Impact of Diet on Gut Microbiota in Diverticular Disease of the Colon: An Exploratory Retrospective Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion Criteria
2.2. Exclusion Criteria
2.3. Endpoints
2.4. Microbial DNA Extraction and 16S rRNA Amplicon Sequencing
2.5. Bioinformatics and Statistical Analysis
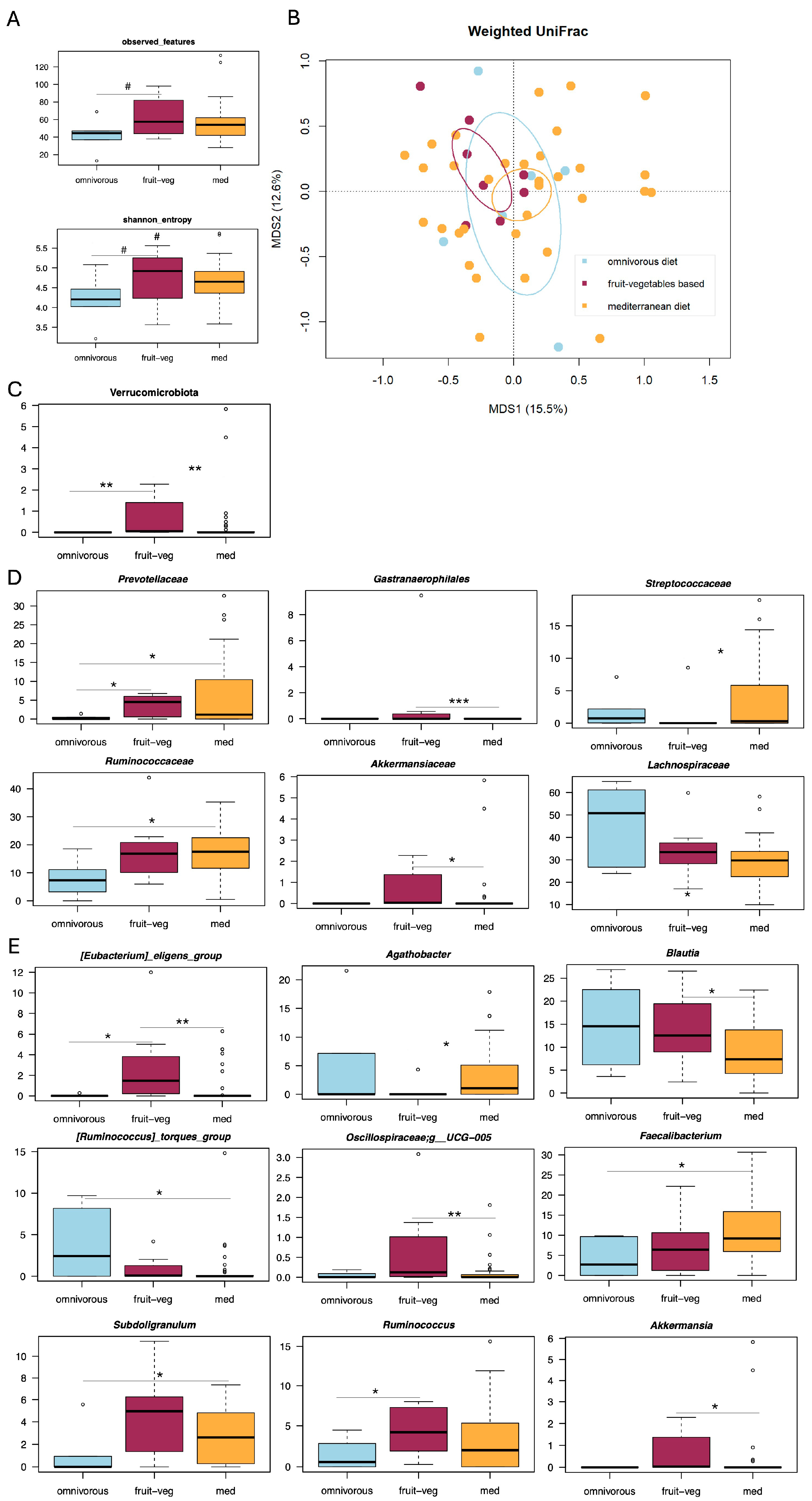
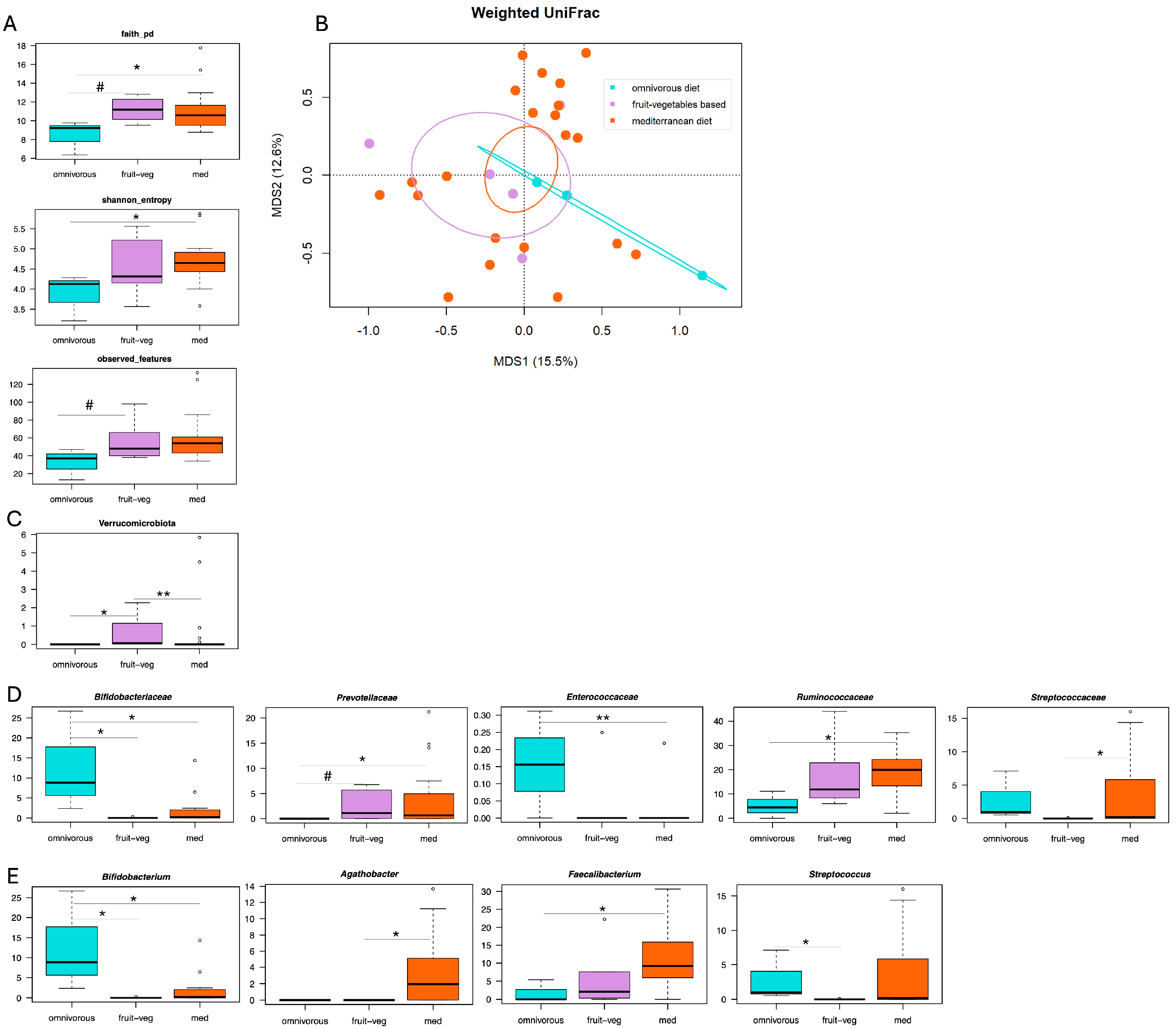
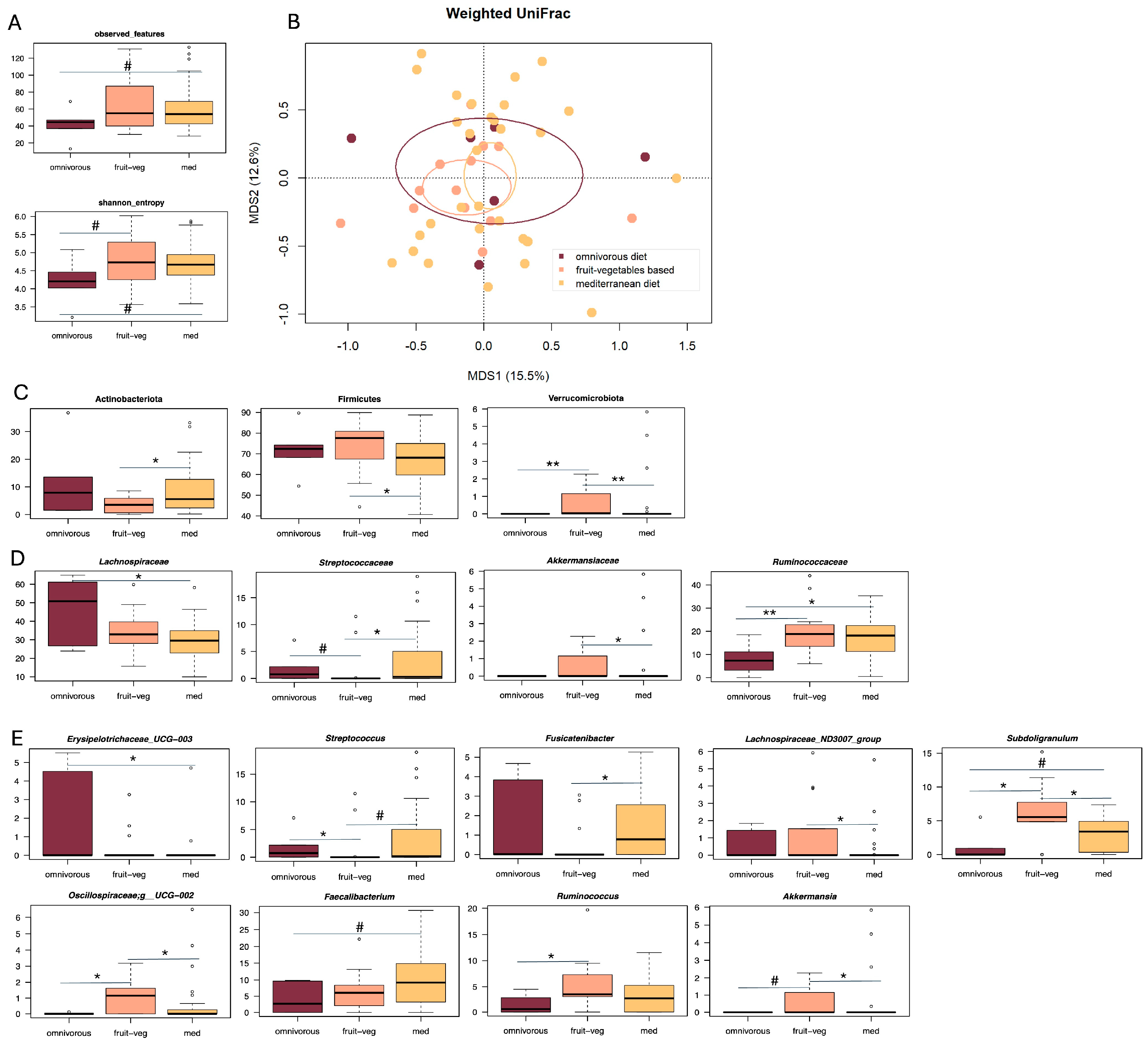
3. Results
3.1. Gut Microbiota Profiling in Patients with Diverticular Disease and Different Dietary Habits
3.2. Correlations Between Gut Microbiota, Dietary Patterns and Abdominal Pain Severity in SUDD
3.3. Correlations Between Gut Microbiota, Dietary Patterns and DICA Classification in SUDD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GM | Gut Microbiota |
| DD | Diverticular Disease |
| DICA | Diverticular Inflammation and Complication Assessment |
| SUDD | Symptomatic Uncomplicated Diverticular Disease |
References
- Tursi, A.; Scarpignato, C.; Strate, L.L.; Lanas, A.; Kruis, W.; Lahat, A.; Danese, S. Colonic diverticular disease. Nat. Rev. Dis. Primers 2020, 6, 20. [Google Scholar] [CrossRef]
- Urabe, M.; Nishida, T.; Shimakoshi, H.; Shimoda, A.; Amano, T.; Sugimoto, A.; Takahashi, K.; Mukai, K.; Matsubara, T.; Yamamoto, M.; et al. Distinct Clinical Factors in Hospitalized Patients with Diverticular Bleeding and Diverticulitis. Digestion 2018, 99, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Carabotti, M.; Morselli Labate, A.M.; Cremon, C.; Cuomo, R.; Pace, F.; Andreozzi, P.; Falangone, F.; Barbara, G.; Annibale, B.; REMAD group. Distinguishing features between patients with acute diverticulitis and diverticular bleeding: Results from the REMAD registry. Dig. Liver Dis. 2021, 53, 202–209. [Google Scholar] [CrossRef]
- Painter, N.S. The cause of diverticular disease of the colon, its symptoms and its complications: Review and hypothesis. J. R. Coll. Surg. Edinb. 1985, 30, 118–122. [Google Scholar]
- The Definition of Dietary Fiber. Available online: https://www.cerealsgrains.org/resources/definitions/Documents/DietaryFiber/DFDef.pdf (accessed on 5 March 2025).
- Slavin, J. Fiber and Prebiotics: Mechanisms and Health Benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef]
- Peery, A.F.; Sandler, R.S.; Ahnen, D.J. Constipation and a low-fiber diet are not associated with diverticulosis. Clin. Gastroenterol. Hepatol. 2013, 1, 1622–1627. [Google Scholar] [CrossRef]
- Peery, A.F.; Barrett, P.R.; Park, D. A high-fiber diet does not protect against asymptomatic diverticulosis. Gastroenterology 2012, 142, 266–272.e1. [Google Scholar] [CrossRef]
- Aldoori, W.H.; Giovannucci, E.L.; Rimm, E.B.; Wing, A.L.; Trichopoulos, D.V.; Willett, W.C. A prospective study of diet and the risk of symptomatic diverticular disease in men. Am. J. Clin. Nutr. 1994, 60, 757–764. [Google Scholar] [CrossRef]
- Cao, Y.; Strate, L.L.; Keeley, B.R.; Tam, I.; Wu, K.; Giovannucci, E.L.; Chan, A.T. Meat intake and risk of diverticulitis among men. Gut 2018, 67, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Tursi, A.; Franceschi, M.; Elisei, W.; Picchio, M.; Di Mario, F.; Brandimarte, G. The natural history of symptomatic uncomplicated diverticular disease: A long-term follow-up study. Ann. Gastroenterol. 2021, 34, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Tursi, A.; Elisei, W.; Picchio, M.; Nasi, G.; Mastromatteo, A.M.; Di Mario, F.; Di Rosa, E.; Brandimarte, M.A.; Brandimarte, G. Impact of diverticular inflammation and complication assessment classification on the burden of medical therapies in preventing diverticular disease complications in Italy. Ann. Transl. Med. 2017, 5, 320. [Google Scholar] [CrossRef]
- Zheng, J.; Sun, Q.; Zhang, M.; Liu, C.; Su, Q.; Zhang, L.; Xu, Z.; Lu, W.; Ching, J.; Tang, W.; et al. Noninvasive, microbiome-based diagnosis of inflammatory bowel disease. Nat. Med. 2024, 30, 3555–3567. [Google Scholar] [CrossRef] [PubMed]
- Tursi, A.; Turroni, S.; De Bastiani, R.; Procaccianti, G.; D’Amico, F.; Allegretta, L.; Antonino, N.; Baldi, E.; Casamassima, C.; Casella, G.; et al. Gut microbiota in symptomatic uncomplicated diverticular disease stratifies by severity of abdominal pain. Eur. J. Gastroenterol. Hepatol. 2025, 37, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Tursi, A.; Procaccianti, G.; De Bastiani, R.; Turroni, S.; D’Amico, F.; Allegretta, L.; Antonino, N.; Baldi, E.; Casamassima, C.; Casella, G.; et al. Micro-encapsulated and colonic-release sodium butyrate modulates gut microbiota and improves abdominal pain in patients with symptomatic uncomplicated diverticular disease. Front. Med. 2025, 12, 1487892. [Google Scholar] [CrossRef]
- Tursi, A.; Procaccianti, G.; Turroni, S.; De Bastiani, R.; D’Amico, F.; Allegretta, L.; Antonino, N.; Baldi, E.; Casamassima, C.; Casella, G.; et al. Gut microbiota perturbations are linked to endoscopic severity of diverticular disease. J. Gastrointestin. Liver Dis. 2025, 34, 317–322. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef]
- Kolodziejczyk, A.; Zeng, D.; Elinav, E. Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- D’Amico, F.; Rinaldi, M.; Pascale, R.; Fabbrini, M.; Morelli, M.C.; Siniscalchi, A.; Laici, C.; Coadonato, S.; Ravaioli, M.; Cescon, M.; et al. Gut microbiome dynamics and Enterobacterales infection in liver transplant recipients: A prospective observational study. JHEP Rep. 2024, 6, 101039. [Google Scholar] [CrossRef]
- Basis, C.M.; Moore, N.M.; Lolans, K.; Seekatz, A.M.; Weinstein, R.A.; Young, V.B.; Hayden, M.K.; CDC Prevention Epicenters Program. Comparison of stool versus rectal swab samples and storage conditions on bacterial community profiles. BMC Microbiol. 2017, 17, 78. [Google Scholar] [CrossRef]
- Tursi, A.; Brandimarte, G.; Di Mario, F.; Andreoli, A.; Annunziata, M.L.; Astegiano, M.; Bianco, M.A.; Buri, L.; Cammarota, G.; Capezzuto, E.; et al. Development and validation of an endoscopic classification of diverticular disease of the colon: The DICA classification. Dig. Dis. 2015, 33, 68–76. [Google Scholar] [CrossRef]
- Tursi, A.; Brandimarte, G.; Di Mario, F.; Elisei, W.; Picchio, M.; Allegretta, L.; Annunziata, M.L.; Bafutto, M.; Bassotti, G.; Bianco, M.A.; et al. Prognostic performance of the ‘DICA’ endoscopic classification and the ‘CODA’ score in predicting clinical outcomes of diverticular disease: An international, multicentre, prospective cohort study. Gut 2022, 71, 1350–1358. [Google Scholar] [CrossRef]
- Tursi, A.; Elisei, W.; Picchio, M.; Giorgetti, G.M.; Brandimarte, G. Moderate to severe and prolonged left-lower abdominal pain is the best symptom characterizing symptomatic uncomplicated diverticular disease of the colon: A comparison with fecal calprotectin in clinical setting. J. Clin. Gastroenterol. 2015, 49, 218–221. [Google Scholar] [CrossRef]
- Craig, W.J.; Mangels, A.R. American Dietetic Association. Position of the American Dietetic Association: Vegetarian diets. J. Am. Diet. Assoc. 2009, 109, 1266–1282. [Google Scholar]
- NHS Choices. The Vegetarian Diet (English). Available online: https://www.nhs.uk/live-well/eat-well/how-to-eat-a-balanced-diet/the-vegetarian-diet/ (accessed on 10 August 2025).
- NHS Choices. Eating a Balanced Diet (English). Available online: https://www.nhs.uk/live-well/eat-well/how-to-eat-a-balanced-diet/eating-a-balanced-diet/ (accessed on 10 August 2025).
- Freedberg, D.E.; Chang, L. Gastrointestinal symptoms in COVID-19: The long and the short of it. Curr. Opin. Gastroenterol. 2022, 38, 555–561. [Google Scholar] [CrossRef]
- Masella, A.P.; Bartram, A.K.; Truszkowski, J.M.; Brown, D.G.; Neufeld, J.D. PANDAseq: Paired-end assembler for illumina sequences. BMC Bioinform. 2012, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.V.; Johnson, A.J.A.; Holmes, A.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahè, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Culhane, A.C.; Thioulouse, J.; Perrière, G.; Higgins, D.G. MADE4: An R package for multivariate analysis of gene expression data. Bioinformatics 2005, 21, 2789–2790. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, W.; Mehta, R.; Nguyen, L.H.; Song, M.; Drew, D.A.; Asnicar, F.; Huttenhower, C.; Segata, N.; Wolf, J.; et al. Diet and gut microbial associations in irritable bowel syndrome according to disease subtype. Gut Microbes 2023, 15, 2262130. [Google Scholar] [CrossRef]
- Christensen, C.; Knudsen, A.; Arnesen, E.K.; Hatlebakk, J.G.; Sletten, I.S.; Fadnes, L.T. Diet, Food, and Nutritional Exposures and Inflammatory Bowel Disease or Progression of Disease: An Umbrella Review. Adv. Nutr. 2024, 15, 100219. [Google Scholar] [CrossRef]
- Yang, J.; Li, Y.; Wen, Z.; Liu, W.; Meng, L.; Huang, H. Oscillospira—A candidate for the next-generation probiotics. Gut Microbes 2021, 13, 1987783. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Ricci, C.; Rizzello, F.; Valerii, M.C.; Spisni, E.; Gionchetti, P.; Turroni, S.; Candela, M.; D’Amico, F.; Spigarelli, R.; Bellocchio, I.; et al. Geraniol Treatment for Irritable Bowel Syndrome: A Double-Blind Randomized Clinical Trial. Nutrients 2022, 14, 4208. [Google Scholar] [CrossRef] [PubMed]
- Malinen, E.; Krogius-Kurikka, L.; Lyra, A.; Nikkilä, J.; Jääskeläinen, A.; Rinttilä, T.; Vilpponen-Salmela, T.; von Wright, A.J.; Palva, A. Association of symptoms with gastrointestinal microbiota in irritable bowel syndrome. World J. Gastroenterol. 2010, 16, 4532–4540. [Google Scholar] [CrossRef]
- Schaus, S.R.; Vasconcelos Pereira, G.; Luis, A.S.; Madlambayan, E.; Terrapon, N.; Ostrowski, M.P.; Jin, C.; Henrissat, B.; Hansson, G.C.; Martens, E.C. Ruminococcus torques is a keystone degrader of intestinal mucin glycoprotein, releasing oligosaccharides used by Bacteroides thetaiotaomicron. mBio 2024, 15, e0003924. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhu, S.; Dong, Y.; Xie, T.; Chai, Z.; Gao, X.; Dai, Y.; Wang, X. The Role of Gut Microbiome in Irritable Bowel Syndrome: Implications for Clinical Therapeutics. Biomolecules 2024, 14, 1643. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O. Insights into the Role of Erysipelotrichaceae in the Human Host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef]
- Tursi, A.; Piovani, D.; Brandimarte, G.; Di Mario, F.; Figlioli, G.; Dumitrascu, D.; Chaves Oliveira, E.; Papagrigoriadis, S.; Stundiene, I.; Reichert, M.C.; et al. Therapeutic strategies for the prevention of acute diverticulitis according to Diverticular Inflammation Complication Assessment (DICA) endoscopic score: Apost-hoc analysis of a prospective international study. Eur. J. Gastroenterol. Hepatol. 2025; in press. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tursi, A.; Procaccianti, G.; D’Amico, F.; De Bastiani, R.; Allegretta, L.; Antonino, N.; Baldi, E.; Casamassima, C.; Casella, G.; Ciuffi, M.; et al. Impact of Diet on Gut Microbiota in Diverticular Disease of the Colon: An Exploratory Retrospective Study. Microorganisms 2025, 13, 2428. https://doi.org/10.3390/microorganisms13112428
Tursi A, Procaccianti G, D’Amico F, De Bastiani R, Allegretta L, Antonino N, Baldi E, Casamassima C, Casella G, Ciuffi M, et al. Impact of Diet on Gut Microbiota in Diverticular Disease of the Colon: An Exploratory Retrospective Study. Microorganisms. 2025; 13(11):2428. https://doi.org/10.3390/microorganisms13112428
Chicago/Turabian StyleTursi, Antonio, Giorgia Procaccianti, Federica D’Amico, Rudi De Bastiani, Leonardo Allegretta, Natale Antonino, Elisabetta Baldi, Carlo Casamassima, Giovanni Casella, Mario Ciuffi, and et al. 2025. "Impact of Diet on Gut Microbiota in Diverticular Disease of the Colon: An Exploratory Retrospective Study" Microorganisms 13, no. 11: 2428. https://doi.org/10.3390/microorganisms13112428
APA StyleTursi, A., Procaccianti, G., D’Amico, F., De Bastiani, R., Allegretta, L., Antonino, N., Baldi, E., Casamassima, C., Casella, G., Ciuffi, M., De Bastiani, M., Lazzarotto, L., Licci, C., Mancuso, M., Penna, A., Pranzo, G., Sanna, G., Tosetti, C., Zamparella, M., ... Turroni, S. (2025). Impact of Diet on Gut Microbiota in Diverticular Disease of the Colon: An Exploratory Retrospective Study. Microorganisms, 13(11), 2428. https://doi.org/10.3390/microorganisms13112428








