Abstract
Staphylococcus aureus is an important human pathogen known for its versatility and ability to cause a wide range of infections. The aim of this study was to isolate and identify S. aureus from skin lesions from human patients, to determine antimicrobial resistance and biofilm formation potential at phenotypic and genotypic levels, as well as to verify the activity of efflux pump production. Out of 51 samples collected from skin lesions of various etiologies, 13 isolates were identified as S. aureus. All isolates showed the ability to form biofilms, which correlated with the presence of the icaABCD, agrA, srtA, clfAB, and fnbAB genes, while the bap gene was absent. The highest rates of resistance were observed for ampicillin (69.2%) and gentamicin (46.2%), as well as for erythromycin and clindamycin (38.5%). The mecA gene was present in two isolates, but phenotypic resistance to methicillin was confirmed in only one of them, suggesting possible heterogeneous expression or regulated activity of resistance mechanisms. The mecC gene was not present in any isolate. Efflux pump production was observed in only three isolates, showing weak to intermediate levels. These findings indicate the high biofilm potential and variable antimicrobial resistance of S. aureus clinical isolates, which pose a challenge for the treatment of emerging skin infections.
1. Introduction
The skin is the largest organ of the human body and performs several key functions, such as protective, thermoregulatory, and sensory functions. Skin diseases encompass a variety of disorders that impact the skin and its related structures and are among the most common diseases in the human population worldwide [1,2]. Among these, a skin lesion—defined as an abnormality or change in the structure, appearance, or colour of the skin that differs from the surrounding tissue—can be superficial or extend into deeper parts. We distinguish between primary (present from birth or arising during life) and secondary (changes in the primary lesion), and between benign and malignant skin lesions. Their emergence is often the result of the interaction of various internal and external factors [1,3].
The most commonly occurring skin lesions are skin and soft tissue infections caused by a wide range of microorganisms [2]. Under normal conditions, the skin hosts a diverse microbiome that is in balance. However, when the skin’s natural defence mechanisms are weakened [4] or the integrity of the skin barrier is compromised, it creates conditions for the colonisation of the affected area by a variety of microorganisms [5]. The most commonly isolated pathogens of skin and soft tissue infections include Staphylococcus aureus, Streptococcus spp., Clostridium spp., Enterococcus spp., Klebsiella pneumoniae, Escherichia coli, Proteus spp., Pseudomonas aeruginosa, Acinetobacter baumannii, Neisseria spp., and Vibrio spp. [6,7].
Staphylococcus aureus (S. aureus) is commonly found in the physiological microbiota of humans and animals but is also a highly adaptable and opportunistic pathogen capable of causing various diseases. Its pathogenicity is driven by a complex of virulence factors that enhance its capacity to adhere to surfaces, invade or evade the host’s immune system, or generate substances that harm tissues [8]. Proteins referred to as MSCRAMM (microbial surface components recognizing adhesive matrix molecules), which include the clustering factors ClfA (clfA), ClfB (clfB), fibronectin-binding proteins FnbA (fnbA), FnbB (fnbB), or proteins composed of serine-aspartate dipeptide repeats (SdrC, SdrD, SdrF), contribute not only to adhesion but also to invasion of host cells or tissues [9]. Concomitantly secreted toxins such as α-hemolysin, Panton-Valentine leukocidin (PVL), or toxic shock syndrome toxin (TSST) contribute to tissue destruction and systemic toxicity [10]. Overexpression of the srtA gene, which encodes the transpeptidase sortase A, enhances the speed at which surface proteins are anchored during cell wall biosynthesis, leading to heightened virulence [9].
S. aureus can form biofilms on medical devices, prosthetic implants, and tissue surfaces, protecting the bacteria from the immune system and antimicrobial treatments and contributing to chronic or recurrent infections [11]. These biofilms are typically encased in a glycocalyx layer composed of polysaccharide intercellular antigen (PIA), synthesised by the icaABCD locus, though some strains form biofilms independently of this locus. Other factors involved in biofilm formation include protein A, fibronectin-binding proteins, autolysin, extracellular DNA, and biofilm matrix proteins such as Bap [10]. The agrA gene encodes a regulatory factor that is part of the agr (accessory gene regulator) quorum-sensing system, a key regulatory mechanism in S. aureus that controls the expression of virulence factors, biofilm formation, and also influences the ability of the bacterium to cause disease [12].
The treatment of infections caused by S. aureus remains challenging due to the rapid development of resistance to antimicrobial agents. The occurrence of methicillin-resistant S. aureus (MRSA) in both healthcare and community environments worsens these issues through its various resistance mechanisms, including the production of penicillin-binding protein 2a (PBP2a), which is encoded by the mecA gene found within the staphylococcal chromosome cassette mec (SCCmec). The global rise of resistant strains, including MRSA, vancomycin-intermediate S. aureus (VISA), and vancomycin-resistant S. aureus (VRSA), underscores the pathogen’s remarkable evolutionary adaptability [13].
Another important mechanism of antimicrobial resistance in S. aureus is the activation of efflux pumps [14]. These pumps are transport proteins found in the cytoplasmic membrane of bacteria and play a critical role in excreting drugs from the cell [15]. They also have a regulatory function—they are involved in the secretion of toxic substances into the external environment [16]. According to the energy source, they are divided into primary, where the source is ATP, and secondary, where the source is an electrochemical gradient [17]. There are six families of efflux pumps; each family is characterised by a group of substrates. In Staphylococcus species, four of these families—ABC (the ATP-binding cassette family), MATE (the multidrug and toxin extrusion family), MFS (the major facilitator superfamily family), and SMR (the small multidrug resistance family)—have been documented. Notably, the first three families are associated with antimicrobial resistance [18,19].
This study aimed to identify and monitor the prevalence of S. aureus from skin lesions of different etiologies obtained from hospitalized patients. The study included monitoring biofilm formation as a significant virulence factor associated with antimicrobial resistance, detecting antimicrobial resistance profiles, assessing the production of efflux pumps, and identifying genes related to antimicrobial resistance as well as genes involved in biofilm formation. The main objective of this study was to obtain and provide new insights into the phenotypic and genotypic profiles of antimicrobial resistance and biofilm formation.
2. Materials and Methods
2.1. Sample Collection, Cultivation, and Identification of Staphylococcus aureus
Over a six-month period, a total of 51 samples from skin lesions of various etiologies were obtained from patients hospitalised at the Department of Dermatovenerology of the University of Pavol Jozef Šafárik and the Louis Pasteur University Hospital in Košice, Slovakia. The samples were taken from lesions with a confirmed infectious etiology, based on clinical evaluation (presence of pus, blisters, rashes, spots, wounds, bleeding, swelling, erythema, local warmth, pain, or itching). One swab was collected from each patient using a sterile cotton swab with Amies Copan 108C transport medium (Copan Italia, Brescia, Italy). Samples were stored at 4 °C before culture on media and processed within 24 h of collection.
All samples were grown on the surface of Columbia Blood Agar with the addition of 5% sterile defibrinated sheep blood, Baird-Parker Agar, and Mannitol Salt Agar (HiMedia Laboratories, Mumbai, India) and incubated under the conditions listed in Table 1. The isolated bacteria were categorised into the Staphylococcus genus based on their growth traits, biochemical characteristics, and Gram staining.

Table 1.
Microbiological media, incubation conditions, and criteria for identification S. aureus.
Species identification was performed using the commercial biochemical kit STAPHYtest 24 (Erba Lachema, Brno, Czech Republic). All identified staphylococci were confirmed at the genus level through multiplex PCR (mPCR) with primers that amplify a segment of the 16S rRNA gene specific to the Staphylococcus genus. Additionally, species-specific primers for S. aureus were used to identify the genes eap (extracellular adhesion protein) and nuc (thermostable nuclease). Finally, all staphylococcal isolates that were biochemically identified and confirmed at the genetic level using mPCR were also identified using a proteomic approach with MALDI-TOF MS (version 2.0, BioTyper Library version 3.0; Bruker Daltonics, Billerica, MA, USA) [20].
2.2. Genomic DNA Extraction
Genomic DNA was extracted from overnight cultures of S. aureus inoculated in modified brain heart infusion broth (mBHI; HiMedia Laboratories, Mumbai, India) containing 1.0% glucose and 2.0% sodium chloride using the High Pure PCR DNA Extraction Kit (Roche Molecular Systems, Inc., Pleasanton, CA, USA). The quantity and purity of DNA were analyzed using a NanoDropTM spectrophotometer with an ND-8000 system (Thermo Fisher Scientific, Waltham, MA, USA) by measuring absorbance at 260 and 280 nm.
2.3. Testing of Biofilm Formation
The ability of S. aureus isolates to form biofilms was evaluated using a modified method based on O’Toole et al. [21]. Each well of a 96-well polystyrene microtiter plate F-type (Brand GMBH + CO KG, Wertheim, Germany) received 100 μL of mBHI and 100 μL of a bacterial suspension at 1 McFarland turbidity. The plates were incubated stationary at 37 °C for 24 h. After incubation, the contents of the wells were aspirated, and 200 μL of a 0.1% crystal violet solution was added to each well, followed by 30-min incubation at room temperature. After washing and drying the wells, crystal violet that had bound to the adherent cells was extracted with 200 μL of 30% acetic acid per well. S. epidermidis CCM 4418, a non-biofilm-forming strain, and S. aureus CCM 4223, a biofilm-forming strain (Czech Collection of Microorganisms, Brno, Czech Republic), were used as reference strains. The negative control was pure mBHI. The optical density was determined spectrophotometrically at a wavelength 550 nm using a BioTek model Synergy 4 reader (Merck, Darmstadt, Germany) in triplicate for each strain tested, and then the mean values were calculated.
2.4. Antimicrobial Susceptibility Testing
The minimum inhibitory concentration (MIC) testing was determined by the colourimetric microdilution method according to Gattringer et al. [22] with automated readout via the Miditech system (Bel-Miditech s.r.o., Bratislava, Slovakia). The following antimicrobials were tested: ampicillin (AMP), ampicillin + sulbactam (SAM), piperacillin + tazobactam (TZP), oxacillin (OXA), cefoxitin (FOX), gentamicin (GEN), ciprofloxacin (CIP), moxifloxacin (MFX), erythromycin (ERY), clindamycin (CLI), linezolid (LNZ), rifampicin (RIF), vancomycin (VAN), teicoplanin (TEC), tetracycline (TET), tigecycline (TGC), chloramphenicol (CHL), trimethoprim (TMP), trimethoprim + sulphonamide (COT), and nitrofurantoin (NIT). The results of the MIC values of each antimicrobial were interpreted according to the clinical breakpoints of EUCAST 2024 (version 14.0) [23].
2.5. Detection of Efflux Pump
Efflux pump production activity was assessed on Mueller-Hinton agar (HiMedia Laboratories, Mumbai, India) containing 2.5 mg/L ethidium bromide in a Petri dish using a cartwheel method based on expelling a fluorescent dye, as described by Martins et al. [24]. The plates were divided into eight sectors by radial lines (cartwheel pattern). A bacterial suspension (0.5 McFarland standard) was prepared from the overnight culture and saline solution. The samples were swabbed onto agar surfaces, beginning at the middle and progressing toward the periphery. Subsequently, the plates were incubated for 24 h at 37 °C, protected from light in aluminium foil. After incubation, the swabbed plates were examined by the UV-Reader Quantum system (source of the UV light; Vilber Lourmat, Collégien, France) and analysed with the VisionCapt digital imaging system (Vilber Lourmat, Collégien, France). S. aureus ATCC 25923EtBr (efflux pump producer) and S. aureus ATCC 25923 (non-efflux pump producer) from the American Type Culture Collection (Manassas, VA, USA) served as reference strains. Saline solution alone was used as a negative (purity) control.
2.6. Gene Detection Using PCR
Genes associated with antimicrobial resistance, specifically mecA and mecC (which encode resistance to beta-lactams), as well as genes involved in biofilm formation (including bap, icaABCD, clfAB, fnbAB, srtA, and agrA), were detected using both simplex and mPCR. Table 2 provides a list of all primers used in the PCR reaction. The PCR reactions were carried out in a Mastercycler® Nexus X2 thermal cycler (Eppendorf, Hamburg, Germany). The composition of the reaction mixture and the PCR conditions used in this study were described by Király et al. [25]. Amplified PCR products were electrophoretically separated in a Wide Mini-Sub® GT Cell electrophoresis system (Bio-Rad, Hercules, CA, USA) on a 2.5% agarose gel using the non-toxic GoodView™ reagent (Amplia s.r.o., Bratislava, Slovakia). Subsequently, PCR products were visualised under UV light of an ultraviolet transilluminator (Bio-Imaging Systems, Modi’in-Maccabim-Re’ut, Israel) and recorded using a Kodak Gel Logic 100 digital imaging system (Kodak, Rochester, NY, USA).

Table 2.
List of primers used for PCR analysis.
2.7. Statistical Analysis
Biofilm formation was evaluated based on the obtained data by statistical analysis with GraphPad Prism program version 8.3.0 (GraphPad Software Inc., San Diego, CA, USA) through One-Way ANOVA, accompanied by Dunnett’s test to assess significance at p < 0.001.
The minimum inhibitory concentration values were further processed using the Miditech Analyser interpretation software (Bel-Miditech s.r.o., Bratislava, Slovakia, cat. n. 002002), which also performs statistical analysis.
3. Results
3.1. Identification of Bacterial Isolates
Out of 51 examined samples collected from skin lesions of hospitalised patients, 29 samples were identified as representatives of the genus Staphylococcus based on culture, biochemical characteristics, and microscopic examination.
Using the STAPHYtest 24 biochemical kit, 29 isolates of Staphylococcus spp. were identified, with 13 classified as S. aureus and 16 as coagulase-negative staphylococci (CoNS).
All staphylococci were subsequently confirmed using mPCR. The 29 isolates were positive for the presence of a section of the 16S rRNA gene specific for the genus Staphylococcus (141 bp). Additionally, fragments of the eap (230 bp) and nuc (103 bp) genes were detected in the identified S. aureus strains (13/29), which were absent in the other Staphylococcus spp. (16/29). This confirmed the results obtained at the phenotypic level.
All staphylococci isolates were also identified using MALDI-TOF MS. According to the obtained score values, the staphylococci were identified as S. aureus (n = 13), while 16 CoNS isolates included the following species: S. epidermidis (8/16), S. haemolyticus (7/16), and S. capitis (1/16).
3.2. Evaluation of Biofilm Formation
The ability to form biofilms was evaluated in all 13 confirmed isolates of S. aureus. All isolates demonstrated significant biofilm-forming ability, alongside a positive control (S. aureus reference strain CCM 4223), in comparison to the non-biofilm-forming reference strain S. epidermidis CCM 4418. One isolate (sample no. 2) exhibited a notably lower ability for biofilm formation than the other isolates (Figure 1).
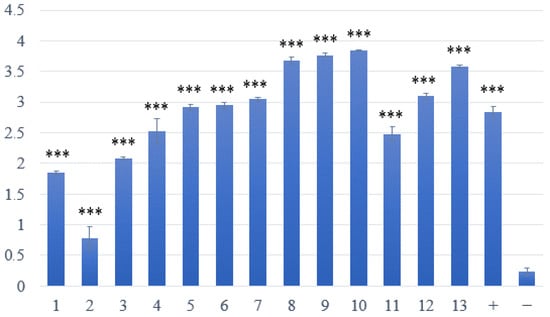
Figure 1.
Assessment of biofilm formation in S. aureus isolated from skin lesions. Samples 1–13—strains with weak or strong production of biofilm; positive (+) control—biofilm-forming S. aureus CCM 4223; negative (−) control—non-biofilm-forming S. epidermidis CCM 4418. *** Significantly higher ability to produce biofilm (p < 0.001).
3.3. Antimicrobial Susceptibility Profiles of S. aureus Isolates
The minimum inhibitory concentration (MIC) of antimicrobials was determined in 13 confirmed isolates of S. aureus (Table 3).

Table 3.
Antimicrobial susceptibility profile of S. aureus isolates.
S. aureus isolates were predominantly susceptible to all antimicrobial agents tested. Among the 13 total isolates, the highest resistance rates were observed to ampicillin (69.2%), gentamicin (46.2%), erythromycin, and clindamycin (38.5%). Resistance to oxacillin, cefoxitin, and chloramphenicol was recorded in one isolate (7.7%).
The MIC xG values for erythromycin were 1.30 mg/L, for clindamycin 0.80 mg/L, and for erythromycin 8.9 mg/L, which exceeded the EUCAST clinical breakpoints.
According to the resistance profile, the system automatically evaluated the resistance mechanisms (Figure 2). It is important to note that multiple resistance mechanisms have been identified in some strains simultaneously. The most common mechanism was resistance to penicillins (69.2%), followed by resistance to aminoglycosides (PH(2″)-AC(6′)!) (46.2%); this represents complications in treatment with these antimicrobial agents and combined enzymatic resistance to gentamicin, tobramycin, netilmicin, and ampicillin. The MLSB (macrolide-lincosamide-streptogramin B) mechanism was also present. The MLSB inducible (MLSB/i) (30.8%), manifested in clinical resistance to lincosamides and streptogramin B induced by 14- and 15-membered macrolides, constitutive (MLSB/c) (7.7%), determining resistance to all MLSB antimicrobials, were found [32]. One isolate (no. 11; 7.7%) was confirmed to have an MRSA mechanism, in which all beta-lactams are clinically ineffective in therapy. Multiresistance was not detected in any of the S. aureus isolates studied.
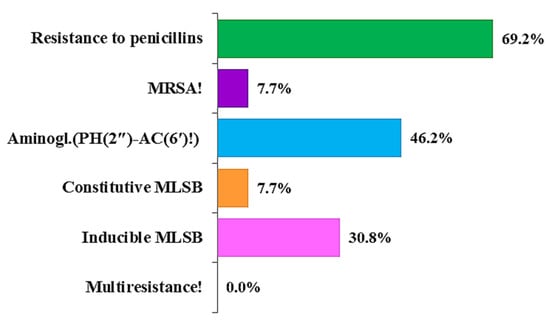
Figure 2.
Overview of phenotypically predicted resistance mechanisms in clinical S. aureus isolates.
Although the system did not evaluate multiresistance in any of the isolates examined, based on the analysis of the susceptibility profile to the tested antimicrobial substances, we found that isolates 4, 5, 7, 12, and 13 exhibited multiresistance (Table 3), which we define as the resistance of bacteria to three or more different groups of antimicrobial substances.
3.4. Evaluation of Efflux Pump Production
Out of the 13 isolates tested, none were classified as strong producers of efflux pumps. One isolate (7.7%) was categorised as an intermediate efflux pump producer. Two isolates (15.4%) were identified as weak efflux pump producers, while the remaining ten isolates (76.9%) showed no signs of efflux pump production (Figure 3 and Figure 4).
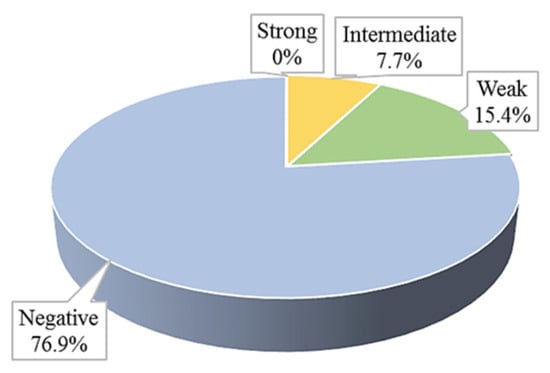
Figure 3.
Efflux pumps production in clinical S. aureus isolates.
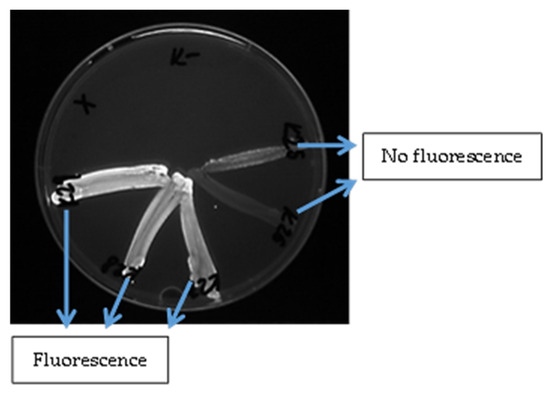
Figure 4.
Evaluation of the fluorescence of S. aureus isolates detected under UV light after adding ethidium bromide to the medium.
3.5. Identification of Antimicrobial Resistance Genes and Genes Related to Biofilm Formation
In our study, we analysed 13 S. aureus isolates and detected the mecA gene in only two clinical samples (isolates no. 4 and 11). Of these, only sample number 11 was identified as MRSA, exhibiting phenotypic resistance to the beta-lactam antibiotics ampicillin, oxacillin, and cefoxitin. Additionally, the mecC gene was not found in any of the isolates we examined, which aligns with its lower prevalence in clinical practice.
A more detailed analysis of biofilm formation was performed in all 13 isolates, not only at the phenotypic level but also at the genotypic level, using mPCR and simplex PCR (gene bap). All S. aureus isolates showed the ability to form biofilms, which correlated with the high presence of icaABCD, agrA, srtA, and also fnbAB and clfAB genes encoding adhesive proteins. The bap gene was absent in all of the isolates; nevertheless, this does not eliminate the possibility of considerable biofilm activity, since biofilm formation in S. aureus is affected by a range of intricate mechanisms. A summary of the biofilm-related genes detected in S. aureus can be found in Table 4. Table 5 presents a detailed evaluation of clinical strains of S. aureus.

Table 4.
Presence and distribution of genetic determinants of biofilm formation in S. aureus isolates.

Table 5.
Comprehensive evaluation of clinical S. aureus strains: phenotypic antimicrobial resistance and biofilm formation, efflux pump data, and genotypic analysis.
4. Discussion
Due to its characteristic properties, human skin has unique properties that make it an unsuitable environment for many microorganisms, effectively acting as a barrier that prevents their entry into the organism. Healthy skin is naturally dry, has a low pH on the surface, and an upper layer that continuously renews itself. The skin also produces antimicrobial peptides that can directly destroy or inhibit the growth of microorganisms. However, certain bacteria, such as Staphylococcus spp., particularly S. aureus, can overcome these defence mechanisms and not only colonise the skin but also be the primary cause of complicating pathogenesis and persistence of skin diseases [33].
In our study, we identified S. aureus in 25.5% (13/51) of clinical samples collected from skin lesions of various etiologies in hospitalised patients. All isolates were confirmed using both molecular and proteomic methods, ensuring high accuracy in identification at the species level. Our findings are consistent with previously published data indicating that S. aureus is a significant etiological agent responsible for skin and soft tissue infections, especially in patients with weakened immunity or compromised skin barrier [5,6]. The rate of S. aureus colonisation of skin lesions in published studies varies not only between countries but also between cities where the studies were conducted. In the study by Khalili et al. [34], the colonisation rate of S. aureus skin lesions was 45%, with results varying between cities (Shiraz 23% vs. Tehran 51%). A higher prevalence is also reported by Vella et al. [35], who also recorded a higher presence in the cities of Chicago (92%), Columbia (44%), and Vanderbilt (40%). The authors state that differences in results could be related to sampling methods, repeated infections, and variations among healthcare facilities. On the other hand, similar or lower prevalence rates were observed in studies from Nigeria (26.6%) [36], Ethiopia (23.6%) [37], and Brazil (20%) [38].
Biofilm formation is a significant virulence factor that complicates the treatment of skin infections caused by S. aureus. The ability of bacteria to form resilient biofilms may lead to their persistent presence on the human skin. Additionally, S. aureus bacteria within biofilms are shielded from the host’s immune response and the effects of antimicrobial agents, which can result in recurrent and chronic infections [39,40]. In a study conducted in Sweden, a total of 160 clinical isolates of S. aureus from patients with various skin diseases were examined, and all of them successfully formed biofilms [41]. Our results showed that all 13 S. aureus isolates analysed also had the ability to form biofilm, which correlates with the presence of key biofilm-related genes.
The icaABCD operon was found in all isolates, underscoring its central function in the production of polysaccharide intercellular adhesin (PIA), a crucial component of the biofilm matrix. Among the genes involved, icaA and icaD are particularly important for the biofilm formation process [11]. Moreover, all isolates were also positive for agrA and srtA, which are genes involved in regulating virulence factor expression and attachment of surface proteins. These findings further support the fact that biofilm formation in S. aureus is multifactorial and carefully regulated by quorum sensing and protein anchoring systems [9]. In our study, we detected the adhesive genes clfA, clfB, fnbA, and fnbB at varying frequencies. These genes play a crucial role in the initial adhesion to host tissues and the accumulation of biofilms [8]. Although the bap gene was absent in all isolates, it is important to note that the presence of the bap gene is not universal across all strains, and this absence does not preclude the possibility of biofilm formation. Biofilm formation can occur independently of Bap, using alternative mechanisms that involve other adhesins such as icaABCD or fnbAB. This suggests that there may be functional redundancy or species-specific pathways in biofilm formation [11,42].
The bap gene encodes the Bap (biofilm-associated protein) protein, a high-molecular-weight protein that was first described in S. aureus isolated from bovine mastitis [43]. Bap is crucial for biofilm formation, particularly in facilitating initial attachment to both living (biotic) and non-living (abiotic) surfaces. The C-terminal region of Bap contains a typical domain involved in cell adhesion. Moreover, it enhances strong intercellular adhesion when the Bap protein experiences partial proteolytic cleavage, which releases fragments that contain the N-terminal region. In this reaction, which is influenced by environmental factors like calcium ion concentration and acidic pH, spontaneous aggregation into amyloid fibers occurs. The gene is commonly located on mobile genetic elements, which leads to its varying distribution among different isolates. All S. aureus isolates that express Bap exhibit high adhesion and strong biofilm-forming ability [11,44,45].
The prevalence of detected biofilm-associated genes (such as bap, icaABCD, clfAB, fnbAB, srtA, and agrA) differs substantially across published studies, reflecting variability in the distribution and detection rates of individual genes [29,46,47,48].
Resistant bacteria, particularly multiresistant strains, pose a major public health problem. Among these, MRSA is one of the most significant regarding resistance to antimicrobial agents [6]. Recent studies indicate a decrease in the prevalence of MRSA in hospitals; however, it remains a significant cause of skin and soft tissue infections. Community-associated MRSA (CA-MRSA) appears to be more virulent and spreads more rapidly than hospital-associated MRSA (HA-MRSA) [49]. Higher levels of resistance among groups of people with poor hygiene, limited access to healthcare, and those engaging in risky behaviours (i.e., injecting drugs) contribute to the persistence and spread of MRSA in the population. Therefore, it is important to focus not only on appropriate therapeutic approaches but also on prevention and education in the areas of hygiene and antimicrobial resistance [50].
In our study, the most dominant resistance of S. aureus isolates was to ampicillin (69.2%) and also to oxacillin (7.7%), which belong to the beta-lactam group. This finding is consistent with other published studies, which suggest a near-universal prevalence due to the widespread production of beta-lactamases encoded mainly by the blaZ gene [51]. Koumaki et al. [52] indicate that the prevalence of S. aureus resistance to beta-lactams continues to be significantly high in both community and hospital settings, frequently surpassing 90%. Similarly, in a study by Ji et al. [51], S. aureus from patients with skin and soft tissue infections also showed dominant resistance to penicillin and oxacillin (100%), followed by cefoxitin (95.2%). They also found resistance to clindamycin and erythromycin in S. aureus, but only in 76.2% of the isolates examined. In contrast, resistance to gentamicin was less than 14%. No isolate was resistant to daptomycin, ceftaroline, linezolid, tigecycline, teicoplanin, or vancomycin.
Our results also showed increased resistance to gentamicin (46.2%) and erythromycin (38.5%), highlighting the need for more cautious use of these antimicrobial agents. The prevalence of clindamycin resistance among S. aureus isolates was 38.5% in our study. A recent systematic review reported that the prevalence of clindamycin resistance in S. aureus isolates varies significantly, ranging from 2.9% to 44%. The highest prevalence, at 44%, was reported in Egypt, while the lowest prevalence of 2.9% was observed in Côte d’Ivoire. These findings highlight the need for a more rational and locally adapted use of this antimicrobial [53,54]. Previous research has shown an increased occurrence of multidrug-resistant S. aureus in both community and hospital settings [6].
According to our results, the most frequently detected resistance mechanisms in S. aureus isolates were resistance to penicillins and to aminoglycosides caused by production of modification enzyme PH(2″)-AC(6′)!. Resistance to penicillins is generally caused by the production of beta-lactamases or the presence of the mecA gene, which encodes the penicillin-binding protein PBP2a. PBP2a prevents beta-lactam from binding to the cell surface due to reduced affinity [55]. In 2011, a new mecC gene was discovered, which shows 63% homology with the mecA gene. The mecC gene encodes the PBP2c protein, which gives staphylococci a similar level of resistance to beta-lactams as PBP2a does. These mechanisms lead to resistance against virtually all beta-lactams, a phenomenon known as MRSA [56]. Our study confirmed the presence of one isolate of MRSA at the phenotypic level, and it was found to carry the mecA gene. This finding aligns with the results of Almuhayawi et al. [7], who reported increased rates of methicillin and vancomycin resistance among S. aureus isolates from skin lesions. On the other hand, the mecC gene was not present in any of the isolates, confirming its lower incidence in clinical settings [13]. In addition to PBP2a/PBP2c synthesis, staphylococcal resistance to beta-lactams may be caused by other mechanisms. Some MRSA strains are associated with so-called resistance mechanisms without mec genes. Examples include strains with BORSA (borderline oxacillin-resistant S. aureus) or MODSA (modified S. aureus) phenotypes [56].
The Miditech system has also identified the mechanism of MLSB resistance—acquired resistance to macrolides, lincosamides, and streptogramin B. This type of resistance results from reduced binding of these drugs to the methylated 50S ribosomal subunit, which is an overlapping binding site for all three groups of antimicrobial agents [57]. There are two types of MLSB resistance: constitutive MLSB phenotype (MLSB/c), in which the bacterial strain constantly produces rRNA methyltransferase, and inducible MLSB (MLSB/i), in which methyltransferase production is observed in the presence of an inducing agent [58]. Establishing the MLSB phenotype accurately is crucial, given that clindamycin is advised for managing uncomplicated skin and soft tissue infections, particularly as an alternative for treating MRSA-related infections [59]. The research conducted by Henry and Khachemoune [60] confirmed the MLSB phenotype in seven strains, of which three exhibited resistance to methicillin at the same time. Current resistance to beta-lactams, macrolides, and lincosamides greatly restricts available treatment options. Likewise, this study identified three isolates that exhibited simultaneous resistance to aminoglycosides along with MLSB resistance, further complicating the management of these infections. However, resistance to rifampicin, tetracycline, and ciprofloxacin was not observed, which are frequently employed for treating skin infections in human medicine [61].
Most articles describing efflux pumps are devoted to reviews, or rather genotypic characterisation, and the impact of new potential efflux pump inhibitors (e.g., [62,63,64]). From the available data about phenotypic identification of efflux pumps, Favour et al. [65] in their study report up to 83% occurrence of efflux pump producers in their samples. However, they were only multiresistant isolates without distinction into weak, intermediate, and strong producers. Nevertheless, it is more compared to our 23.1%. Patel et al. [66] identified efflux pumps in 47.7% of isolates, which is also more than we managed to do. The reason may also be the larger number of clinical isolates of S. aureus. Abdi et al. [67] described a 10% incidence of clinical isolates of S. aureus producing efflux pumps.
5. Conclusions
The findings of our study substantiate the clinical relevance of Staphylococcus aureus as a cause of skin infections of various etiologies, particularly due to its strong ability to form biofilms and increasing antimicrobial resistance. All isolates displayed phenotypic capacity for biofilm formation, which is closely related to the presence of key genes such as icaABCD, agrA, srtA, clfAB, and fnbAB. While the bap gene was absent, biofilm formation was nonetheless strong, underscoring the intricate nature of regulatory mechanisms. The observed antimicrobial resistance, including multiresistant and MRSA strains, highlights the need for targeted diagnostics and treatment. The results support the importance of combining phenotypic and genotypic methods in assessing the virulence and resistance of clinical isolates of S. aureus.
Author Contributions
Conceptualization: J.K., N.D. and V.H.; methodology: J.K., N.D., V.H., S.H., G.G. and M.N.; validation: J.K., N.D. and G.G.; formal analysis: J.K., N.D., V.H., G.G., P.H., S.H., M.N. and Z.F.; investigation: J.K., N.D., V.H., G.G., P.H., S.H., M.N. and Z.F.; resources: Z.F.; data curation: J.K., N.D., V.H., G.G., M.N., P.H. and S.H.; writing—original draft preparation: J.K., N.D., V.H. and S.H.; writing—review and editing: J.K., N.D. and G.G.; visualization: J.K., N.D., V.H. and G.G.; supervision: G.G. and E.P.; funding acquisition: G.G., J.K. and Z.F. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Slovak Research and Development Agency under the contract number APVV-23-0488. This publication was also supported by the Cultural and Educational Grant Agency (KEGA) and the Scientific Grant Agency (VEGA) of the Ministry of Education, Research, Development, and Youth of the Slovak Republic, with project numbers KEGA 018UVLF-4/2025, KEGA 009UVLF-4/2025 and VEGA 1/0446/22.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Louis Pasteur University Hospital in Košice (2022/EK10084, approval date is: 1 January 2022).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| agr | accessory gene regulator |
| AMP | ampicillin |
| BORSA | borderline oxacillin-resistant Staphylococcus aureus |
| CA-MRSA | community-associated methicillin-resistant Staphylococcus aureus |
| CIP | ciprofloxacin |
| CLI | clindamycin |
| CoNS | coagulase-negative staphylococci |
| COT | trimethoprim + sulphonamide |
| DNA | deoxyribonucleic acid |
| ERY | erythromycin |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| FOX | cefoxitin |
| GEN | gentamicin |
| HA-MRSA | hospital-associated methicillin-resistant Staphylococcus aureus |
| CHL | chloramphenicol |
| LNZ | linezolid |
| MALDI-TOF MS | matrix-assisted laser desorption ionization–time of flight mass spectrometry |
| mBHI | modified brain heart infusion broth |
| MFX | moxifloxacin |
| MIC | minimum inhibitory concentration |
| MLSB | macrolide-lincosamide-streptogramin B resistance |
| MODSA | modified Staphylococcus aureus |
| MRSA | methicillin-resistant Staphylococcus aureus |
| MSCRAMM | microbial surface components recognizing adhesive matrix molecules |
| NIT | nitrofurantoin |
| OXA | oxacillin |
| PCR | polymerase chain reaction |
| PIA | polysaccharide intercellular antigen |
| PVL | Panton-Valentine leucocidin |
| RIF | rifampicin |
| SAM | ampicillin + sulbactam |
| SCCmec | staphylococcal cassette chromosome mec |
| TEC | teicoplanin |
| TET | tetracycline |
| TGC | tigecycline |
| TMP | trimethoprim |
| TSST | toxic shock syndrome toxin |
| TZP | piperacillin + tazobactam |
| VAN | vancomycin |
| VISA | vancomycin-intermediate Staphylococcus aureus |
| VRSA | vancomycin-resistant Staphylococcus aureus |
References
- Ali, M.U.; Khalid, M.; Alshanbari, H.; Zafar, A.; Lee, S.W. Enhancing Skin Lesion Detection: A Multistage Multiclass Convolutional Neural Network-Based Framework. Bioengineering 2023, 10, 1430. [Google Scholar] [CrossRef]
- Skuhala, T.; Trkulja, V.; Rimac, M.; Dragobratović, A.; Desnica, B. Analysis of Types of Skin Lesions and Diseases in Everyday Infectious Disease Practice—How Experienced Are We? Life 2022, 12, 978. [Google Scholar] [CrossRef]
- Osmosis. Available online: https://www.osmosis.org/answers/skin-lesions (accessed on 2 July 2025).
- Pătrașcu, A.-I.; Vâță, D.; Temelie-Olinici, D.; Mocanu, M.; Guguluș, D.-L.; Marinescu, M.; Stafie, L.; Tarcău, B.-M.; Creţu, I.; Popescu, I.-A.; et al. Skin Lesions with Loss of Tissue and Cutaneous-Onset Sepsis: The Skin Infection–Sepsis Relationship. Diagnostics 2024, 14, 659. [Google Scholar] [CrossRef] [PubMed]
- Sandru, F.; Poenaru, E.; Stoleru, S.; Radu, A.-M.; Roman, A.-M.; Ionescu, C.; Zugravu, A.; Nader, J.M.; Băicoianu-Nițescu, L.-C. Microbial Colonization and Antibiotic Resistance Profiles in Chronic Wounds: A Comparative Study of Hidradenitis Suppurativa and Venous Ulcers. Antibiotics 2025, 14, 53. [Google Scholar] [CrossRef]
- Tsige, Y.; Tadesse, S.; G/Eyesus, T.; Tefera, M.M.; Amsalu, A.; Menberu, M.A.; Gelaw, B. Prevalence of Methicillin-Resistant Staphylococcus aureus and Associated Risk Factors among Patients with Wound Infection at Referral Hospital, Northeast Ethiopia. J. Pathog. 2020, 2020, 3168325. [Google Scholar] [CrossRef]
- Almuhayawi, M.S.; Alruhaili, M.H.; Gattan, H.S.; Alharbi, M.T.; Nagshabandi, M.; Jaouni, S.A.; Selim, S.; Alanazi, A.; Alruwaili, Y.; Faried, O.A.; et al. Staphylococcus aureus induced wound infections which antimicrobial resistance, methicillin-and vancomycin-resistant: Assessment of emergence and cross sectional study. Infect. Drug Resist. 2023, 2023, 5335–5346. [Google Scholar] [CrossRef]
- Touaitia, R.; Mairi, A.; Ibrahim, N.A.; Basher, N.S.; Idres, T.; Touati, A. Staphylococcus aureus: A Review of the Pathogenesis and Virulence Mechanisms. Antibiotics 2025, 14, 470. [Google Scholar] [CrossRef]
- Sato’o, Y. Staphylococcus aureus Pathogenesis Based on Genetic Background. In Staphylococcus Aureus: Interplay Between Bacteria and Hosts; Nakane, A., Asano, K., Eds.; Springer: Singapore, 2024; pp. 119–150. [Google Scholar] [CrossRef]
- Linz, M.S.; Mattappallil, A.; Finkel, D.; Parker, D. Clinical Impact of Staphylococcus aureus Skin and Soft Tissue Infections. Antibiotics 2023, 12, 557. [Google Scholar] [CrossRef]
- Peng, Q.; Tang, X.; Dong, W.; Sun, N.; Yuan, W. A Review of Biofilm Formation of Staphylococcus aureus and Its Regulation Mechanism. Antibiotics 2022, 12, 12. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Ito, T.; Tamai, M.; Nakagawa, S.; Nakamura, Y. The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections. Inflamm. Regener. 2024, 44, 9. [Google Scholar] [CrossRef]
- Hulme, J. Harnessing Ultrasonic Technologies to Treat Staphylococcus Aureus Skin Infections. Molecules 2025, 30, 512. [Google Scholar] [CrossRef]
- Ahmad, A.A.M.; Abdelgalil, S.Y.; Khamis, T.; Abdelwahab, A.M.O.; Atwa, D.N.; Elmowalid, G.A. Thymoquinone’ potent impairment of multidrug-resistant Staphylococcus aureus NorA efflux pump activity. Sci. Rep. 2024, 14, 16483. [Google Scholar] [CrossRef]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef] [PubMed]
- Dashtbani-Roozbehani, A.; Brown, M.H. Efflux Pump Mediated Antimicrobial Resistance by Staphylococci in Health-Related Environments: Challenges and the Quest for Inhibition. Antibiotics 2021, 10, 1502. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, M.; Nabi, B.; Daswani, M.; Viquar, I.; Pal, N.; Sharma, P.; Tiwari, S.; Sarma, D.K.; Shubham, S.; Kumar, M. Role of bacterial efflux pump proteins in antibiotic resistance across microbial species. Microb. Pathog. 2023, 181, 106182. [Google Scholar] [CrossRef] [PubMed]
- Garcia, Í.R.; de Oliveira Garcia, F.A.; Pereira, P.S.; Coutinho, H.D.M.; Siyadatpanah, A.; Norouzi, R.; Wilairatana, P.; de Lourdes Pereira, M.; Nissapatorn, V.; Tintino, S.R.; et al. Microbial resistance: The role of efflux pump superfamilies and their respective substrates. Life Sci. 2022, 295, 120391. [Google Scholar] [CrossRef]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- Bessède, E.; Angla-gre, M.; Delagarde, Y.; Hieng, S.S.; M’enard, A.; M’egraud, F. Matrix-assisted laser-desorption/ionization biotyper: Experience in the routine of a university hospital. Clin. Microbiol. Infect. 2011, 17, 533–538. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Pratt, L.A.; Watnick, P.I.; Newman, D.K.; Weaver, V.B.; Kolter, R. Genetic approaches to study of biofilms. Methods Enzym. 1999, 310, 91–109. [Google Scholar]
- Gattringer, R.; Niks, M.; Ostertag, R.; Schwarz, K.; Medvedovic, H.; Graninger, W.; Georgopoulos, A. Evaluation of MIDITECH automated colorimetric MIC reading for antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2002, 49, 651–659. [Google Scholar] [CrossRef]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 14.0. 2024. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_14.0_Breakpoint_Tables.pdf (accessed on 1 January 2024).
- Martins, M.; Viveiros, M.; Couto, I.; Costa, S.S.; Pacheco, T.; Fanning, S.; Pagès, J.M.; Amaral, L. Identification of efflux pump-mediated multidrug-resistant bacteria by the ethidium bromide-agar cartwheel method. In vivo 2011, 25, 171–178. [Google Scholar]
- Király, J.; Hajdučková, V.; Gregová, G.; Szabóová, T.; Pilipčinec, E. Resistant S. aureus Isolates Capable of Producing Biofilm from the Milk of Dairy Cows with Subclinical Mastitis in Slovakia. Agriculture 2024, 14, 571. [Google Scholar] [CrossRef]
- Kryvtsova, M.V.; Király, J.; Koščová, J.; Kostenko, Y.Y.; Bubnov, R.V.; Spivak, M.Y. Determination of biofilm formation and associated gene detection in Staphylococcus genus isolated from the oral cavity under inflammatory periodontal diseases. Stud. Biol. 2020, 14, 49–64. [Google Scholar] [CrossRef]
- Hussain, M.; von Eiff, C.; Sinha, B.; Joost, I.; Herrmann, M.; Peters, G.; Becker, K. eap Gene as novel target for specific identification of Staphylococcus aureus. J. Clin. Microbiol. 2008, 46, 470–476. [Google Scholar] [CrossRef]
- Iqbal, Z.; Seleem, M.N.; Hussain, H.I.; Huang, L.; Hao, H.; Yuan, Z. Comparative virulence studies and transcriptome analysis of Staphylococcus aureus strains isolated from animals. Sci. Rep. 2016, 6, 35442. [Google Scholar] [CrossRef]
- Nourbakhsh, F.; Namvar, A.E. Detection of genes involved in biofilm formation in Staphylococcus aureus isolates. GMS Hyg. Infect. Control 2016, 11, Doc07. [Google Scholar] [CrossRef]
- Pereyra, E.A.L.; Picech, F.; Renna, M.S.; Baravalle, C.; Andreotti, C.S.; Russi, R.; Calvinho, L.F.; Diez, C.; Dallard, B.E. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet. Microbiol. 2016, 183, 69–77. [Google Scholar] [CrossRef]
- Englerová, K.; Bedlovičová, Z.; Nemcová, R.; Király, J.; Maďar, M.; Hajdučková, V.; Styková, E.; Mucha, R.; Reiffová, K. Bacillus amyloliquefaciens-Derived Lipopeptide Biosurfactants Inhibit Biofilm Formation and Expression of Biofilm-Related Genes of Staphylococcus aureus. Antibiotics 2021, 10, 1252. [Google Scholar] [CrossRef]
- Regecová, I.; Výrostková, J.; Zigo, F.; Gregová, G.; Pipová, M.; Jevinová, P.; Becová, J. Detection of Resistant and Enterotoxigenic Strains of Staphylococcus warneri Isolated from Food of Animal Origin. Foods 2022, 11, 1496. [Google Scholar] [CrossRef]
- Gehrke, A.-K.E.; Giai, C.; Gómez, M.I. Staphylococcus aureus Adaptation to the Skin in Health and Persistent/Recurrent Infections. Antibiotics 2023, 12, 1520. [Google Scholar] [CrossRef]
- Khalili, H.; Najar-Peerayeh, S.; Mahrooghi, M.; Mansouri, P.; Bakhshi, B. Methicillin-resistant Staphylococcus aureus colonization of infectious and non-infectious skin and soft tissue lesions in patients in Tehran. BMC Microbiol. 2021, 21, 282. [Google Scholar] [CrossRef]
- Vella, V.; Galgani, I.; Polito, L.; Arora, A.K.; Creech, C.B.; David, M.Z.; Lowy, F.D.; Macesic, N.; Ridgway, J.P.; Uhlemann, A.C.; et al. Staphylococcus aureus skin and soft tissue infection recurrence rates in outpatients: A retrospective database study at 3 US medical centers. Clin. Infect. Dis. 2021, 73, 1045–1053. [Google Scholar] [CrossRef]
- Ghebremedhin, B.; Olugbosi, M.O.; Raji, A.M.; Layer, F.; Bakare, R.A.; König, B.; König, W. Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J. Clin. Microbiol. 2009, 47, 2975–2980. [Google Scholar] [CrossRef]
- Godebo, G.; Kibru, G.; Tassew, H. Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann. Clin. Microbiol. Antimicrob. 2013, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Almeida, G.C.M.; dos Santos, M.M.; Lima, N.G.M.; Cidral, T.A.; Melo, M.C.N.; Lima, K.C. Prevalence and factors associated with wound colonization by Staphylococcus spp. and Staphylococcus aureus in hospitalized patients in inland northeastern Brazil: A cross-sectional study. BMC Infect. Dis. 2014, 14, 328. [Google Scholar] [CrossRef]
- Rasquel-Oliveira, F.S.; Ribeiro, J.M.; Martelossi-Cebinelli, G.; Costa, F.B.; Nakazato, G.; Casagrande, R.; Verri, W.A. Staphylococcus aureus in Inflammation and Pain: Update on Pathologic Mechanisms. Pathogens 2025, 14, 185. [Google Scholar] [CrossRef]
- Sivori, F.; Cavallo, I.; Truglio, M.; De Maio, F.; Sanguinetti, M.; Fabrizio, G.; Licursi, V.; Francalancia, M.; Fraticelli, F.; La Greca, I.; et al. Staphylococcus aureus colonizing the skin microbiota of adults with severe atopic dermatitis exhibits genomic diversity and convergence in biofilm traits. Biofilm 2024, 8, 100222. [Google Scholar] [CrossRef]
- Kwiecinski, J.; Kahlmeter, G.; Jin, T. Biofilm Formation by Staphylococcus aureus Isolates from Skin and Soft Tissue Infections. Curr. Microbiol. 2015, 70, 698–703. [Google Scholar] [CrossRef]
- Vergara-Irigaray, M.; Valle, J.; Merino, N.; Latasa, C.; García, B.; Ruiz de los Mozos, I.; Toledo-Arana, A.; Penadés, J.R.; Lasa, I. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect. Immun. 2009, 77, 3978–3991. [Google Scholar] [CrossRef]
- Cucarella, C.; Solano, C.; Valle, J.; Amorena, B.; Lasa, I.; Penadés, J.R. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 2001, 183, 2888–2896. [Google Scholar] [CrossRef]
- Valle, J.; Fang, X.; Lasa, I. Revisiting Bap Multidomain Protein: More Than Sticking Bacteria Together. Front. Microbiol. 2020, 11, 613581. [Google Scholar] [CrossRef] [PubMed]
- Matilla-Cuenca, L.; Gil, C.; Cuesta, S.; Rapún-Araiz, B.; Žiemytè, M.; Mira, A.; Lasa, I.; Valle, J. Antibiofilm activity of flavonoids on staphylococcal biofilms through targeting BAP amyloids. Sci. Rep. 2020, 10, 18968. [Google Scholar] [CrossRef] [PubMed]
- Rimi, S.S.; Ashraf, M.N.; Sigma, S.H.; Ahammed, M.T.; Siddique, M.P.; Zinnah, M.A.; Rahman, M.T.; Islam, M.S. Biofilm formation, agr typing and antibiotic resistance pattern in methicillin-resistant Staphylococcus aureus isolated from hospital environments. PLoS ONE 2024, 19, e0308282. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.H.; Wong, M.Y.; Huang, T.Y.; Kao, C.C.; Lin, Y.H.; Lu, C.H.; Huang, Y.K. Exploration of agr types, virulence-associated genes, and biofilm formation ability in Staphylococcus aureus isolates from hemodialysis patients with vascular access infections. Front. Cell Infect. Microbiol. 2024, 14, 1367016. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Humphreys, H.; Robinson, D.A.; Loughman, A.; Foster, T.J.; O’Gara, J.P. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008, 190, 3835–3850. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Invasive Staphylococcus aureus Infection Surveillance. Available online: https://www.cdc.gov/healthcareassociatedinfections/php/haiceip/invasivestaphylococcus.html. (accessed on 17 April 2024).
- Mediavilla, J.R.; Chen, L.; Mathema, B.; Kreiswirth, B.N. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 2012, 15, 588–595. [Google Scholar] [CrossRef]
- Ji, X.; Zhu, W.; Lu, H.; Wu, Z.; Chen, H.; Lin, C.H.; Zeng, Z.; You, C.; Li, L. Antibiotic Resistance Profiles and MLST Typing of Staphylococcus Aureus Clone Associated with Skin and Soft Tissue Infections in a Hospital of China. Infect. Drug, Resist. 2024, 17, 2555–2566. [Google Scholar] [CrossRef]
- Koumaki, D.; Maraki, S.; Evangelou, G.; Koumaki, V.; Gregoriou, S.; Kouloumvakou, S.; Petrou, D.; Rovithi, E.; Zografaki, K.; Doxastaki, A.; et al. Clinical features and Antibiotic Susceptibility of Staphylococcus aureus-infected dermatoses. J. Clin. Med. 2025, 14, 1084. [Google Scholar] [CrossRef]
- Bawankar, N.S.; Agrawal, G.N.; Zodpey, S.S. Revealing inducible clindamycin resistance in methicillin-resistant S. aureus: A vital diagnostic imperative for effective treatment. J. Postgrad. Med. 2024, 70, 223–226. [Google Scholar] [CrossRef]
- Assefa, M. Inducible Clindamycin-Resistant Staphylococcus aureus Strains in Africa: A Systematic Review. Int. J. Microbiol. 2022, 2022, 1835603. [Google Scholar] [CrossRef]
- Michalik, M.; Podbielska-Kubera, A.; Dmowska-Koroblewska, A. Antibiotic Resistance of Staphylococcus aureus Strains—Searching for New Antimicrobial Agents—Review. Pharmaceuticals 2025, 18, 81. [Google Scholar] [CrossRef] [PubMed]
- Brdová, D.; Ruml, T.; Viktorová, J. Mechanisms of staphylococcal resistance to clinically relevant antibiotics. Drug Resist. Updat. 2024, 77, 101147. [Google Scholar] [CrossRef]
- Thumu, S.C.; Halami, P.M. Acquired resistance to macrolide-lincosamide-streptogramin antibiotics in lactic Acid bacteria of food origin. Indian. J. Microbiol. 2012, 52, 530–537. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leclercq, R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002, 34, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Saderi, H.; Emadi, B.; Owlia, P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med. Sci. Monit. 2011, 17, BR48–BR53. [Google Scholar] [CrossRef]
- Henry, T.; Khachemoune, A. Dermatologic conditions and risk factors in people experiencing homelessness (PEH): Systematic review. Arch. Dermatol. Res. 2023, 315, 2795–2803. [Google Scholar] [CrossRef]
- Cohen, P.R. Community-acquired methicillin-resistant Staphylococcus aureus skin infections: A review of epidemiology, clinical features, management, and prevention. Int. J. Dermatol. 2007, 46, 1–11. [Google Scholar] [CrossRef]
- Kosmidis, C.; Schindler, B.D.; Jacinto, P.L.; Patel, D.; Bains, K.; Seo, S.M.; Kaatz, G.W. Expression of multidrug resistance efflux pump genes in clinical and environmental isloates of Staphylococcus aureus. Int. J. Antimicrob. Agents 2012, 40, 204–209. [Google Scholar] [CrossRef]
- Floyd, J.L.; Smith, K.P.; Kumar, S.H.; Floyd, J.T.; Varela, M.F. LmrS Is a Multidrug Efflux Pump of the Major Facilitator Superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother. 2010, 54, 5406–5412. [Google Scholar] [CrossRef]
- Lekshmi, M.; Stephen, J.; Ojha, M.; Kumar, S.; Varela, M. Staphylococcus aureus antimicrobial efflux pumps and their inhibitors: Recent developments. AIMS Med. Sci. 2022, 9, 367–393. [Google Scholar] [CrossRef]
- Favour, E.N.; Isaac, A.A. Phenotypic characterization of biofilm formation and efflux pump activity in multi-drug resistant staphylococcus species isolated from asymptomatic students. J. Microbiol. Exp. 2020, 8, 223–229. [Google Scholar] [CrossRef]
- Patel, D.; Kosmidis, C.; Seo, S.M.; Kaatz, G.W. Ethidium Bromide MIC Screening for Enhanced Efflux Pump Gene Expresion or Efflux Activity in Staphylococcus aureus. AAC 2010, 54, 5070–5073. [Google Scholar] [CrossRef]
- Abdi, P.; Mahdavi Ourtakand, M.; Honarmand Jahromy, S. The Effect of Matricaria chamomilla alcoholic extract on phenotype detection of efflux pumps of methicillin resistant Staphylococcus aureus (MRSA) isolated from skin lesions. Iran. J. Med. Microbiol. 2019, 13, 220–231. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).