Microbial Role in Straw Organic Matter Depolymerization to Dissolved Organic Nitrogen Under Nitrogen Fertilizer Reduction in Coastal Saline Paddy Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description
2.2. Experimental Design and Soil Sampling
2.3. Measurements and Methods
2.3.1. Analysis of Straw Properties
2.3.2. Straw N Hydrolase Activity Analysis
2.3.3. DNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.3.4. High-Throughput Sequencing and Bioinformatics Analyses
2.4. Statistical Analysis
3. Results
3.1. Changes in Straw Properties Under Different Decomposition Stages and N Application Rates
3.2. Changes in Straw apr and chiA Gene Levels, α-Diversity, and Enzyme Activities Under Varying Decomposition Stages and N Application Rates
3.3. Key Factors Regulating Straw N Hydrolase Enzymes
3.4. Changes in the Community Structures of Bacteria Involved in Straw Alkaline Protein and Chitin Degradation Under Different Decomposition Stages and N Application Rates
3.5. Co-Occurrence Network of Straw N-Mineralization Bacteria
3.6. PLS-PM Analysis
4. Discussion
4.1. Moderate N Input Enhances Microbial Cooperation and Enzyme Activities
4.2. Keystone Taxa Drive N Depolymerization at Different Decomposition Stages
4.3. Straw Properties and Temperature Are Primary Regulators of Enzyme-Mediated DON Production
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DON | Dissolved organic nitrogen |
| TC | Total carbon |
| TN | Total nitrogen |
| IN | Inorganic nitrogen |
| TP | Total phosphorus |
| TK | Total potassium |
| C/N | TC to TN ratio |
| EC | Electrical conductivity |
| AP | Alkaline protease |
| CAS | Chitinase |
| LAP | L-leucine aminopeptidase |
| NAG | N-acetylglucosaminidase |
| ACC | Average clustering coefficient |
| APL | Average path length |
| ND | Network density |
References
- Zhao, Y.; Wang, L.; Zhao, H.-L.; Chen, X.-B. Research status and prospects of saline-alkali land amelioration in the coastal region of China. Chin. Agric. Sci. Bull. 2022, 38, 67–74. [Google Scholar]
- Li, H.-Q.; Yao, R.-J.; Yang, J.-S.; Wang, X.-P.; Zheng, F.-L.; Chen, Q.; Xie, W.-P.; Zhang, X. Influencing mechanism of soil salinization on nitrogen transformation processes and efficiency improving methods for high efficient utilization of nitrogen in salinized farmland. Chin. J. Appl. Ecol. 2020, 31, 3915–3924. [Google Scholar]
- Dai, X.; Song, D.; Zhou, W.; Liu, G.; Liang, G.; He, P.; Sun, G.; Yuan, F.; Liu, Z.; Yao, Y.; et al. Partial substitution of chemical N with organic N improves rice yield, soil biochemical indictors and microbial composition in a double rice cropping system in south China. Soil Tillage Res. 2021, 205, 104753. [Google Scholar] [CrossRef]
- Zhou, W.; Ai, C.; Yi, K.-K. Research focus of plant nutrition and fertilizer science in new stage. J. Plant Nutr. Fertil. 2024, 30, 1243–1252. [Google Scholar]
- Ding, W.; Xu, X.; He, P.; Ullah, S.; Zhang, J.; Cui, Z.; Zhou, W. Improving yield and N use efficiency through alternative fertilization options for rice in China: A meta-analysis. Field Crops Res. 2018, 227, 11–18. [Google Scholar] [CrossRef]
- Chen, K.; Ma, T.; Ding, J.; Yu Se Dai, Y.; He, P.; Ma, T. Effects of straw return with N Fertilizer reduction on rice (Oryza sativa L.) morphology, photosynthetic capacity, yield and water–N use efficiency traits under different water regimes. Agronomy 2023, 13, 133. [Google Scholar] [CrossRef]
- Li, T.; He, C.-E.; Ge, X.-Y.; Ou, Y.-Z. Responses of soil mineral N contents, enzyme activities and crop yield to different C/N ratio mediated by straw retention and N fertilization. Chin. J. Ecol. Agric. 2016, 24, 1633–1642. [Google Scholar]
- Farzadfar, S.; Knight, J.D.; Congreves, K.A. Soil organic nitrogen: An overlooked but potentially significant contribution to crop nutrition. Plant Soil 2021, 462, 7–23. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, J.; Bai, S.H.; Teng, Y.; Xu, Z. Evaluating the effects of phytoremediation with biochar additions on soil nitrogen mineralization enzymes and fungi. Environ. Sci. Pollut. Res. 2018, 25, 23106–23116. [Google Scholar] [CrossRef]
- Kurth, J.K.; Albrecht, M.; Karsten, U.; Glaser, K.; Schloter, M.; Schulz, S. Correlation of the abundance of bacteria catalyzing phosphorus and N turnover in biological soil crusts of temperate forests of Germany. Biol. Fertil. Soils 2021, 57, 179–192. [Google Scholar] [CrossRef]
- Gianfreda, L.; Rao, M.A. Potential of extra cellular enzymes in remediation of polluted soils: A review. Enzyme Microb. Technol. 2004, 35, 339–354. [Google Scholar] [CrossRef]
- Bach, H.J.; Hartmann, A.; Schloter, M.; Munch, J.C. PCR primers and functional probes for amplification and detection of bacterial genes for extracellular peptidases in single strains and in soil. J. Microbiol. Methods 2001, 44, 173–182. [Google Scholar] [CrossRef]
- Baraniya, D.; Puglisi, E.; Ceccherini, M.T.; Pietramellara, G.; Giagnoni, L.; Arenella, M.; Nannipieri, P.; Renella, G. Protease encoding microbial communities and protease activity of the rhizosphere and bulk soils of two maize lines with different N uptake efficiency. Soil Biol. Biochem. 2016, 96, 176–179. [Google Scholar] [CrossRef]
- Kumar, C.G.; Takagi, H. Microbial alkaline proteases: From a bioindustrial viewpoint. Biotechnol. Adv. 1999, 17, 561–594. [Google Scholar] [CrossRef]
- Mokashe, N.; Chaudhari, A.; Patil, U. Optimal production and characterization of alkaline protease from newly isolated halotolerant Jeotgalicoccus sp. Biocatal. Agric. Biotechnol. 2015, 4, 235–243. [Google Scholar] [CrossRef]
- Xiao, X.; Yin, X.; Lin, J.; Sun, L.; You, Z.; Wang, P.; Wang, F. Chitinase genes in lake sediments of Ardley Island, Antarctica. Appl. Environ. Microbiol. 2005, 71, 7904–7909. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Lu, Z.; Jie, W.; Xiang-tao, W.; Li-rong, L.; Qian, W.; Guo-bin, L.; Chao, Z. Effect of restoration types on the community structure of microbes harboring nifH and chiA genes in alpine meadow. Chin. J. Appl. Ecol. 2021, 32, 4349–4358. [Google Scholar]
- Ali, S.; Liu, K.; Ahmed, W.; Jing, H.; Qaswar, M.; Kofi Anthonio, C.; Maitlo, A.A.; Lu, Z.; Liu, L.; Zhang, H. Nitrogen mineralization, soil microbial biomass and extracellular enzyme activities regulated by long-term N fertilizer inputs: A comparison study from upland and paddy soils in a red soil region of China. Agronomy 2021, 11, 2057. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Microbial community structure and its functional implications. Nature 2009, 459, 193–199. [Google Scholar] [CrossRef]
- Jia, M.; Gao, Z.; Gu, H.; Zhao, C.; Liu, M.; Liu, F.; Xie, L.; Wang, L.; Zhang, G.; Liu, Y.; et al. Effects of precipitation change and N addition on the composition, diversity, and molecular ecological network of soil bacterial communities in a desert steppe. PLoS ONE 2021, 16, e0248194. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G.A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Song, D.; Guo, Q.; Zhou, W.; Liu, G.; Ma, R.; Liang, G.; He, P.; Sun, G.; Yuan, F.; et al. Predicting the influence of fertilization regimes on potential N fixation through their effect on free-living diazotrophic community structure in double rice cropping systems. Soil Biol. Biochem. 2021, 156, 108220. [Google Scholar] [CrossRef]
- Wang, E.; Lin, X.; Tian, L.; Wang, X.; Ji, L.; Jin, F.; Tian, C. Effects of short-term rice straw return on the soil microbial community. Agriculture 2021, 11, 561. [Google Scholar] [CrossRef]
- Wang, F.; Liu, H.; Yao, H.; Zhang, B.; Li, Y.; Jin, S.; Cao, H. Reducing Application of N fertilizer increases soil bacterial diversity and drives co-occurrence networks. Microorganisms 2024, 12, 1434. [Google Scholar] [CrossRef]
- IUSS Working Group WRB. World Reference Base for Soil Resources. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps, 4th ed.; International Union of Soil Sciences (IUSS): Vienna, Austria, 2022. [Google Scholar]
- Lai, J.; Zou, Y.; Zhang, J.; Peres-Neto, P.R. Generalizing hierarchical and variation partitioning in multiple regression and canonical analyses using the rdacca.hp R package. Methods Ecol. Evol. 2022, 13, 782–788. [Google Scholar] [CrossRef]
- Statnikov, A.; Henaff, M.; Narendra, V.; Konganti, K.; Li, Z.; Yang, L.; Pei, Z.; Blaser, M.J.; Aliferis, C.F.; Alekseyenko, A.V. A comprehensive evaluation of multicategory classification methods for microbiomic data. Microbiome 2013, 1, 11. [Google Scholar] [CrossRef]
- Mai, Y.; Bu, R.; HanShang Lei, Z.; Li, M.; Wang, H.; Cheng, W.; Tang, S.; Wu, J.; Zhu, L. Effects of adding different exogenous N on rice straw decomposition and nutrient release. Trans. Chin. Soc. Agric. Eng. 2021, 37, 210–219. [Google Scholar]
- Ullah, S.; Ai, C.; Huang, S.; Zhang, J.; Jia, L.; Ma, J.; Zhou, W.; He, P. The responses of extracellular enzyme activities and microbial community composition under N addition in an upland soil. PLoS ONE 2019, 14, e0223026. [Google Scholar] [CrossRef]
- Frankenberger, W.T., Jr.; Bingham, F.T. Influence of salinity on soil enzyme activities. Soil Sci. Soc. Am. J. 2022, 46, 1173–1177. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R. Regulation of extracellular protease activity in soil in response to different sources and concentrations of N and carbon. Soil Biol. Biochem. 2008, 40, 3040–3048. [Google Scholar] [CrossRef]
- Cui, Y.; Moorhead, D.L.; Guo, X.; Peng, S.; Wang, Y.; Zhang, X.; Fang, L. Stoichiometric models of microbial metabolic limitation in soil systems. Glob. Ecol. Biogeogr. 2021, 30, 2297–2311. [Google Scholar] [CrossRef]
- Hao, C.; Dungait, J.A.J.; Wei, X.; Ge, T.; Kuzyakov, Y.; Cui, Z.; Tian, J.; Zhang, F. Maize root exudate composition alters rhizosphere bacterial community to control hotspots of hydrolase activity in response to N supply. Soil Biol. Biochem. 2022, 170, 108717. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Deng, Y.; Peng, Y. Long-term ammonium nitrate addition strengthens soil microbial cross-trophic interactions in a Tibetan alpine steppe. Ecology 2025, 106, e70057. [Google Scholar] [CrossRef]
- Philippot, L.; Griffiths, B.S.; Langenheder, S. Microbial Community Resilience across Ecosystems and Multiple Disturbances. Microbiol. Mol. Biol. Rev. 2021, 85, e00026-20. [Google Scholar] [CrossRef]
- Cornell, C.R.; Zhang, Y.; Ning, D.; Xiao, N.; Wagle, P.; Xiao, X.; Zhou, J. Land use conversion increases network complexity and stability of soil microbial communities in a temperate grassland. ISME J. 2023, 17, 2210–2220. [Google Scholar] [CrossRef]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Yang, C.X.; Wang, T.; Gao, L.N.; Yin, H.J.; Lü, X. Isolation, identification and characterization of lignin-degrading bacteria from Qinling, China. J. Appl. Microbiol. 2017, 123, 1447–1460. [Google Scholar] [CrossRef]
- Nakbanpote, W.; Natthawoot, P.; Aphidech, S.; Narongrit, S.; Pawinee, S.; Pimthong, A. Salt-tolerant and plant growth-promoting bacteria isolated from Zn/Cd contaminated soil: Identification and effect on rice under saline conditions. J. Plant Interact. 2014, 9, 379–387. [Google Scholar] [CrossRef]
- Xing, Y.; Luan, C.; Zhang, Z.; Han, B.; Zhang, H.; Li, L.; Ruan, Y.; Zhang, J.; Jia, Z. Deciphering the driving force of straw-decomposing microbiomes in two native forest soils under biogeographically contrasting conditions. Acta Microbiol. Sin. 2024, 64, 2901–2917. [Google Scholar]
- Ninkuu, V.; Liu, Z.; Qin, A.; Xie, Y.; Song, X.; Sun, X. Impact of straw returning on soil ecology and crop yield: A review. Heliyon 2025, 11, e41651. [Google Scholar] [CrossRef] [PubMed]
- Reid, T.E.; Kavamura, V.N.; Abadie, M.; Torres-Ballesteros, A.; Pawlett, M.; Clark, I.M.; Harris, J.; Mauchline, T.H. Inorganic chemical fertilizer application to wheat reduces the abundance of putative plant growth-promoting rhizobacteria. Front. Microbiol. 2021, 12, 642587. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.-J.; Wu, X.-H.; Huang, L.-M.; Xie, W.-W.; Chen, Y.; El-Kassaby, Y.A.; Feng, J.-L. Soil bacteria to regulate phoebe bournei seedling growth and sustainable soil utilization under NPK fertilization. Plants 2021, 10, 1868. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.-D.; Cao, X.-J.; Wu, J.-Y.; Hou, N.; Gao, S.-H. Effects of fermentation time on fermentation quality and microbial community of oat silage. Feed Res. 2022, 45, 106–110. [Google Scholar]
- Tian, X.; Zhang, X.; Yang, G.; Wang, B.; Song, J.; Liu, Q.; Yu, F. Serratia plymuthica F-06 promotes the growth of maize seedlings under salt stress. Microbiol. China 2024, 51, 4545–4559. [Google Scholar]
- Chen, Z.; Zhang, X.; Li, P.; Li, K.; Wang, J. Research progress on functional endophytic bacteria for reducing organic contaminant risks in crops. Acta Microbiol. Sin. 2025, 65, 1885–1904. [Google Scholar]
- Grimont, F.; Grimont, P.A.D. Serratia. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–22. [Google Scholar]
- Luo, L.; Wan, L.; Chen, H.; Wen, C.; Xu, F.; Jia, W.; Nie, Y.; Yuan, H. Enrichment culturing and bacterial community structures analysis of a cold-adapted lignocellulose degrading microflora. J. Agric. Biotechnol. 2015, 23, 727–737. [Google Scholar]
- Zaim, S.; Bekkar, A.A. Advances in research on the use of Brevundimonas spp. to improve crop and soil fertility and for soil bioremediation. Alger. J. Biosci. 2023, 4, 045–051. [Google Scholar] [CrossRef]
- Friedrich, I.; Klassen, A.; Neubauer, H.; Schneider, D.; Hertel, R.; Daniel, R. Living in a Puddle of Mud: Isolation and Characterization of Two Novel Caulobacteraceae Strains Brevundimonas pondensis sp. nov. and Brevundimonas goettingensis sp. nov. Appl. Microbiol. 2021, 1, 38–59. [Google Scholar] [CrossRef]
- Xu, R.; Zhang, Y.; Li, Y.; Song, J.; Liang, Y.; Chen, F.; Wei, X.; Li, C.; Liu, W.; Rensing, C.; et al. Linking bacterial life strategies with the distribution pattern of antibiotic resistance genes in soil aggregates after straw addition. J. Hazard. Mater. 2024, 471, 134355. [Google Scholar] [CrossRef]
- Chernogor, L.; Bakhvalova, K.; Belikova, A.; Belikov, S. Isolation and properties of the bacterial strain Janthinobacterium sp. SLB01. Microorganisms 2022, 10, 1071. [Google Scholar] [CrossRef]
- Brady, C.; Asselin, J.A.; Beer, S.; Brurberg, M.B.; Crampton, B.; Venter, S.; Arnold, D.; Denman, S. Rahnella perminowiae sp. nov., Rahnella bonaserana sp. nov., Rahnella rivi sp. nov. and Rahnella ecdela sp. nov., isolated from diverse environmental sources, and emended description of the genus Rahnella. Int. J. Syst. Evol. Microbiol. 2022, 72, 005190. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yang, M.; Li, X.; Yang, F. The metaproteome of a cellulose degrading microbiot. J. Dalian Polytech. Univ. 2022, 41, 319–325. [Google Scholar]
- Yabe, S.; Sakai, Y.; Yokota, A. Thermosporothrix narukonensis sp. nov., belonging to the class Ktedonobacteria, isolated from fallen leaves on geothermal soil, and emended description of the genus Thermosporothrix. Int. J. Syst. Evol. Microbiol. 2016, 66, 2152–2157. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Revathi, K.; Khanna, S. Biodegradation of cellulosic and lignocellulosic waste by Pseudoxanthomonas sp R-28. Carbohydr. Polym. 2015, 134, 761–766. [Google Scholar] [CrossRef]
- Hou, L.; Jiang, J.; Jiang, J.; Xu, Z.; Liang, Z. Isolation of Pseudoxanthomonas sp. J1 and bioinformatics analysis of lignocellulose-degrading genes. J. Nanjing Agric. Univ. 2016, 39, 573–581. [Google Scholar]
- Liu, Y.; Zhou, L.; Yang, X.; Li, P.; Dai, J.; Xie, Y.; Wang, X.; Wang, Z.; Su, Z.; Feng, L. Lysobacter chinensis sp. nov., a cellulose-degrading strain isolated from cow dung compost. Antonie Leeuwenhoek 2022, 115, 1031–1040. [Google Scholar] [CrossRef]
- Zhou, S.-S.; Huang, Y.-L.; Huang, J.-Z.; Li, S.-R. Research progress in bioactive natural products from Lysobacter. Biotechnol. Bull. 2023, 39, 41–49. [Google Scholar]
- Yue, H.; Miller, A.L.; Khetrapal, V.; Jayaseker, V.; Wright, S.; Du, L. Biosynthesis, regulation, and engineering of natural products from Lysobacter. Nat. Prod. Rep. 2022, 39, 842–874. [Google Scholar] [CrossRef]
- Dong, Y.; Chen, R.; Petropoulos, E.; Yu, B.; Zhang, J.; Lin, X.; Gao, M.; Feng, Y. Interactive effects of salinity and SOM on the ecoenzymatic activities across coastal soils subjected to a saline gradient. Geoderma 2022, 406, 115519. [Google Scholar] [CrossRef]
- Sharma, S.; Gupta, N.; Chakkal, A.S.; Sharma, N.; Alamri, S.; Siddiqui, M.H.; Haider, F.U. Changes in Enzyme Activities in Salt-Affected Soils during Incubation Study of Diverse Particle Sizes of Rice Straw. Agriculture 2023, 13, 1694. [Google Scholar] [CrossRef]
- Roy, S.; Karapurkar, J.; Baidya, P.; Jose, M.; Bagchi, S. Community composition, and not species richness, of microbes influences decomposer functional diversity in soil. Soil Biol. Biochem. 2023, 187, 109225. [Google Scholar] [CrossRef]
- Stegen, J.C.; Bottos, E.M.; Jansson, J.K. A unified conceptual framework for prediction and control of microbiomes. Curr. Opin. Microbiol. 2018, 44, 20–27. [Google Scholar] [CrossRef]
- Wallenstein, M.; Allison, S.D.; Ernakovich, J.; Steinweg, J.M.; Sinsabaugh, R. Controls on the Temperature Sensitivity of Soil Enzymes: A Key Driver of In Situ Enzyme Activity Rates; Shukla, G., Varma, A., Eds.; Soil Enzymology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 245–258. [Google Scholar]
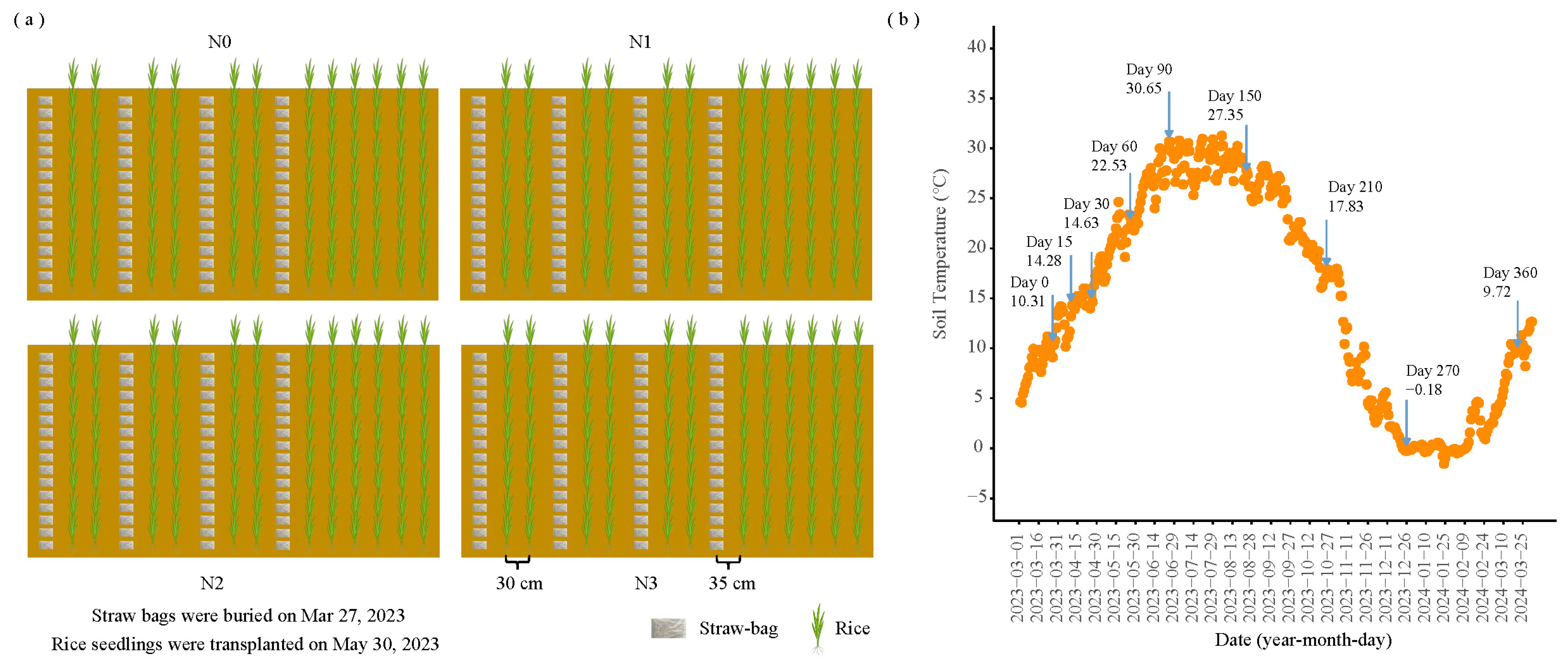
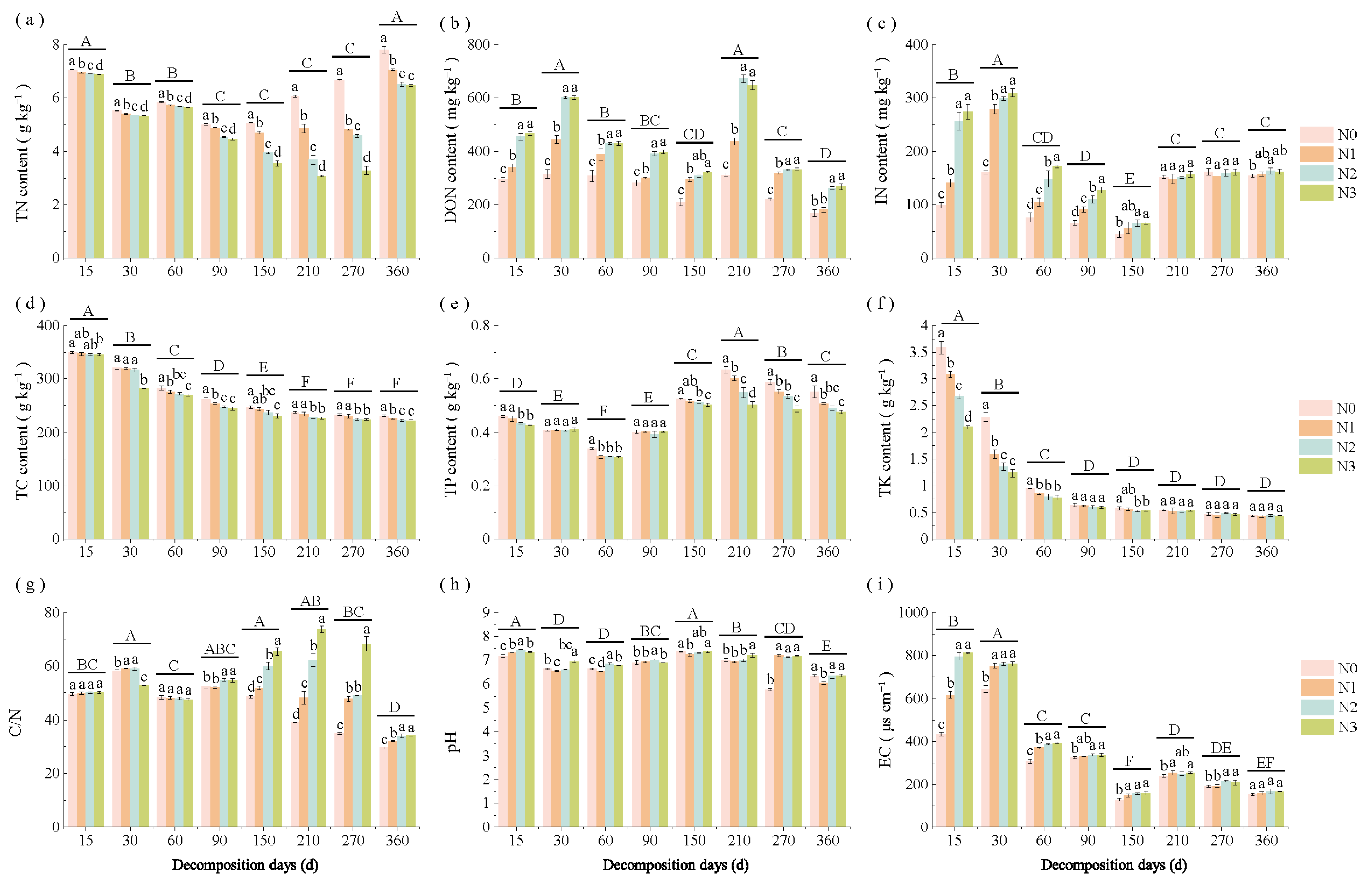
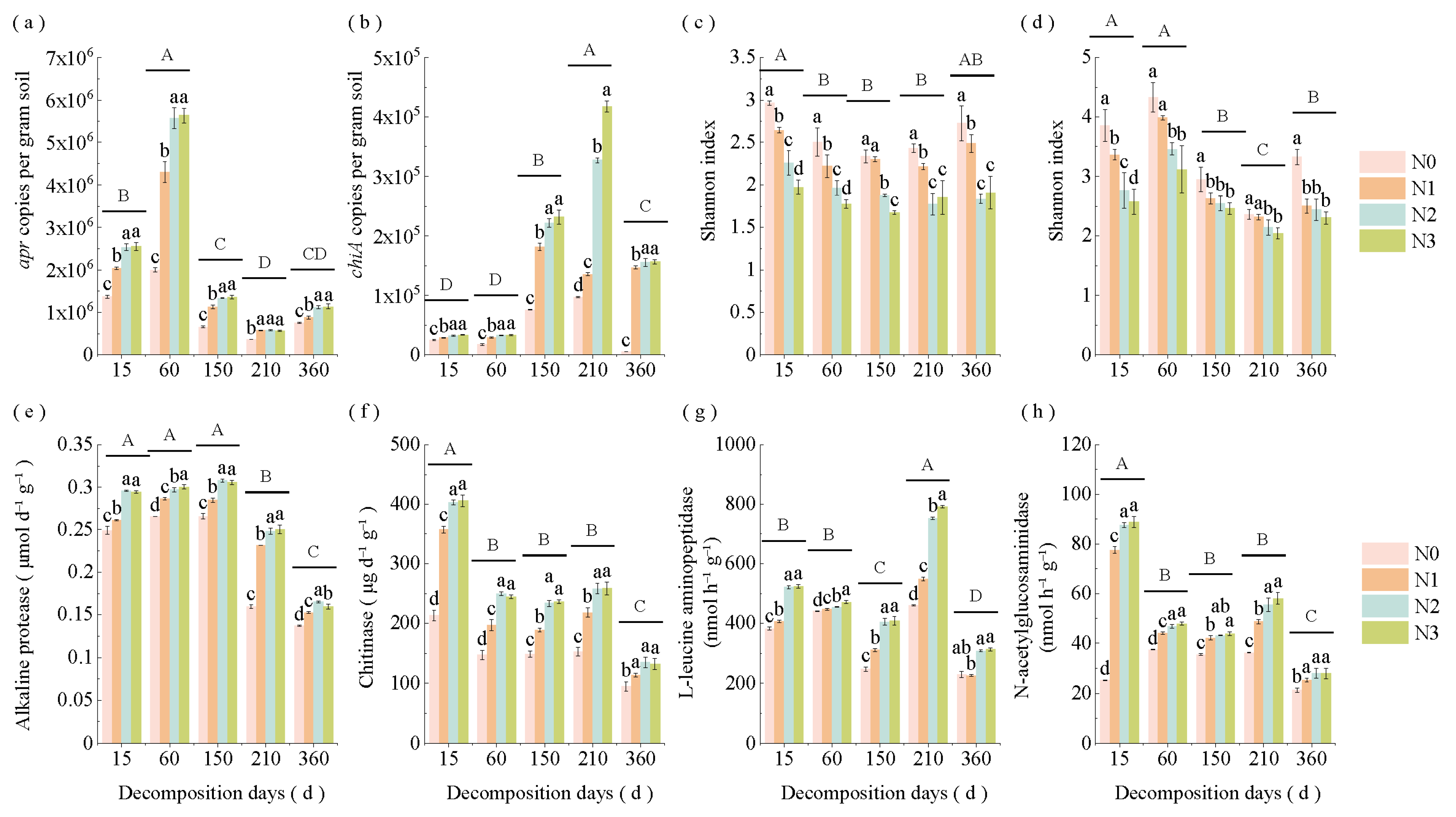
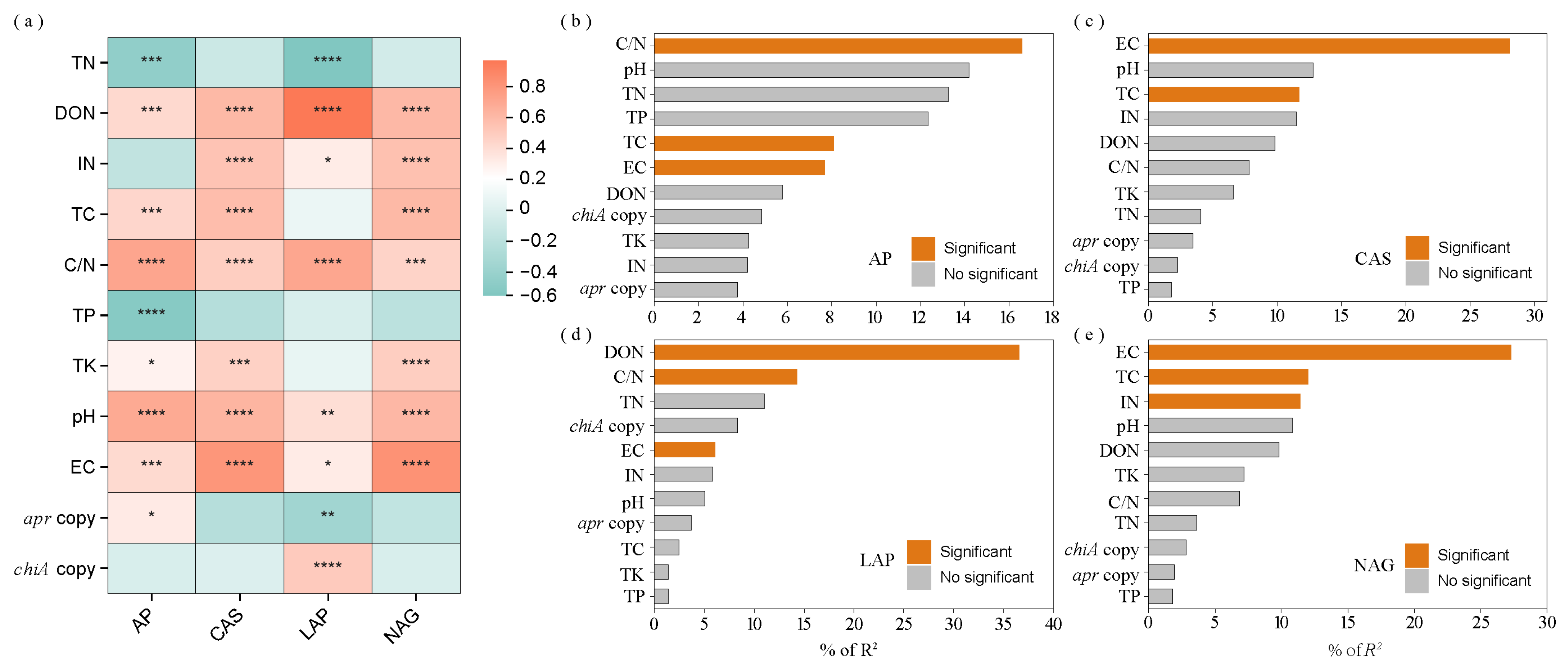
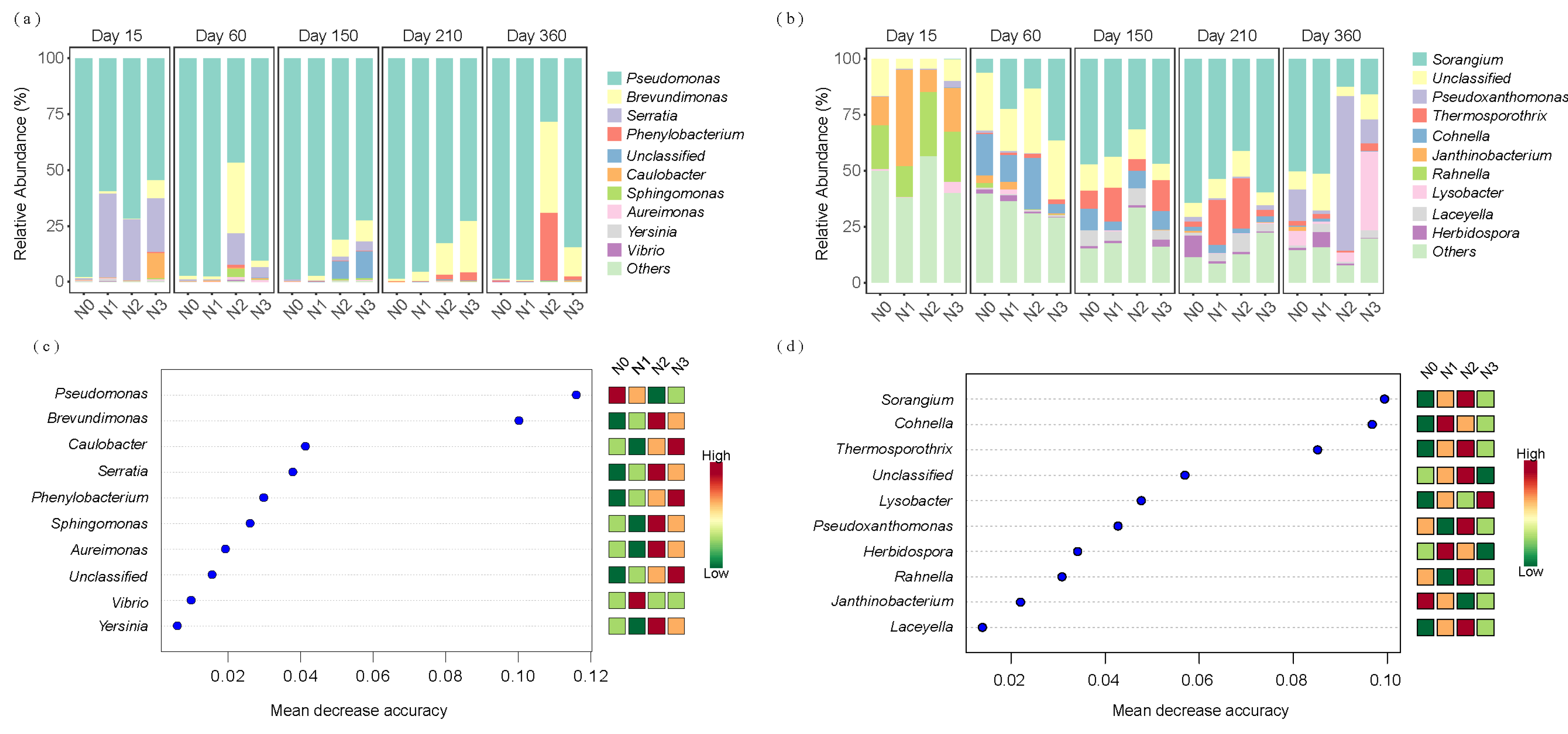
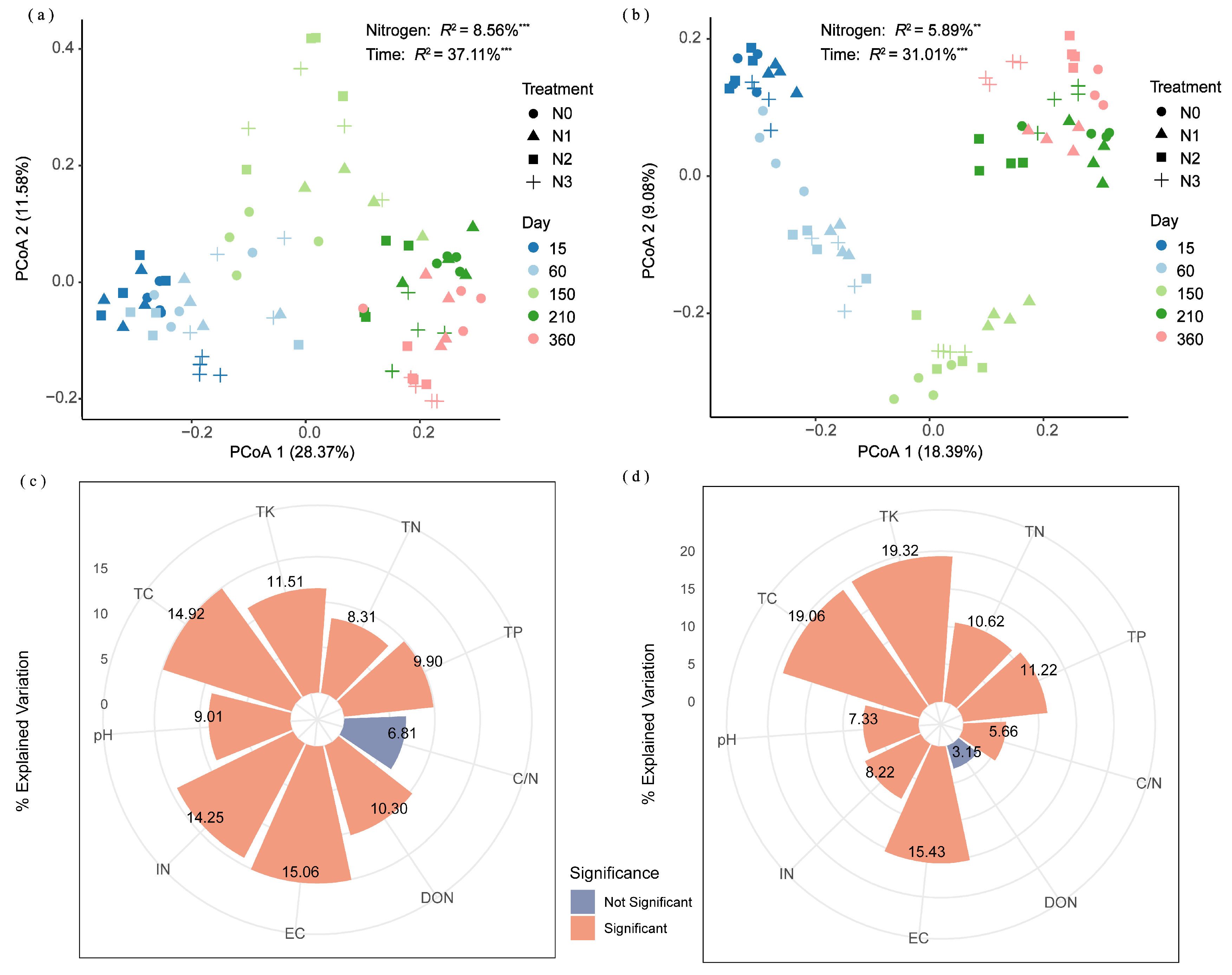
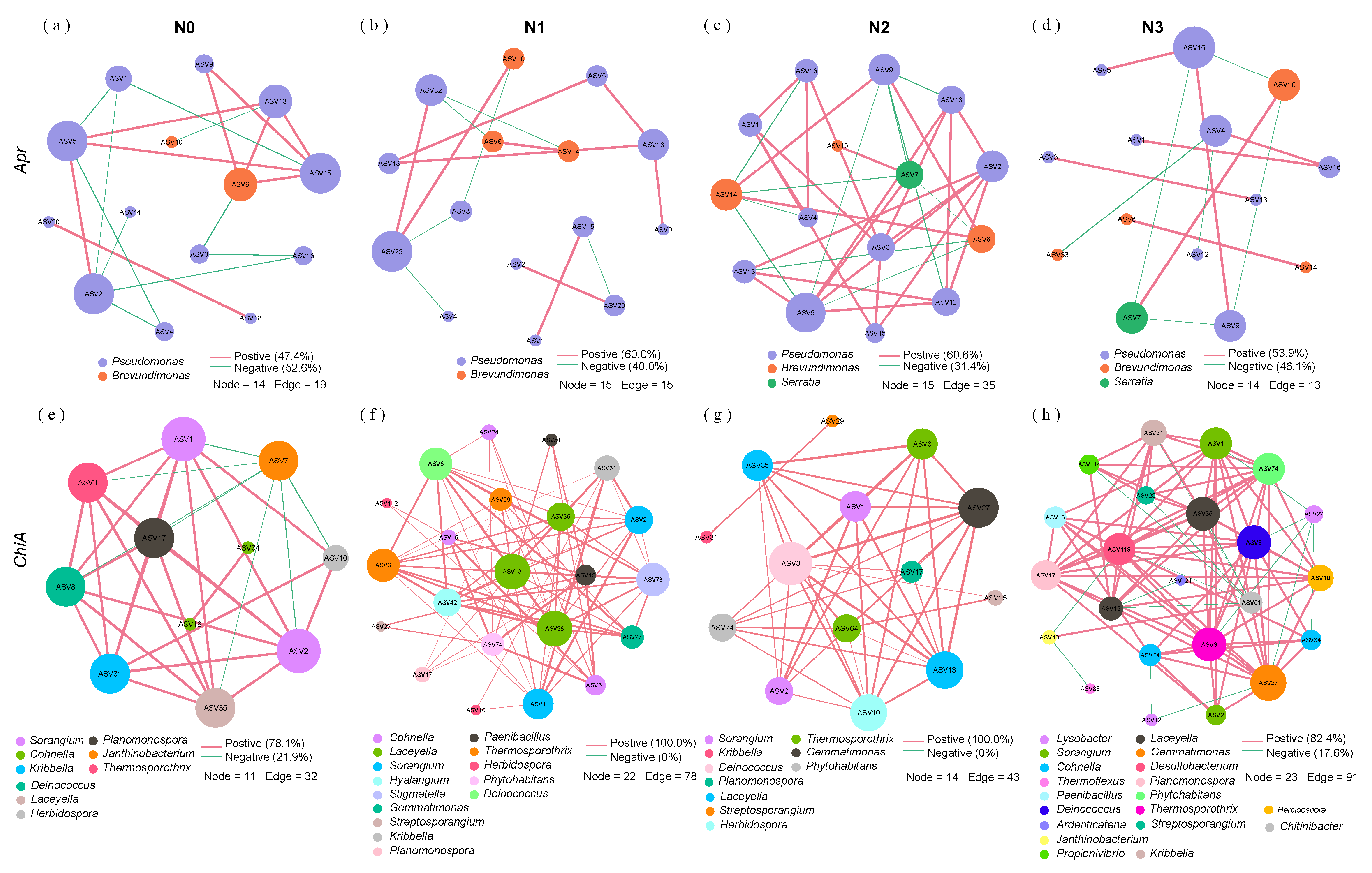
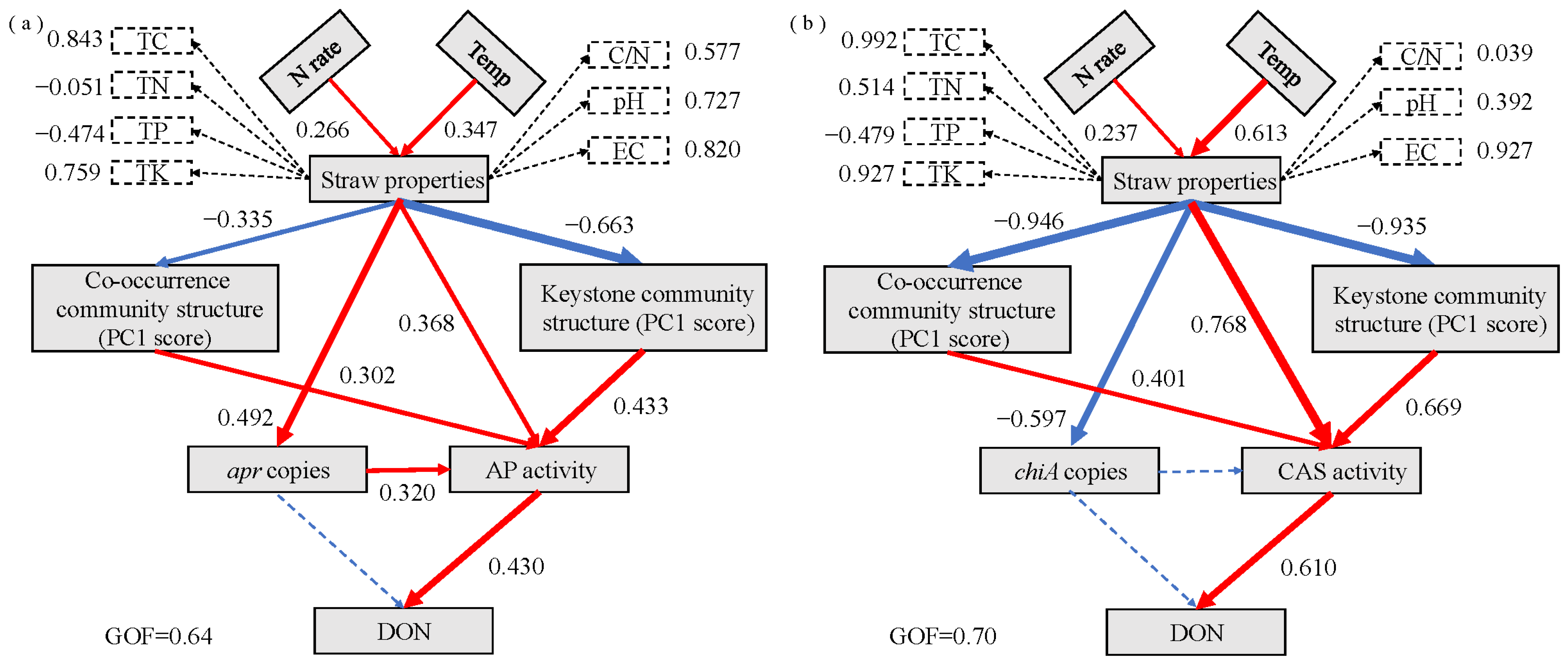
| Soil Type | Organic C (g/kg) | TN (g/kg) | Alkaline N (mg/kg) | Available P (mg/kg) | Available K (mg/kg) | pH | EC (μs/cm) | Soil Salt Content (g/kg) |
| Saline paddy soil | 7.09 | 0.84 | 34.6 | 15.4 | 243.2 | 8.13 | 365.8 | 1.17 |
| Straw Type | TC (g/kg) | TN (g/kg) | TP (g/kg) | TK (g/kg) | Cellulose (%) | Hemicellulose (%) | Lignin (%) | pH |
| Rice straw | 384.3 | 8.78 | 0.87 | 12.3 | 36.7 | 24.0 | 13.4 | 6.83 |
| Treatment | ACC | Change vs. N0 (%) | APL | Change vs. N0 (%) | ND | Change vs. N0 (%) | |
|---|---|---|---|---|---|---|---|
| Alkaline protein-degrading bacteria | N0 | 0.42 | - | 1.99 | - | 0.29 | - |
| N1 | 0.43 | 2.38 | 1.79 | −10.1 | 0.33 | 13.8 | |
| N2 | 0.68 | 61.9 | 1.58 | −20.6 | 0.49 | 69.0 | |
| N3 | 0.50 | 19.1 | 2.26 | 13.5 | 0.28 | −3.45 | |
| Chitin-degrading bacteria | N0 | 0.78 | - | 1.60 | - | 0.52 | - |
| N1 | 0.74 | −5.13 | 1.61 | 0.63 | 0.48 | −7.69 | |
| N2 | 0.83 | 6.41 | 1.35 | −15.6 | 0.68 | 30.8 | |
| N3 | 0.74 | −5.13 | 1.50 | −6.25 | 0.54 | 3.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, X.; Sun, J.; Li, H.; Zhao, Z.; Ma, R.; Liu, Y.; Shan, N.; Yao, Y.; Xue, Z. Microbial Role in Straw Organic Matter Depolymerization to Dissolved Organic Nitrogen Under Nitrogen Fertilizer Reduction in Coastal Saline Paddy Soil. Microorganisms 2025, 13, 2333. https://doi.org/10.3390/microorganisms13102333
Dai X, Sun J, Li H, Zhao Z, Ma R, Liu Y, Shan N, Yao Y, Xue Z. Microbial Role in Straw Organic Matter Depolymerization to Dissolved Organic Nitrogen Under Nitrogen Fertilizer Reduction in Coastal Saline Paddy Soil. Microorganisms. 2025; 13(10):2333. https://doi.org/10.3390/microorganisms13102333
Chicago/Turabian StyleDai, Xianglin, Jianping Sun, Hao Li, Zijing Zhao, Ruiping Ma, Yahui Liu, Nan Shan, Yutao Yao, and Zhizhong Xue. 2025. "Microbial Role in Straw Organic Matter Depolymerization to Dissolved Organic Nitrogen Under Nitrogen Fertilizer Reduction in Coastal Saline Paddy Soil" Microorganisms 13, no. 10: 2333. https://doi.org/10.3390/microorganisms13102333
APA StyleDai, X., Sun, J., Li, H., Zhao, Z., Ma, R., Liu, Y., Shan, N., Yao, Y., & Xue, Z. (2025). Microbial Role in Straw Organic Matter Depolymerization to Dissolved Organic Nitrogen Under Nitrogen Fertilizer Reduction in Coastal Saline Paddy Soil. Microorganisms, 13(10), 2333. https://doi.org/10.3390/microorganisms13102333







