.; Flórez, L.V. How It All Begins: Bacterial Factors Mediating the Colonization of Invertebrate Hosts by Beneficial Symbionts. Microbiol. Mol. Biol. Rev. 2022, 86, e00126-21. [Google Scholar] [CrossRef]
- Dittami, S.M.; Arboleda, E.; Auguet, J.-C.; Bigalke, A.; Briand, E.; Cárdenas, P.; Cardini, U.; Decelle, J.; Engelen, A.H.; Eveillard, D.; et al. A Community Perspective on the Concept of Marine Holobionts: Current Status, Challenges, and Future Directions. PeerJ 2021, 9, e10911. [Google Scholar] [CrossRef]
- Fronk, D.C.; Sachs, J.L. Symbiotic Organs: The Nexus of Host–Microbe Evolution. Trends Ecol. Evol. 2022, 37, 599–610. [Google Scholar] [CrossRef]
- Sehnal, L.; Brammer-Robbins, E.; Wormington, A.M.; Blaha, L.; Bisesi, J.; Larkin, I.; Martyniuk, C.J.; Simonin, M.; Adamovsky, O. Microbiome Composition and Function in Aquatic Vertebrates: Small Organisms Making Big Impacts on Aquatic Animal Health. Front. Microbiol. 2021, 12, 567408. [Google Scholar] [CrossRef]
- Diwan, A.D.; Harke, S.N.; Panche, A.N. Host-Microbiome Interaction in Fish and Shellfish: An Overview. Fish Shellfish Immunol. Rep. 2023, 4, 100091. [Google Scholar] [CrossRef]
- Nyholm, S.V. In the Beginning: Egg–Microbe Interactions and Consequences for Animal Hosts. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190593. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, K.; Huang, L.; Dong, P.; Wang, S.; Chen, H.; Lu, Z.; Hou, D.; Zhang, D. Fine-Scale Succession Patterns and Assembly Mechanisms of Bacterial Community of Litopenaeus vannamei Larvae across the Developmental Cycle. Microbiome 2020, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, S.; Prasad, A. Microflora in Fish Digestive Tract Plays Significant Role in Digestion and Metabolism. Rev. Fish Biol. Fish. 2012, 22, 11–16. [Google Scholar] [CrossRef]
- Koll, R.; Hauten, E.; Theilen, J.; Bang, C.; Bouchard, M.; Thiel, R.; Möllmann, C.; Woodhouse, J.N.; Fabrizius, A. Spatio-Temporal Plasticity of Gill Microbiota in Estuarine Fish. Sci. Total Environ. 2024, 957, 177505. [Google Scholar] [CrossRef]
- Bordenstein, S.R.; Theis, K.R. Host Biology in Light of the Microbiome: Ten Principles of Holobionts and Hologenomes. PLoS Biol. 2015, 13, e1002226. [Google Scholar] [CrossRef]
- Jochum, L.; Stecher, B. Label or Concept—What Is a Pathobiont? Trends Microbiol. 2020, 28, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Chandra, H.; Sharma, K.K.; Tuovinen, O.H.; Sun, X.; Shukla, P. Pathobionts: Mechanisms of Survival, Expansion, and Interaction with Host with a Focus on Clostridioides Difficile. Gut Microbes 2021, 13, 1979882. [Google Scholar] [CrossRef] [PubMed]
- Rajeev, R.; Adithya, K.K.; Kiran, G.S.; Selvin, J. Healthy Microbiome: A Key to Successful and Sustainable Shrimp Aquaculture. Rev. Aquac. 2021, 13, 238–258. [Google Scholar] [CrossRef]
- Iebba, V.; Totino, V.; Gagliardi, A.; Santangelo, F.; Cacciotti, F.; Trancassini, M.; Mancini, C.; Cicerone, C.; Corazziari, E.; Pantanella, F.; et al. Eubiosis and Dysbiosis: The Two Sides of the Microbiota SuMMAry. New Microbiol. 2016, 39, 1–12. [Google Scholar] [PubMed]
- Hou, K.; Wu, Z.-X.; Chen, X.-Y.; Wang, J.-Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Fan, Y.; Pedersen, O. Gut Microbiota in Human Metabolic Health and Disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef]
- Hu, C.; Patil, Y.; Gong, D.; Yu, T.; Li, J.; Wu, L.; Liu, X.; Yu, Z.; Ma, X.; Yong, Y.; et al. Heat Stress-Induced Dysbiosis of Porcine Colon Microbiota Plays a Role in Intestinal Damage: A Fecal Microbiota Profile. Front. Vet. Sci. 2022, 9, 686902. [Google Scholar] [CrossRef]
- He, G.; Zhang, B.; Yi, K.; Chen, T.; Shen, C.; Cao, M.; Wang, N.; Zong, J.; Wang, Y.; Liu, K.; et al. Heat Stress-Induced Dysbiosis of the Gut Microbiota Impairs Spermatogenesis by Regulating Secondary Bile Acid Metabolism in the Gut. Sci. Total Environ. 2024, 937, 173305. [Google Scholar] [CrossRef]
- Krkosek, M. Host Density Thresholds and Disease Control for Fisheries and Aquaculture. Aquac. Environ. Interact. 2010, 1, 21–32. [Google Scholar] [CrossRef]
- Bouwmeester, M.M.; Goedknegt, M.A.; Poulin, R.; Thieltges, D.W. Collateral Diseases: Aquaculture Impacts on Wildlife Infections. J. Appl. Ecol. 2021, 58, 453–464. [Google Scholar] [CrossRef]
- Dantan, L.; Toulza, E.; Petton, B.; Montagnani, C.; Degremont, L.; Morga, B.; Fleury, Y.; Mitta, G.; Gueguen, Y.; Vidal-Dupiol, J.; et al. Microbial Education for Marine Invertebrate Disease Prevention in Aquaculture. Rev. Aquac. 2024, 16, 1229–1243. [Google Scholar] [CrossRef]
- Zheng, Y.; Yu, M.; Liu, J.; Qiao, Y.; Wang, L.; Li, Z.; Zhang, X.-H.; Yu, M. Bacterial Community Associated with Healthy and Diseased Pacific White Shrimp (Litopenaeus vannamei) Larvae and Rearing Water across Different Growth Stages. Front. Microbiol. 2017, 8, 1362. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Yu, M.; Liu, Y.; Su, Y.; Xu, T.; Yu, M.; Zhang, X.-H. Comparison of Cultivable Bacterial Communities Associated with Pacific White Shrimp (Litopenaeus vannamei) Larvae at Different Health Statuses and Growth Stages. Aquaculture 2016, 451, 163–169. [Google Scholar] [CrossRef]
- Boopathi, S.; Meenatchi, R.; Brindangnanam, P.; Sudhakaran, G.; Coumar, M.S.; Arockiaraj, J. Microbiome Analysis of Litopenaeus vannamei Reveals Vibrio as Main Risk Factor of White Faeces Syndrome. Aquaculture 2023, 576, 739829. [Google Scholar] [CrossRef]
- Chang, Y.-T.; Ko, H.-T.; Wu, P.-L.; Kumar, R.; Wang, H.-C.; Lu, H.-P. Gut Microbiota of Pacific White Shrimp (Litopenaeus vannamei) Exhibits Distinct Responses to Pathogenic and Non-Pathogenic Vibrio parahaemolyticus. Microbiol. Spectr. 2023, 11, e01180-23. [Google Scholar] [CrossRef]
- Fu, X.; He, J.; Wang, J.; Shen, F.; Qiu, J.; Chen, C.; Zhang, D.; Guo, H. Specific Gut Bacterial Taxa Inhabited in Healthy Shrimp (Penaeus vannamei) Confer Protection against Vibrio parahaemolyticus Challenge. Aquaculture 2024, 579, 740192. [Google Scholar] [CrossRef]
- Fan, L.; Li, Q.X. Characteristics of Intestinal Microbiota in the Pacific White Shrimp Litopenaeus vannamei Differing Growth Performances in the Marine Cultured Environment. Aquaculture 2019, 505, 450–461. [Google Scholar] [CrossRef]
- Hasyimi, W.; Widanarni, W.; Yuhana, M. Growth Performance and Intestinal Microbiota Diversity in Pacific White Shrimp Litopenaeus vannamei Fed with a Probiotic Bacterium, Honey Prebiotic, and Synbiotic. Curr. Microbiol. 2020, 77, 2982–2990. [Google Scholar] [CrossRef]
- Li, E.; Xu, C.; Wang, X.; Wang, S.; Zhao, Q.; Zhang, M.; Qin, J.G.; Chen, L. Gut Microbiota and Its Modulation for Healthy Farming of Pacific White Shrimp Litopenaeus vannamei. Rev. Fish. Sci. Aquac. 2018, 26, 381–399. [Google Scholar] [CrossRef]
- Callac, N.; Giraud, C.; Pham, D.; Ansquer, D.; Wabete, N.; Boulo, V. Active Microbiota of Penaeus stylirostris Larvae: Partially Shaped via Vertical and Horizontal Transmissions and Larval Ontogeny. Microorganisms 2024, 12, 608. [Google Scholar] [CrossRef]
- Callac, N.; Giraud, C.; Boulo, V.; Wabete, N.; Pham, D. Microbial Biomarker Detection in Shrimp Larvae Rearing Water as Putative Bio-Surveillance Proxies in Shrimp Aquaculture. PeerJ 2023, 11, e15201. [Google Scholar] [CrossRef] [PubMed]
- Giraud, C.; Wabete, N.; Lemeu, C.; Selmaoui-Folcher, N.; Pham, D.; Boulo, V.; Callac, N. Environmental Factors and Potential Probiotic Lineages Shape the Active Prokaryotic Communities Associated with Healthy Penaeus stylirostris Larvae and Their Rearing Water. FEMS Microbiol. Ecol. 2024, 100, fiae156. [Google Scholar] [CrossRef]
- Callac, N.; Boulo, V.; Giraud, C.; Beauvais, M.; Ansquer, D.; Ballan, V.; Maillez, J.-R.; Wabete, N.; Pham, D. Microbiota of the Rearing Water of Penaeus stylirostris Larvae Influenced by Lagoon Seawater and Specific Key Microbial Lineages of Larval Stage and Survival. Microbiol. Spectr. 2022, 10, e04241-22. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.; Ansquer, D.; Chevalier, A.; Dauga, C.; Peyramale, A.; Wabete, N.; Labreuche, Y. Selection and Characterization of Potential Probiotic Bacteria for Litopenaeus stylirostris Shrimp Hatcheries in New Caledonia. Aquaculture 2014, 432, 475–482. [Google Scholar] [CrossRef]
- Xiong, J.B.; Sha, H.N.; Chen, J. Updated Roles of the Gut Microbiota in Exploring Shrimp Etiology, Polymicrobial Pathogens, and Disease Incidence. Zool. Res. 2024, 45, 910–923. [Google Scholar] [CrossRef]
- Pham, D.; Charmantier, G.; Wabete, N.; Boulo, V.; Broutoi, F.; Mailliez, J.-R.; Peignon, J.-M.; Charmantier-Daures, M. Salinity Tolerance, Ontogeny of Osmoregulation and Zootechnical Improvement in the Larval Rearing of the Caledonian Blue Shrimp, Litopenaeus stylirostris (Decapoda, Penaeidae). Aquaculture 2012, 362–363, 10–17. [Google Scholar] [CrossRef]
- Giraud, C.; Callac, N.; Beauvais, M.; Mailliez, J.-R.; Ansquer, D.; Selmaoui-Folcher, N.; Pham, D.; Wabete, N.; Boulo, V. Potential Lineage Transmission within the Active Microbiota of the Eggs and the Nauplii of the Shrimp Litopenaeus stylirostris: Possible Influence of the Rearing Water and More. PeerJ 2021, 9, e12241. [Google Scholar] [CrossRef]
- Giraud, C.; Callac, N.; Boulo, V.; Lam, J.-S.; Pham, D.; Selmaoui-Folcher, N.; Wabete, N. The Active Microbiota of the Eggs and the Nauplii of the Pacific Blue Shrimp Litopenaeus stylirostris Partially Shaped by a Potential Vertical Transmission. Front. Microbiol. 2022, 13, 886752. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Hugerth, L.W.; Wefer, H.A.; Lundin, S.; Jakobsson, H.E.; Lindberg, M.; Rodin, S.; Engstrand, L.; Andersson, A.F. DegePrime, a Program for Degenerate Primer Design for Broad-Taxonomic-Range PCR in Microbial Ecology Studies. Appl. Environ. Microbiol. 2014, 80, 5116–5123. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S.D.; Lin, M.H. “Package ‘ANCOMBC’”. Available online: https://bioconductor.statistik.tu-dortmund.de/packages/3.13/bioc/manuals/ANCOMBC/man/ANCOMBC.pdf (accessed on 13 August 2021).
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, D.; Dong, Q. microbiomeMarker an R Package for MicroBiome Marker Identification and VisualizaTion. Bioinformatics 2022, 38, 4027–4029. [Google Scholar] [CrossRef] [PubMed]
- Biau, G.; Scornet, E. A Random Forest Guided Tour. Test 2016, 25, 197–227. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling Function and Taxonomy in the Global Ocean Microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef] [PubMed]
- Louca, S.; Jacques, S.M.S.; Pires, A.P.F.; Leal, J.S.; Srivastava, D.S.; Parfrey, L.W.; Farjalla, V.F.; Doebeli, M. High Taxonomic Variability despite Stable Functional Structure across Microbial Communities. Nat. Ecol. Evol. 2016, 1, 0015. [Google Scholar] [CrossRef]
- Sansupa, C.; Wahdan, S.F.M.; Hossen, S.; Disayathanoowat, T.; Wubet, T.; Purahong, W. Can We Use Functional Annotation of Prokaryotic Taxa (FAPROTAX) to Assign the Ecological Functions of Soil Bacteria? Appl. Sci. 2021, 11, 688. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a Package of R Functions for Community Ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Reyes, G.; Betancourt, I.; Andrade, B.; Panchana, F.; Román, R.; Sorroza, L.; Trujillo, L.E.; Bayot, B. Microbiome of Penaeus vannamei Larvae and Potential Biomarkers Associated with High and Low Survival in Shrimp Hatchery Tanks Affected by Acute Hepatopancreatic Necrosis Disease. Front. Microbiol. 2022, 13, 838640. [Google Scholar] [CrossRef]
- Xue, M.; Wu, L.; He, Y.; Liang, H.; Wen, C. Biases during DNA Extraction Affect Characterization of the Microbiota Associated with Larvae of the Pacific White Shrimp, Litopenaeus vannamei. PeerJ 2018, 6, e5257. [Google Scholar] [CrossRef]
- García-López, R.; Cornejo-Granados, F.; Lopez-Zavala, A.A.; Sánchez-López, F.; Cota-Huízar, A.; Sotelo-Mundo, R.R.; Guerrero, A.; Mendoza-Vargas, A.; Gómez-Gil, B.; Ochoa-Leyva, A. Doing More with Less: A Comparison of 16S Hypervariable Regions in Search of Defining the Shrimp Microbiota. Microorganisms 2020, 8, 134. [Google Scholar] [CrossRef]
- Qin, Q.-L.; Li, Y.; Sun, L.-L.; Wang, Z.-B.; Wang, S.; Chen, X.-L.; Oren, A.; Zhang, Y.-Z. Trophic Specialization Results in Genomic Reduction in Free-Living Marine Idiomarina Bacteria. MBio 2019, 10, 10-1128. [Google Scholar] [CrossRef]
- Angthong, P.; Uengwetwanit, T.; Uawisetwathana, U.; Koehorst, J.J.; Arayamethakorn, S.; Schaap, P.J.; Santos, V.M.D.; Phromson, M.; Karoonuthaisiri, N.; Chaiyapechara, S.; et al. Investigating Host-Gut Microbial Relationship in Penaeus Monodon upon Exposure to Vibrio harveyi. Aquaculture 2023, 567, 739252. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, Y.; Liu, Q.; Dong, H.; Li, H.; Xiong, D.; Zhang, J. Changes in the Intestine Microbial, Digestion and Immunity of Litopenaeus vannamei in Response to Dietary Resistant Starch. Sci. Rep. 2019, 9, 6464. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Hwang, Y.M.; Baik, K.S.; Choi, K.S.; Ka, J.O.; Seong, C.N. Mesoflavibacter aestuarii Sp. Nov., a Zeaxanthinproducing Marine Bacterium Isolated from Seawater. Int. J. Syst. Evol. Microbiol. 2014, 64, 1932–1937. [Google Scholar] [CrossRef] [PubMed]
- Vinay, T.N.; Patil, P.K.; Aravind, R.; Anand, P.S.S.; Baskaran, V.; Balasubramanian, C.P. Microbial Community Composition Associated with Early Developmental Stages of the Indian White Shrimp, Penaeus indicus. Mol. Genet. Genom. 2022, 297, 495–505. [Google Scholar] [CrossRef]
- Sandhya, S.V.; Sandeep, K.P.; Vijayan, K.K. In Vivo Evaluation of Microbial Cocktail of Microalgae-Associated Bacteria in Larval Rearing from Zoea I to Mysis I of the Indian White Shrimp, Penaeus indicus. J. Appl. Phycol. 2020, 32, 3949–3954. [Google Scholar] [CrossRef]
- Ooi, M.C.; Goulden, E.F.; Trotter, A.J.; Smith, G.G.; Bridle, A.R. Aquimarina Sp. Associated with a Cuticular Disease of Cultured Larval Palinurid and Scyllarid Lobsters. Front. Microbiol. 2020, 11, 573588. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, J.; Han, Z.; Chen, F.; Lv, A.; Hu, X.; Sun, X.; Qi, H.; Guo, Y. Vibrio parahaemolyticus Alters the Community Composition and Function of Intestinal Microbiota in Pacific White Shrimp, Penaeus Vannamei. Aquaculture 2021, 544, 737061. [Google Scholar] [CrossRef]
- Reyes, G.; Andrade, B.; Betancourt, I.; Panchana, F.; Solórzano, R.; Preciado, C.; Sorroza, L.; Trujillo, L.E.; Bayot, B. Microbial Signature Profiles of Penaeus vannamei Larvae in Low-Survival Hatchery Tanks Affected by Vibriosis. PeerJ 2023, 11, e15795. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, J.; Chen, F.; Qi, H.; Chen, L.; Sung, Y.Y.; Huang, Y.; Lv, A.; Hu, X. Phenotypic and Genomic Characterization of a Vibrio parahaemolyticus Strain Causing Disease in Penaeus Vannamei Provides Insights into Its Niche Adaptation and Pathogenic Mechanism. Microb. Genom. 2021, 7, 000549. [Google Scholar] [CrossRef] [PubMed]
- Bergen, N.; Krämer, P.; Romberg, J.; Wichels, A.; Gerlach, G.; Brinkhoff, T. Shell Disease Syndrome Is Associated with Reduced and Shifted Epibacterial Diversity on the Carapace of the Crustacean Cancer pagurus. Microbiol. Spectr. 2022, 10, e03419-22. [Google Scholar] [CrossRef]
- Restrepo, L.; Domínguez-Borbor, C.; Bajaña, L.; Betancourt, I.; Rodríguez, J.; Bayot, B.; Reyes, A. Microbial Community Characterization of Shrimp Survivors to AHPND Challenge Test Treated with an Effective Shrimp Probiotic (Vibrio diabolicus). Microbiome 2021, 9, 88. [Google Scholar] [CrossRef]
- Dong, C.; Wei, L.; Wang, J.; Lai, Q.; Huang, Z.; Shao, Z. Genome-Based Taxonomic Rearrangement of Oceanobacter-Related Bacteria Including the Description of Thalassolituus hydrocarbonoclasticus Sp. Nov. and Thalassolituus pacificus Sp. Nov. and Emended Description of the Genus Thalassolituus. Front. Microbiol. 2022, 13, 1051202. [Google Scholar] [CrossRef]
- Angthong, P.; Uengwetwanit, T.; Arayamethakorn, S.; Chaitongsakul, P.; Karoonuthaisiri, N.; Rungrassamee, W. Bacterial Analysis in the Early Developmental Stages of the Black Tiger Shrimp (Penaeus monodon). Sci. Rep. 2020, 10, 4896. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Luo, X.; Ye, X.; Chen, Y.; Wang, H.; Wang, L.; Wang, Y.; Yang, Y.; Li, Z.; Cao, H.; et al. Predatory Myxococcales Are Widely Distributed in and Closely Correlated with the Bacterial Community Structure of Agricultural Land. Appl. Soil Ecol. 2020, 146, 103365. [Google Scholar] [CrossRef]
- Wang, H.; Huang, J.; Wang, P.; Li, T. Insights into the Microbiota of Larval and Postlarval Pacific White Shrimp (Penaeus vannamei) along Early Developmental Stages: A Case in Pond Level. Mol. Genet. Genom. 2020, 295, 1517–1528. [Google Scholar] [CrossRef]
- Liñan-Vidriales, M.A.; Peña-Rodríguez, A.; Tovar-Ramírez, D.; Elizondo-González, R.; Barajas-Sandoval, D.R.; Ponce-Gracía, E.I.; Rodríguez-Jaramillo, C.; Balcázar, J.L.; Quiroz-Guzmán, E. Effect of Rice Bran Fermented with Bacillus and Lysinibacillus Species on Dynamic Microbial Activity of Pacific White Shrimp (Penaeus vannamei). Aquaculture 2021, 531, 735958. [Google Scholar] [CrossRef]
- Panigrahi, A.; Das, R.R.; Sivakumar, M.R.; Saravanan, A.; Saranya, C.; Sudheer, N.S.; Vasagam, K.P.K.; Mahalakshmi, P.; Kannappan, S.; Gopikrishna, G. Bio-Augmentation of Heterotrophic Bacteria in Biofloc System Improves Growth, Survival, and Immunity of Indian White Shrimp Penaeus indicus. Fish Shellfish Immunol. 2020, 98, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Masri, M.; Sukmawaty, E.; Aditia, L. Novel Chitinolytic Bacteria from the Shrimp Shell Processing Waste. Biodiversitas 2021, 22, 2672–2681. [Google Scholar] [CrossRef]
- Dimkić, I.; Bhardwaj, V.; Carpentieri-Pipolo, V.; Kuzmanović, N.; Degrassi, G. The Chitinolytic Activity of the Curtobacterium Sp. Isolated from Field-Grown Soybean and Analysis of Its Genome Sequence. PLoS ONE 2021, 16, e0259465. [Google Scholar] [CrossRef] [PubMed]
- Vasquez, Y.M.S.-C.; Cueva-Yesquen, L.G.; Duarte, A.W.F.; Rosa, L.H.; Valladão, R.; Lopes, A.R.; Bonugli-Santos, R.C.; Oliveira, V.M. de Genomics, Proteomics, and Antifungal Activity of Chitinase from the Antarctic Marine Bacterium Curtobacterium Sp. CBMAI 2942. Int. J. Mol. Sci. 2024, 25, 9250. [Google Scholar] [CrossRef]
- Campa-Córdova, Á.I.; Aguirre-Guzman, G.; Méndez-Martínez, Y.; Medina-Félix, D.; Ceseña, C.E.; García-Armenta, J.; Valenzuela-Chávez, J.A. Immunochemical Response and Gene Expression in Juvenile Shrimp (Litopenaeus vannamei) Exposed to Microorganisms Isolated from Marine Sediment. Vet. Mex. OA 2024, 11, 1–16. [Google Scholar] [CrossRef]
- Huang, L.; Guo, H.; Chen, C.; Huang, X.; Chen, W.; Bao, F.; Liu, W.; Wang, S.; Zhang, D. The Bacteria from Large-Sized Bioflocs Are More Associated with the Shrimp Gut Microbiota in Culture System. Aquaculture 2020, 523, 735159. [Google Scholar] [CrossRef]
- Vega-Carranza, A.S.; Escamilla-Montes, R.; Fierro-Coronado, J.A.; Diarte-Plata, G.; Guo, X.; García-Gutiérrez, C.; Luna-González, A. Investigating the Effect of Bacilli and Lactic Acid Bacteria on Water Quality, Growth, Survival, Immune Response, and Intestinal Microbiota of Cultured Litopenaeus vannamei. Animals 2024, 14, 2676. [Google Scholar] [CrossRef]
- Pedrosa-Gerasmio, I.R.; Nozaki, R.; Kondo, H.; Hirono, I. Gut Bacterial Community Profile in Pacific White Shrimp Litopenaeus vannamei Following 5-Aminolevulinic Acid Supplementation. Aquac. Res. 2020, 51, 4075–4086. [Google Scholar] [CrossRef]
- Mo, B.; Li, J.; Liao, G.; Wang, L.; Fan, L. Toxic Effects of Glyphosate on Histopathology and Intestinal Microflora of Juvenile Litopenaeus vannamei. Aquat. Toxicol. 2023, 255, 106399. [Google Scholar] [CrossRef] [PubMed]
- Lobb, B.; Hodgson, R.; Lynch, M.D.J.; Mansfield, M.J.; Cheng, J.; Charles, T.C.; Neufeld, J.D.; Craig, P.M.; Doxey, A.C. Time Series Resolution of the Fish Necrobiome Reveals a Decomposer Succession Involving Toxigenic Bacterial Pathogens. mSystems 2020, 5, e00145-20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wang, Y.; Hilal, M.G.; Yu, Q.; Feng, T.; Li, H. Temporal Succession of Water Microbiomes and Resistomes during Carcass Decomposition in a Fish Model. J. Hazard. Mater. 2021, 403, 123795. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Callac, N.; Giraud, C.; Perez, V.; Ansquer, D.; Lam, J.-S.; Boulo, V.; Pham, D.; Wabete, N. Potential Bacterial Biomarkers Associated with Penaeus stylirostris Shrimp Larvae to Infer Holobiont Health and Dysbiosis Across Larvae Stages. Microorganisms 2025, 13, 2452. https://doi.org/10.3390/microorganisms13112452
Callac N, Giraud C, Perez V, Ansquer D, Lam J-S, Boulo V, Pham D, Wabete N. Potential Bacterial Biomarkers Associated with Penaeus stylirostris Shrimp Larvae to Infer Holobiont Health and Dysbiosis Across Larvae Stages. Microorganisms. 2025; 13(11):2452. https://doi.org/10.3390/microorganisms13112452
Chicago/Turabian StyleCallac, Nolwenn, Carolane Giraud, Valérie Perez, Dominique Ansquer, Jean-Sébastien Lam, Viviane Boulo, Dominique Pham, and Nelly Wabete. 2025. "Potential Bacterial Biomarkers Associated with Penaeus stylirostris Shrimp Larvae to Infer Holobiont Health and Dysbiosis Across Larvae Stages" Microorganisms 13, no. 11: 2452. https://doi.org/10.3390/microorganisms13112452
APA StyleCallac, N., Giraud, C., Perez, V., Ansquer, D., Lam, J.-S., Boulo, V., Pham, D., & Wabete, N. (2025). Potential Bacterial Biomarkers Associated with Penaeus stylirostris Shrimp Larvae to Infer Holobiont Health and Dysbiosis Across Larvae Stages. Microorganisms, 13(11), 2452. https://doi.org/10.3390/microorganisms13112452





