Changes in Gut Phageome and Bacteriome Following Fecal Microbiota Transfer in Patients with Intestinal Graft-Versus-Host Disease and Crohn’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Patients and Study Design
2.3. Pretransplant Examination of Microbiota Donors
2.4. Fecal Microbiota Transfer
2.5. Clinical Laboratory Studies
2.6. NGS Procedures
2.7. Bioinformatic Analysis of the 16S rRNA NGS Data
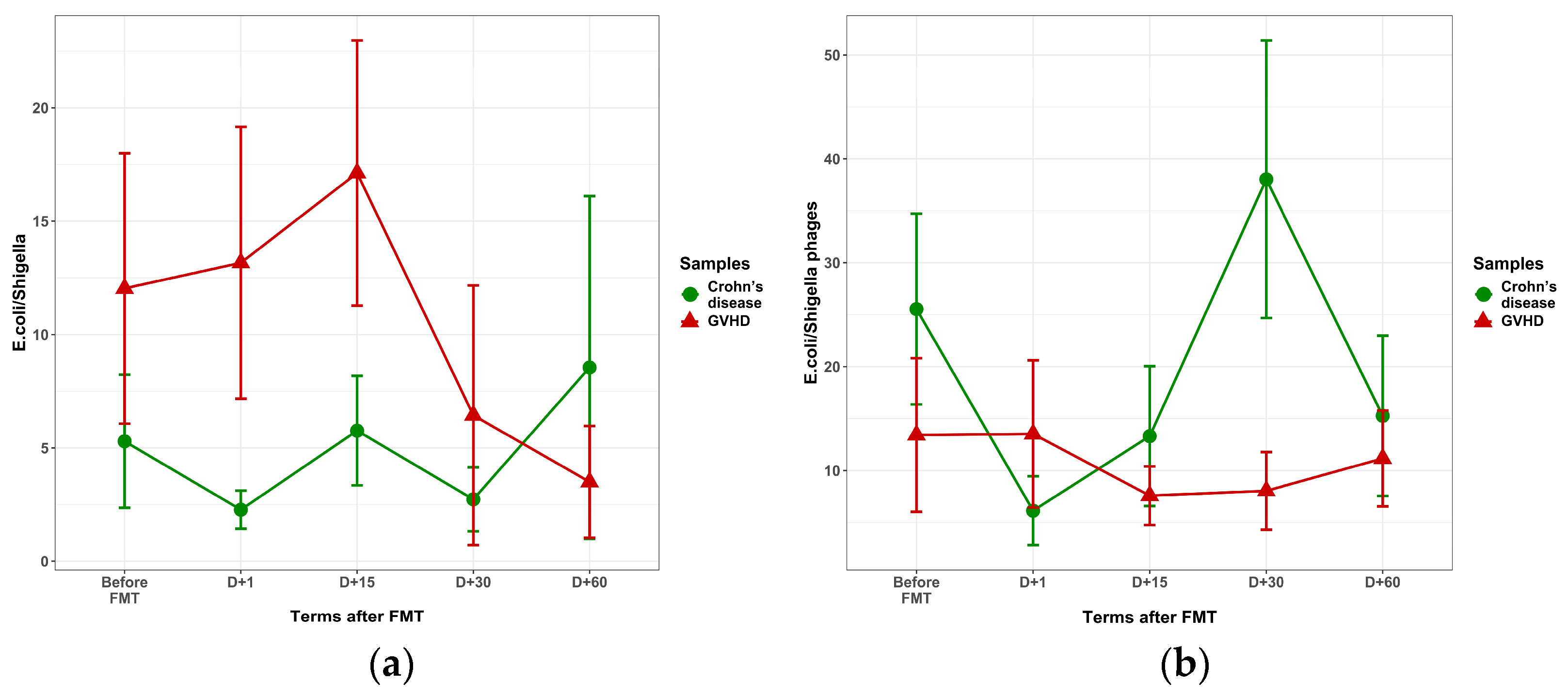
3. Results
3.1. Clinical Effects of FMT in GVHD and Crohn’s Disease
3.2. Relative Contents of Fecal Bacteriome Before and After FMT
3.2.1. Ratios of Bacterial Genera Before FMT
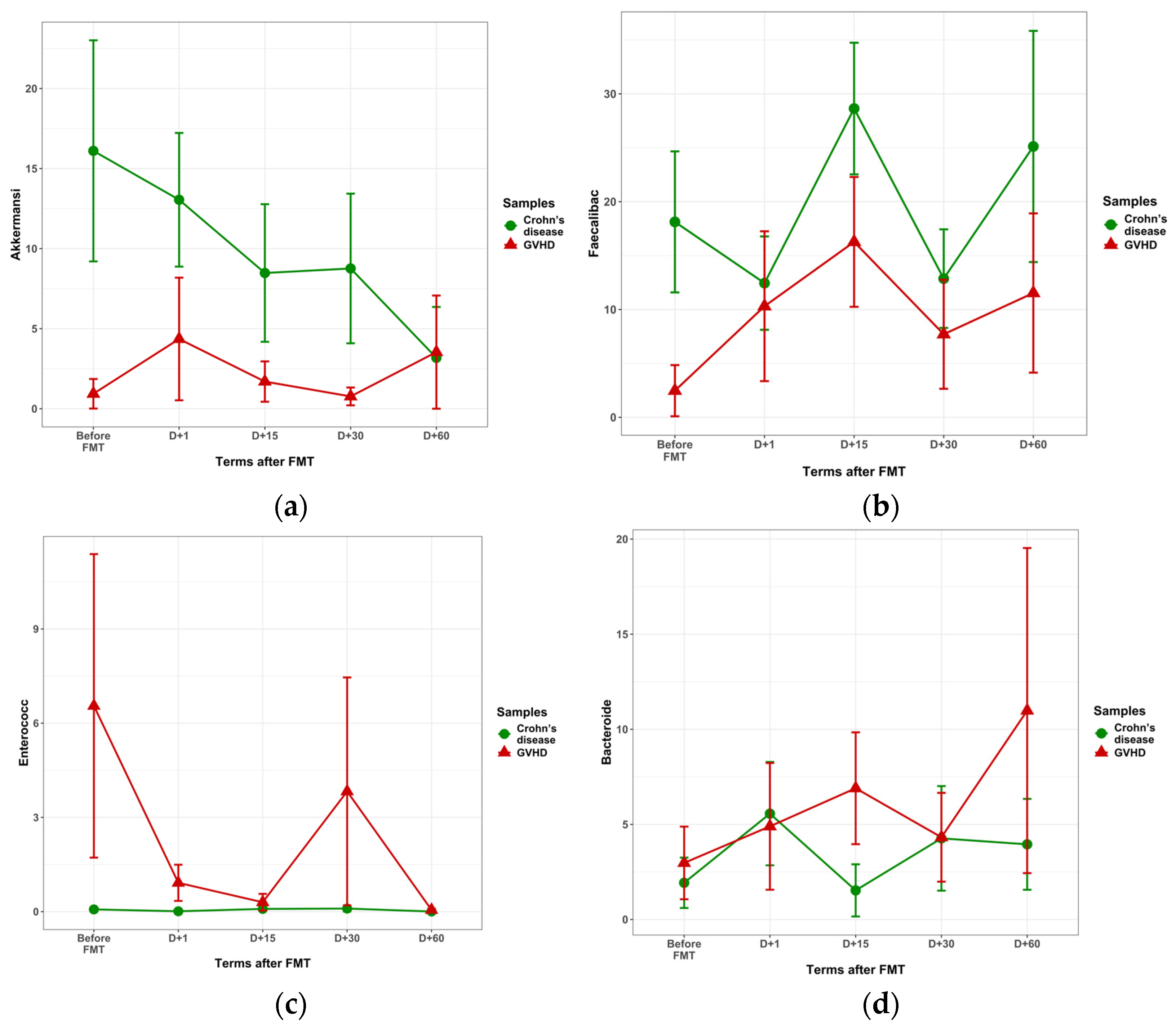
3.2.2. Bacteriome in Crohn’s Disease After FMT
3.2.3. Bacteriome Changes in GVHD Following FMT
3.3. Fecal Virome in the Course of FMT
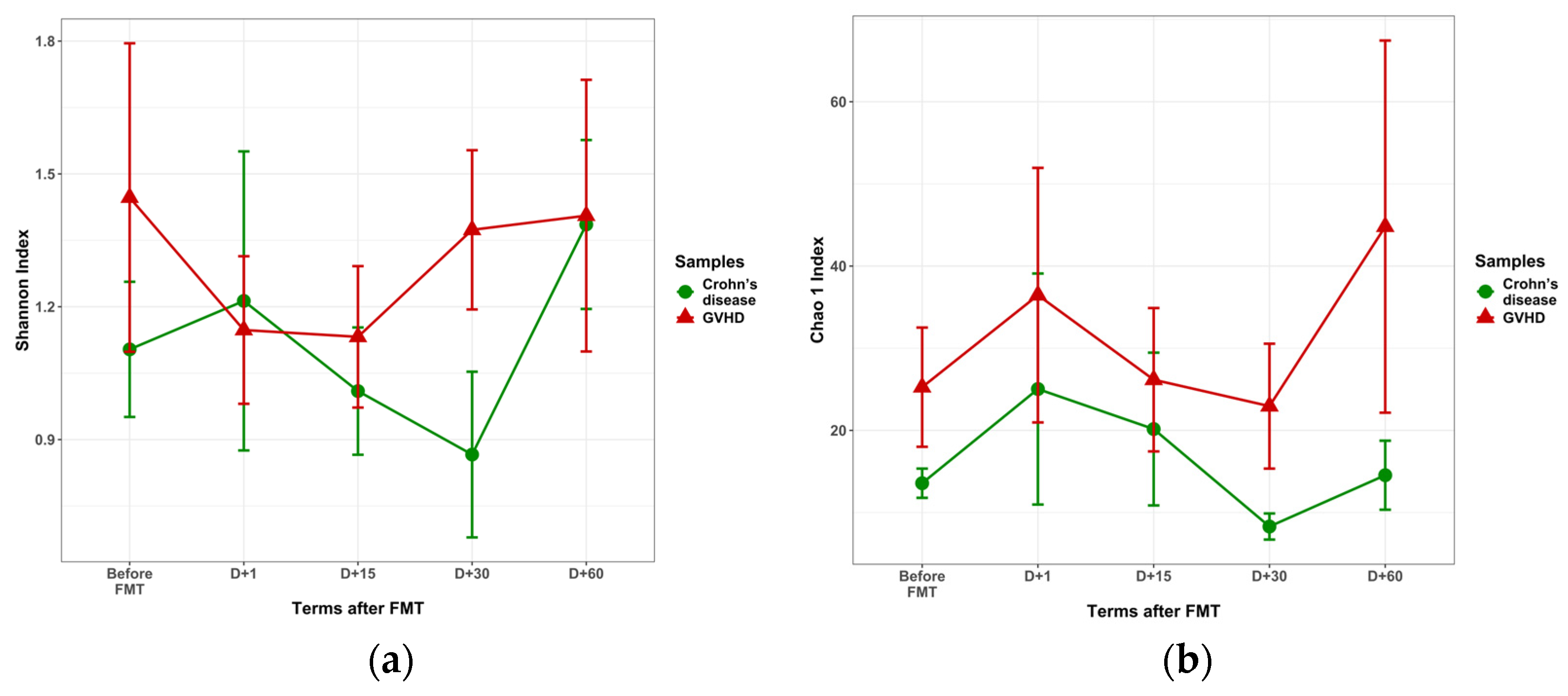
3.3.1. Pre-FMT Diversity of Viral Populations
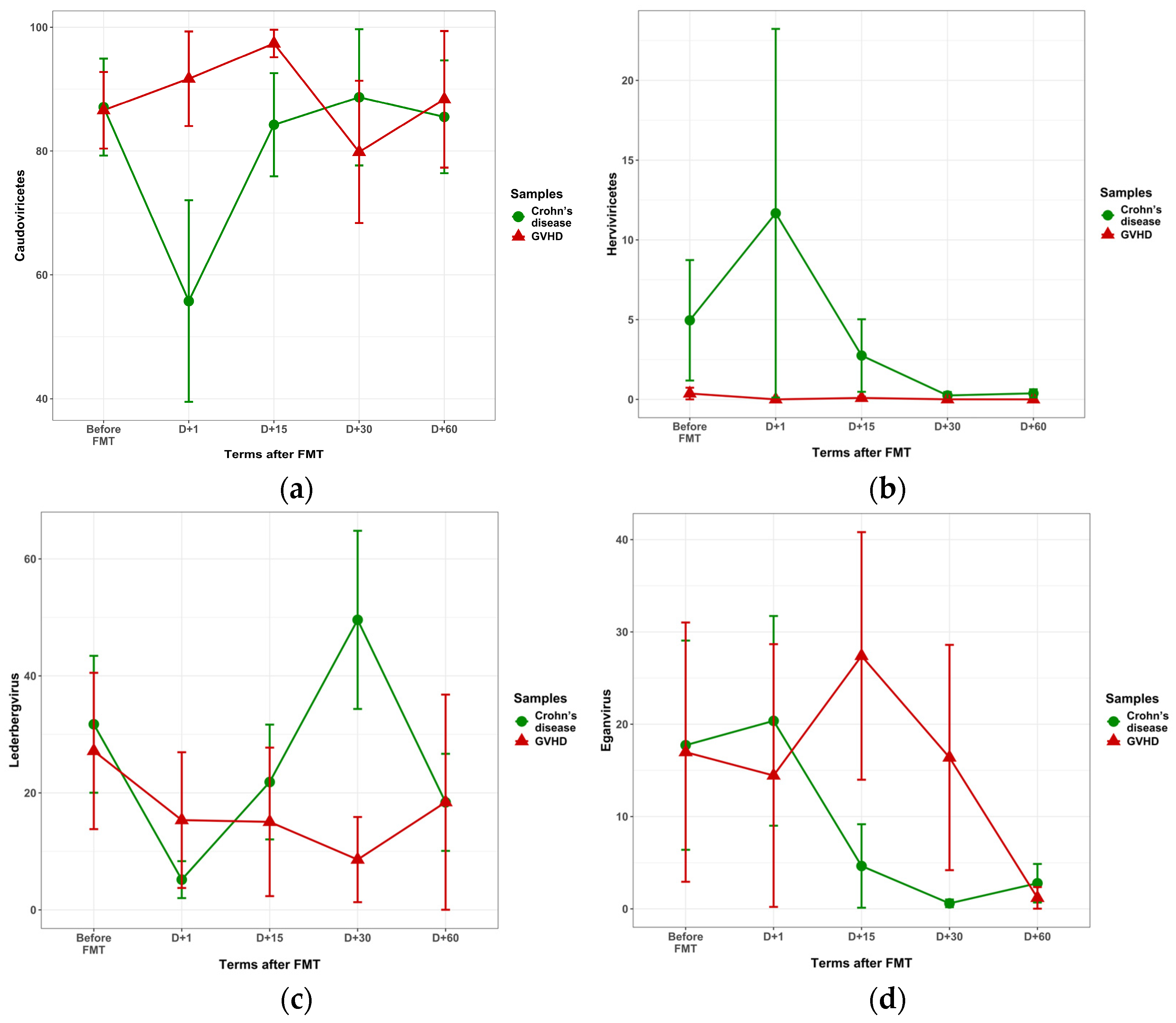
3.3.2. Post-FMT Changes in Fecal Virome
3.4. Potential Interactions Between Fecal Bacteria and Phages
3.4.1. Differential Clustering of Fecal Bacteria and Phages
3.4.2. Correlations Between Phage Genera and Their Host Bacteria
4. Discussion
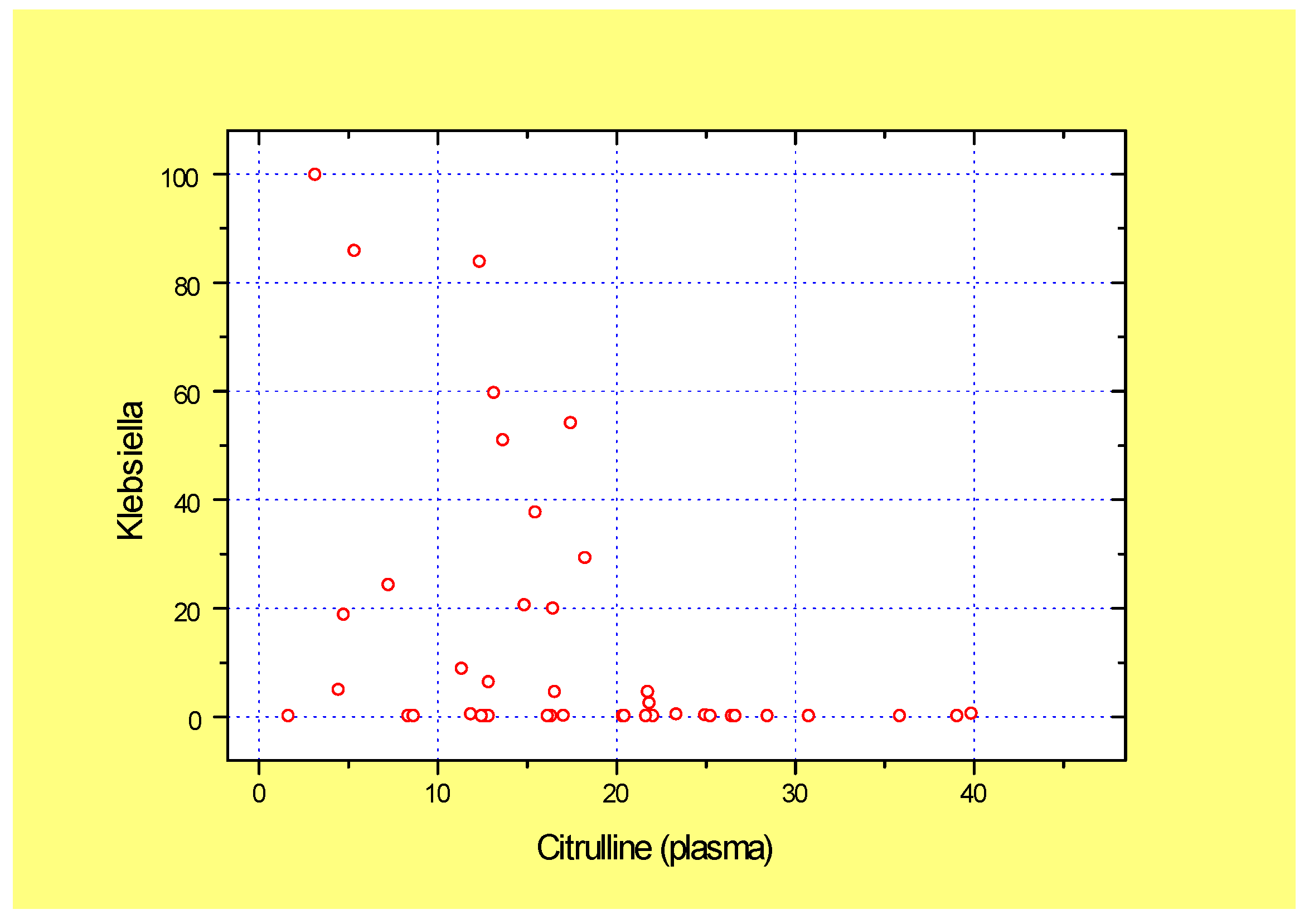
4.1. Intestinal Damage and Plasma Citrulline in GVHD
4.2. FMT-Associated Bacteriome Changes
4.3. Phageome Changes After FMT
4.4. Possible Interaction Between E. coli and Bacteriophages
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weber, D.; Meedt, E.; Poeck, H.; Thiele-Orberg, E.; Hiergeist, A.; Gessner, A.; Holler, E. Fecal microbiota transfer in acute graft-versus-host disease following allogeneic stem cell transplantation. Visc. Med. 2024, 40, 210–216. [Google Scholar] [CrossRef]
- Levy, O.; Teixeira-Pinto, A.; White, M.L.; Carroll, S.F.; Lehmann, L.; Wypij, D.; Guinan, E. Endotoxemia and elevation of lipopolysaccharide-binding protein after hematopoietic stem cell transplantation. Pediatr. Infect. Dis. J. 2003, 22, 978–981. [Google Scholar] [CrossRef]
- Dekker, I.M.; Bruggink, H.; van der Meij, B.S.; Wierdsma, N.J. State of the art: The role of citrulline as biomarker in patients with chemotherapy- or graft-versus-host-disease-induced mucositis. Curr. Opin. Clin. Nutr. Metab. Care 2021, 24, 416–427. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, G.R.; Loftus, E.V.; Isaacs, K.L.; Regueiro, M.D.; Gerson, L.B.; Sands, B.E. ACG Clinical Guideline: Management of Crohn’s Disease in Adults. Am. J. Gastroenterol. 2018, 113, 481–517. [Google Scholar] [CrossRef] [PubMed]
- Rahaman, M.M.; Wangchuk, P.; Sarker, S. A systematic review on the role of gut microbiome in inflammatory bowel disease: Spotlight on virome and plant metabolites. Microb. Pathog. 2025, 205, 107608. [Google Scholar] [CrossRef] [PubMed]
- Fetter, K.; Weigel, M.; Ott, B.; Fritzenwanker, M.; Stricker, S.; de Laffolie, J.; Hain, T. The microbiome landscape in pediatric Crohn’s disease and therapeutic implications. Gut Microbes 2023, 15, 2247019. [Google Scholar] [CrossRef]
- Sharma, B.; Agriantonis, G.; Twelker, K.; Ebelle, D.; Kiernan, S.; Siddiqui, M.; Soni, A.; Cheerasarn, S.; Simon, W.; Jiang, W.; et al. Gut microbiota serves as a crucial independent biomarker in inflammatory bowel disease (IBD). Int. J. Mol. Sci. 2025, 26, 2503. [Google Scholar] [CrossRef]
- Bernardi, F.; Ungaro, F.; D’Amico, F.; Zilli, A.; Parigi, T.L.; Massimino, L.; Allocca, M.; Danese, S.; Furfaro, F. The role of viruses in the pathogenesis of immune-mediated gastro-intestinal diseases. Int. J. Mol. Sci. 2024, 25, 8301. [Google Scholar] [CrossRef]
- Federici, S.; Kviatcovsky, D.; Valdés-Mas, R.; Elinav, E. Microbiome-phage interactions in inflammatory bowel disease. Clin. Microbiol. Infect. 2023, 29, 682–688. [Google Scholar] [CrossRef]
- Fehily, S.R.; Basnayake, C.; Wright, E.K.; Kamm, M.A. Fecal microbiota transplantation therapy in Crohn’s disease: Systematic review. J. Gastroenterol. Hepatol. 2021, 36, 2672–2686. [Google Scholar] [CrossRef]
- Youngster, I.; Eshel, A.; Geva, M.; Danylesko, I.; Henig, I.; Zuckerman, T.; Fried, S.; Yerushalmi, R.; Shem-Tov, N.; Fein, J.A.; et al. Fecal microbiota transplantation in capsules for the treatment of steroid refractory and steroid dependent acute graft vs. host disease: A pilot study. Bone Marrow Transplant. 2024, 59, 409–416. [Google Scholar] [CrossRef]
- Legoff, J.; Resche-Rigon, M.; Bouquet, J.; Robin, M.; Naccache, S.N.; Mercier-Delarue, S.; Federman, S.; Samayoa, E.; Rousseau, C.; Piron, P.; et al. The eukaryotic gut virome in hematopoietic stem cell transplantation: New clues in enteric graft-versus-host disease. Nat. Med. 2017, 23, 1080–1085. [Google Scholar] [CrossRef] [PubMed]
- Zanella, M.C.; Cordey, S.; Kaiser, L. Beyond cytomegalovirus and Epstein-Barr virus: A review of viruses composing the blood virome of solid organ transplant and hematopoietic stem cell transplant recipients. Clin. Microbiol. Rev. 2020, 33, e00027-20. [Google Scholar] [CrossRef] [PubMed]
- Jansen, S.A.; Nijhuis, W.; Leavis, H.L.; Riezebos-Brilman, A.; Lindemans, C.A.; Schuurman, R. Broad virus detection and variant discovery in fecal samples of hematopoietic transplant recipients using targeted sequence capture metagenomics. Front. Microbiol. 2020, 11, 560179. [Google Scholar] [CrossRef]
- Goloshchapov, O.V.; Shvetsov, A.N.; Chukhlovin, A.B.; Spiridonova, A.A.; Vladovskaya, M.D.; Zubarovskaya, L.S.; Kulagin, A.D. Incidence of common herpesviruses in colonic mucosal biopsies following hematopoietic stem cell transplantation. Microorganisms 2022, 10, 2128. [Google Scholar] [CrossRef]
- Liu, Q.; Xu, Z.; Dai, M.; Su, Q.; Leung Chan, F.K.; Ng, S.C. Faecal microbiota transplantations and the role of bacteriophages. Clin. Microbiol. Infect. 2023, 29, 689–694. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, C.; Liu, Y.; Yao, J.; Yang, X.; Wu, S.; Du, J.; Yang, X. Beyond faecal microbiota transplantation, the non-negligible role of faecal virome or bacteriophage transplantation. J. Microbiol. Immunol. Infect. 2023, 56, 893–908. [Google Scholar] [CrossRef]
- Lam, S.; Bai, X.; Shkoporov, A.N.; Park, H.; Wu, X.; Lan, P.; Zuo, T. Roles of the gut virome and mycobiome in faecal microbiota transplantation. Lancet Gastroenterol. Hepatol. 2022, 7, 472–484. [Google Scholar] [CrossRef]
- Zhang, F.; Zuo, T.; Yeoh, Y.K.; Cheng, F.W.T.; Liu, Q.; Tang, W.; Cheung, K.C.Y.; Yang, K.; Cheung, C.P.; Mo, C.C.; et al. Longitudinal dynamics of gut bacteriome, mycobiome and virome after fecal microbiota transplantation in graft-versus-host disease. Nat. Commun. 2021, 12, 65. [Google Scholar] [CrossRef] [PubMed]
- Holler, E.; Greinix, H.; Zeiser, R. Acute Graft-Versus-Host Disease. In The EBMT Handbook: Hematopoietic Stem Cell Transplantation and Cellular Therapies, 7th ed.; Carreras, E., Dufour, C., Mohty, M., Eds.; Springer: Cham, Switzerland, 2019; Chapter 43. [Google Scholar] [CrossRef]
- Przepiorka, D.; Weisdorf, D.; Martin, P.; Klingemann, H.G.; Beatty, P.; Hows, J.; Thomas, E.D. 1994 Consensus Conference on Acute GVHD Grading. Bone Marrow Transplant. 1995, 15, 825–828. [Google Scholar] [PubMed]
- Lewis, S.J.; Heaton, K.W. Stool form scale as a useful guide to intestinal transit time. Scand. J. Gastroenterol. 1997, 32, 920–924. [Google Scholar] [CrossRef]
- Myerson, D.; Steinbach, G.; Gooley, T.A.; Shulman, H.M. Graft-versus-host disease of the gut: A histologic activity grading system and validation. Biol. Blood Marrow Transplant. 2017, 23, 1573–1579. [Google Scholar] [CrossRef]
- Moiseev, I.S.; Balashov, D.N.; Bronin, G.O.; Vahonina, L.V.; Budaeva, I.G.; Dinikina, Y.V.; Drokov, M.Y.; Zorina, N.A.; Feducova, Y.; Ziganshina, S.E.; et al. Recommendations of Russian Society for Hematopoietic Stem Cell Transplantation, Gene and Cell therapy for diagnosis and treatment of graft-versus-host disease. Cell Ther. Transplant. 2025, 14, 55–69. [Google Scholar] [CrossRef]
- Goloshchapov, O.V.; Chukhlovin, A.B.; Polev, D.E.; Eismont, Y.A.; Bug, D.S.; Kusakin, A.V.; Kosarev, O.V.; Klementeva, R.V.; Gostev, V.V.; Ageevets, V.A.; et al. Time-Dependent Shifts in Intestinal Bacteriome, Klebsiella Colonization and Incidence of Antibiotic-Resistance Genes after Allogeneic Hematopoietic Stem Cell Transplantation. Biomedicines 2024, 12, 1566. [Google Scholar] [CrossRef] [PubMed]
- Chukhlovin, A.B.; Dudurich, V.V.; Kusakin, A.V.; Polev, D.E.; Ermachenko, E.D.; Aseev, M.V.; Zakharov, Y.A.; Eismont, Y.A.; Danilov, L.G.; Glotov, O.S. Evaluation of gut microbiota in healthy persons and type 1 diabetes mellitus patients in North-Western Russia. Microorganisms 2023, 11, 1813. [Google Scholar] [CrossRef] [PubMed]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilić-Stojanović, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Goloshchapov, O.V.; Shchukina, O.B.; Kusakin, A.V.; Tsai, V.V.; Kalinin, R.S.; Eismont, Y.A.; Glotov, O.S.; Chukhlovin, A.B. Next-generation sequencing-based monitoring of intestinal bacteria and bacteriophages following fecal microbiota transplantation in inflammatory bowel diseases. Pathogens 2023, 12, 1438. [Google Scholar] [CrossRef]
- Bartolomeo, M.P.; Maisano, F. Validation of a reversed-phase HPLC method for quantitative amino acid analysis. J. Biomol. Tech. 2006, 17, 131–137. [Google Scholar] [PubMed]
- Andrews, S. Fast QC: A Quality Control Tool for High Throughput Sequence Data; FastQC: San Diego, CA, USA, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 2020, 36, 1303–1304. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Zhloba, A.A.; Subbotina, T.F. Disturbances of citrulline homeostasis in patients with arterial hypertension. Nephrologia 2024, 28, 47–54. [Google Scholar] [CrossRef]
- Harris, A.C.; Young, R.; Devine, S.; Hogan, W.J.; Ayuk, F.; Bunworasate, U.; Chanswangphuwana, C.; Efebera, Y.A.; Holler, E.; Litzow, M.; et al. International, Multicenter Standardization of Acute Graft-versus-Host Disease Clinical Data Collection: A Report from the Mount Sinai Acute GVHD International Consortium. Biol. Blood Marrow Transplant. 2016, 22, 4–10. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Kim, H.T.; Spyrou, N.; Katsivelos, N.; Kowalyk, S.; Eng, G.; Kasikis, S.; Beheshti, R.; Baez, J.; Akahoshi, Y.; et al. The MAGIC algorithm probability predicts treatment response and long-term outcomes to second-line therapy for acute GVHD. Blood Adv. 2024, 8, 3488–3496. [Google Scholar] [CrossRef]
- Bayraktar, E.; Graf, T.; Ayuk, F.A.; Beutel, G.; Penack, O.; Luft, T.; Brueder, N.; Castellani, G.; Reinhardt, H.C.; Kröger, N.; et al. Data-driven grading of acute graft-versus-host disease. Nat. Commun. 2023, 14, 7799. [Google Scholar] [CrossRef]
- Rashidi, A.; Shanley, R.; Holtan, S.G.; MacMillan, M.L.; Blazar, B.R.; Khoruts, A.; Weisdorf, D.J. Pretransplant Serum Citrulline Predicts Acute Graft-versus-Host Disease. Biol. Blood Marrow Transplant. 2018, 24, 2190–2196. [Google Scholar] [CrossRef]
- Geboes, K. What histologic features best differentiate Crohn’s disease from ulcerative colitis? Inflamm. Bowel Dis. 2008, 14, S168–S169. [Google Scholar] [CrossRef]
- Zeiser, R.; Warnatz, K.; Rosshart, S.; Sagar; Tanriver, Y. GVHD, IBD, and primary immunodeficiencies: The gut as a target of immunopathology resulting from impaired immunity. Eur. J. Immunol. 2022, 52, 1406–1418. [Google Scholar] [CrossRef]
- Eshel, A.; Sharon, I.; Nagler, A.; Bomze, D.; Danylesko, I.; Fein, J.A.; Geva, M.; Henig, I.; Shimoni, A.; Zuckerman, T.; et al. Origins of bloodstream infections following fecal microbiota transplantation: A strain-level analysis. Blood Adv. 2022, 6, 568–573. [Google Scholar] [CrossRef] [PubMed]
- van Lier, Y.F.; Vos, J.; Blom, B.; Hazenberg, M.D. Allogeneic hematopoietic cell transplantation, the microbiome, and graft-versus-host disease. Gut Microbes 2023, 15, 2178805. [Google Scholar] [CrossRef]
- Gupta, M.K.; Srivastava, R. Gut microbiome interventions: From dysbiosis to next-generation probiotics (NGPs) for disease management. Probiotics Antimicrob. Proteins 2025, 17, 2629–2652. [Google Scholar] [CrossRef]
- Magro, D.O.; Santos, A.; Guadagnini, D.; de Godoy, F.M.; Silva, S.H.M.; Lemos, W.J.F.; Vitulo, N.; Torriani, S.; Pinheiro, L.V.; Martinez, C.A.R.; et al. Remission in Crohn’s disease is accompanied by alterations in the gut microbiota and mucins production. Sci. Rep. 2019, 9, 13263. [Google Scholar] [CrossRef] [PubMed]
- Taylor, H.; McDonald Kathrin, J.H.; Serrano-Contreras, J.I.; Li, J. Deep remission in paediatric Crohn’s disease is associated with increased abundance of Dialister species and increased valerate. J. Crohn’s Colitis 2020, 14 (Suppl. 1), S045–S046. [Google Scholar] [CrossRef]
- Townsend, E.M.; Kelly, L.; Muscatt, G.; Box, J.D.; Hargraves, N.; Lilley, D.; Jameson, E. The human gut phageome: Origins and roles in the human gut microbiome. Front. Cell. Infect. Microbiol. 2021, 11, 643214. [Google Scholar] [CrossRef]
- Guo, J.; Zhong, Y.; Wang, Y.; Liu, P.; Jin, H.; Wang, Y.; Shi, L.; Wang, P.; Li, W. Phylogenetic Relationships and Evolution of the Genus Eganvirus (186-Type) Yersinia pestis Bacteriophages. Viruses 2024, 16, 748. [Google Scholar] [CrossRef]
- Górski, A.; Jończyk-Matysiak, E.; Międzybrodzki, R.; Weber-Dąbrowska, B.; Borysowski, J. “Phage transplantation in allotransplantation”: Possible treatment in graft-versus-host disease? Front. Immunol. 2018, 9, 941. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J.; Waetzig, G.H.; Rehman, A.; Moltzau-Anderson, J.; Bharti, R.; Grasis, J.A.; Cassidy, L.; Tholey, A.; Fickenscher, H.; Seegert, D.; et al. Efficacy of sterile fecal filtrate transfer for treating patients with Clostridium difficile infection. Gastroenterology 2017, 152, 799–811.e7. [Google Scholar] [CrossRef] [PubMed]
- Chukhlovin, A.B. Laboratory tools for assessment of gut dysbiosis in oncohematological patients. Cell. Ther. Transplant. 2024, 13, 55–58. [Google Scholar] [CrossRef]


| Microbial Genera | GVHD (n = 12) | Crohn’s Disease (CD), n = 15 | Control Group (n = 148) | GVHD vs. CD, p Values | GVHD vs. Control Group, p Values | CD vs. Control Group, p Values |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| Akkermansia | 0.90 ± 0.85 | 15.03 ± 6.52 | 2.11 ± 0.41 | 0.090 | 0.03 | 0.557 |
| Alistipes | 0.03 ± 0.02 | 0.43 ± 0.23 | 3.17 ± 0.23 | 0.50 | <0.001 | <0.001 |
| Bacteroides | 2.77 ± 1.77 | 2.04 ± 1.24 | 23.79 ± 1.07 | 0.497 | <0.001 | <0.001 |
| Bifidobacterium | 0.07 ± 0.04 | 8.41 ± 5.24 | 1.95 ± 0.27 | 0.044 | <0.001 | 0.130 |
| Blautia | 0.19 ± 0.16 | 0.13 ± 0.10 | 0.69 ± 0.07 | 0.792 | <0.001 | <0.001 |
| Clostridium XIVa | 3.71 ± 3.36 | 0.27 ± 0.14 | 0.08 ± 0.02 | 0.649 | 0.775 | 0.832 |
| Collinsella | 1.28 ± 1.27 | 0.71 ± 0.71 | 0.78 ± 0.10 | 0.820 | <0.001 | <0.001 |
| Desulfovibrio | 0.00 | 1.67 ± 1.66 | 0.16 ± 0.06 | 0.180 | 0.021 | 0.193 |
| Dialister | 0.10 ± 0.07 | 3.75 ± 1.91 | 1.53 ± 0.20 | 0.026 | 0.007 | 0.451 |
| Dorea | 0.12 ± 0.11 | 0.60 ± 0.39 | 0.48 ± 0.05 | 0.261 | <0.001 | 0.017 |
| Enterobacter | 3.040 ± 3.04 | 0.002 ± 0.002 | 0.110 ± 0.073 | 0.877 | 0.363 | 0.233 |
| Enterococcus | 8.560 ± 4.88 | 0.07 ± 0.06 | 0.040 ± 0.014 | 0.007 | <0.001 | 0.739 |
| E. coli/Shigella | 12.04 ± 5.97 | 5.29 ± 2.94 | 0.63 ± 0.12 | 0.722 | 0.535 | 0.561 |
| Faecalibacterium | 3.00 ± 2.25 | 16.92 ± 6.21 | 6.09 ± 0.29 | 0.203 | <0.001 | 0.764 |
| Fusobacterium | 1.43 ± 1.41 | 4.76 ± 2.73 | 0.18 ± 0.13 | 0.331 | 0.255 | 0.003 |
| Haemophilus | 1.16 ± 1.15 | 0.12 ± 0.11 | 0.16 ± 0.06 | 0.862 | 0.208 | 0.144 |
| Klebsiella | 30.19 ± 9.54 | 12.10 ± 6.75 | 0.12 ± 0.05 | 0.285 | <0.001 | 0.001 |
| Lachnospiracea | 2.55 ± 1.78 | 0.47 ± 0.26 | 0.12 ± 0.05 | 0.415 | 0.035 | <0.001 |
| Lactobacillus | 0.06 ± 0.03 | 1.86 ± 1.66 | 0.22 ± 0.16 | 0.412 | 0.028 | <0.001 |
| Parabacteroides | 1.28 ± 1.12 | 2.09 ± 1.95 | 2.53 ± 0.18 | 0.880 | <0.001 | <0.001 |
| Phascolarctobacter | 0.02 ± 0.02 | 0.01 ± 0.01 | 2.57 ± 0.23 | 0.893 | <0.001 | <0.001 |
| Prevotella | 0.06 ± 0.02 | 4.18 ± 2.66 | 7.28 ± 0.91 | 0.686 | 0.004 | 0.148 |
| Romboutsia | 0.40 ± 0.40 | 0.84 ± 0.66 | 0.10 ± 0.02 | 0.175 | 0.002 | 0.276 |
| Roseburia | 0.13 ± 0.09 | 0.33 ± 0.19 | 1.19 ± 0.12 | 0.331 | <0.001 | <0.001 |
| Ruminococcus | 0.02 ± 0.01 | 0.71 ± 0.68 | 1.73 ± 0.21 | 0.284 | <0.001 | <0.001 |
| Staphylococcus | 2.90 ± 2.90 | 0.01 ± 0.004 | 0.005 ± 0.004 | 0.879 | 0.008 | 0.165 |
| Sutterella | 0.01 ± 0.01 | 0.31 ± 0.31 | 1.08 ± 0.11 | 0.959 | <0.001 | <0.001 |
| Veillonella | 3.17 ± 1.65 | 4.95 ± 2.50 | 0.09 ± 0.03 | 0.701 | 0.090 | 0.489 |
| Viral Genes: Classes | GVHD (% of Total DNA) | Crohn’s Disease (% of Total DNA) | p Values |
|---|---|---|---|
| Caudoviricetes | 86.59 + 6.19 | 87.09 + 7.83 | 0.524 |
| Herviviricetes | 0.37 + 0.37 | 4.96 + 3.87 | 0.728 |
| Megaviricetes | 1.51 + 1.15 | 0.39 + 0.32 | 0.164 |
| Pokkesviricetes | 8.89 + 4.16 | 7.45 + 6.58 | 0.236 |
| Tectiliviricetes | 0.00 + 0.00 | 0.026 + 0.025 | 0.143 |
| Viral genes: Genera | |||
| Bievrevirus | 1.91 + 1.87 | 6.17 + 5.26 | 0.197 |
| Brunovirus | 2.01 + 1.16 | 0.39 + 0.34 | 0.411 |
| Carjivirus | 0.11 + 0.11 | 0.68 + 0.59 | 0.330 |
| Cytomegalovirus | 0.37+ 0.37 | 4.96 + 3.78 | 0.728 |
| Eganvirus | 16.98 + 14.05 | 17.73 + 11.33 | 0.854 |
| Irrigatiovirus | 1.73 + 1.73 | 0.068 + 0.058 | 0.944 |
| Jouyvirus | 0.02 + 0.02 | 0.35 + 0.35 | 0.804 |
| Lambdavirus | 1.07 + 0.68 | 12.33 + 7.91 | 0.433 |
| Lederbergvirus | 27.17 + 17.36 | 31.73 + 11.70 | 0.586 |
| Lymphoc dis virus | 1.51 + 1.15 | 0.39 + 0.32 | 0.164 |
| Mastadenovirus | 0.00 + 0.00 | 0.026 + 0.023 | 0.143 |
| Novemvirus | 1.74 + 1.74 | 2.98 + 2.57 | 0.576 |
| Oryzopoxvirus | 8.89 + 4.16 | 7.45 + 6.58 | 0.236 |
| Senquatrovirus | 0.037 + 0.037 | 0.089 + 0.065 | 0.576 |
| Shamshuipovirus | 0.042 + 0.042 | 0.077 + 0.052 | 0.340 |
| Tequatrovirus | 4.41 + 3.18 | 3.36 + 3.17 | 0.553 |
| Traversvirus | 0.19 + 0.09 | 0.79 + 0.24 | 0.026 |
| Wanchaivirus | 0.11 + 0.10 | 0.37 + 0.29 | 0.503 |
| Biodiversity Indexes | GVHD | Crohn’s Disease | p Values |
|---|---|---|---|
| Shannon index | 1.45 + 0.35 | 1.10 + 0.15 | 0.615 |
| Simpson index | 0.60 + 0.10 | 0.49 + 0.07 | 0.366 |
| Chao 1 | 25.26 + 7.26 | 13.57 + 1.80 | 0.085 |
| Se_chao 1 | 7.51 + 1.81 | 3.32 + 0.87 | 0.111 |
| GVHD, All Time Points (n = 41) | Crohn’s Disease, All Time Points (n = 55) | |||
|---|---|---|---|---|
| r | p | r | p | |
| Eganvirus EtG | −0.521 | 2.4 × 10−4 | −0.018 | 0.450 |
| Eganvirus ev186 | −0.527 | 2.0 × 10−4 | −0.018 | 0.450 |
| Eganvirus PsP3 | −0.535 | 1.6 × 10−4 | 0.073 | 0.298 |
| Eganvirus SEN1 | −0.511 | 3.2 × 10−4 | −0.106 | 0.221 |
| Salmonella phage BIS20 | −0.553 | 9.0 × 10−5 | 0.063 | 0.324 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chukhlovin, A.B.; Goloshchapov, O.V.; Shchukina, O.B.; Kharitidis, A.M.; Zhloba, A.A.; Subbotina, T.F.; Kusakin, A.V.; Kosarev, O.V.; Tsai, V.V.; Kalinin, R.S.; et al. Changes in Gut Phageome and Bacteriome Following Fecal Microbiota Transfer in Patients with Intestinal Graft-Versus-Host Disease and Crohn’s Disease. Microorganisms 2025, 13, 2337. https://doi.org/10.3390/microorganisms13102337
Chukhlovin AB, Goloshchapov OV, Shchukina OB, Kharitidis AM, Zhloba AA, Subbotina TF, Kusakin AV, Kosarev OV, Tsai VV, Kalinin RS, et al. Changes in Gut Phageome and Bacteriome Following Fecal Microbiota Transfer in Patients with Intestinal Graft-Versus-Host Disease and Crohn’s Disease. Microorganisms. 2025; 13(10):2337. https://doi.org/10.3390/microorganisms13102337
Chicago/Turabian StyleChukhlovin, Alexei B., Oleg V. Goloshchapov, Oksana B. Shchukina, Aleksandra M. Kharitidis, Alexander A. Zhloba, Tatiana F. Subbotina, Aleksey V. Kusakin, Oleg V. Kosarev, Viktoria V. Tsai, Roman S. Kalinin, and et al. 2025. "Changes in Gut Phageome and Bacteriome Following Fecal Microbiota Transfer in Patients with Intestinal Graft-Versus-Host Disease and Crohn’s Disease" Microorganisms 13, no. 10: 2337. https://doi.org/10.3390/microorganisms13102337
APA StyleChukhlovin, A. B., Goloshchapov, O. V., Shchukina, O. B., Kharitidis, A. M., Zhloba, A. A., Subbotina, T. F., Kusakin, A. V., Kosarev, O. V., Tsai, V. V., Kalinin, R. S., Eismont, Y. A., & Glotov, O. S. (2025). Changes in Gut Phageome and Bacteriome Following Fecal Microbiota Transfer in Patients with Intestinal Graft-Versus-Host Disease and Crohn’s Disease. Microorganisms, 13(10), 2337. https://doi.org/10.3390/microorganisms13102337






