Comprehensive Genomic and Phenotypic Characterization of Escherichia coli O78:H9 Strain HPVN24 Isolated from Diarrheic Poultry in Vietnam
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Hemolytic Activity
2.3. Antibiotic Susceptibility
2.4. DNA Extraction and Genome Sequencing
2.5. Genome Assembly
2.6. Genomic Analysis
2.6.1. Multilocus Sequence Typing and Serotyping
2.6.2. Genome Annotation and Visualization
2.6.3. Functional Genomic Analysis
2.6.4. Detection of Virulence and Antimicrobial Resistance Genes
2.6.5. Comparative Genomics and Phylogenetic Analysis
2.6.6. Pan-Genome and Cross-Strains Genetic Profiles Analyses
3. Results
3.1. Hemolytic Activity of the E. coli HPVN24 Strain
3.2. Antibiotic Resistance Profile
3.3. Genome Assembly and Annotation
3.4. Serotyping and Multilocus Sequence Typing
3.5. Functional Genomics Analysis
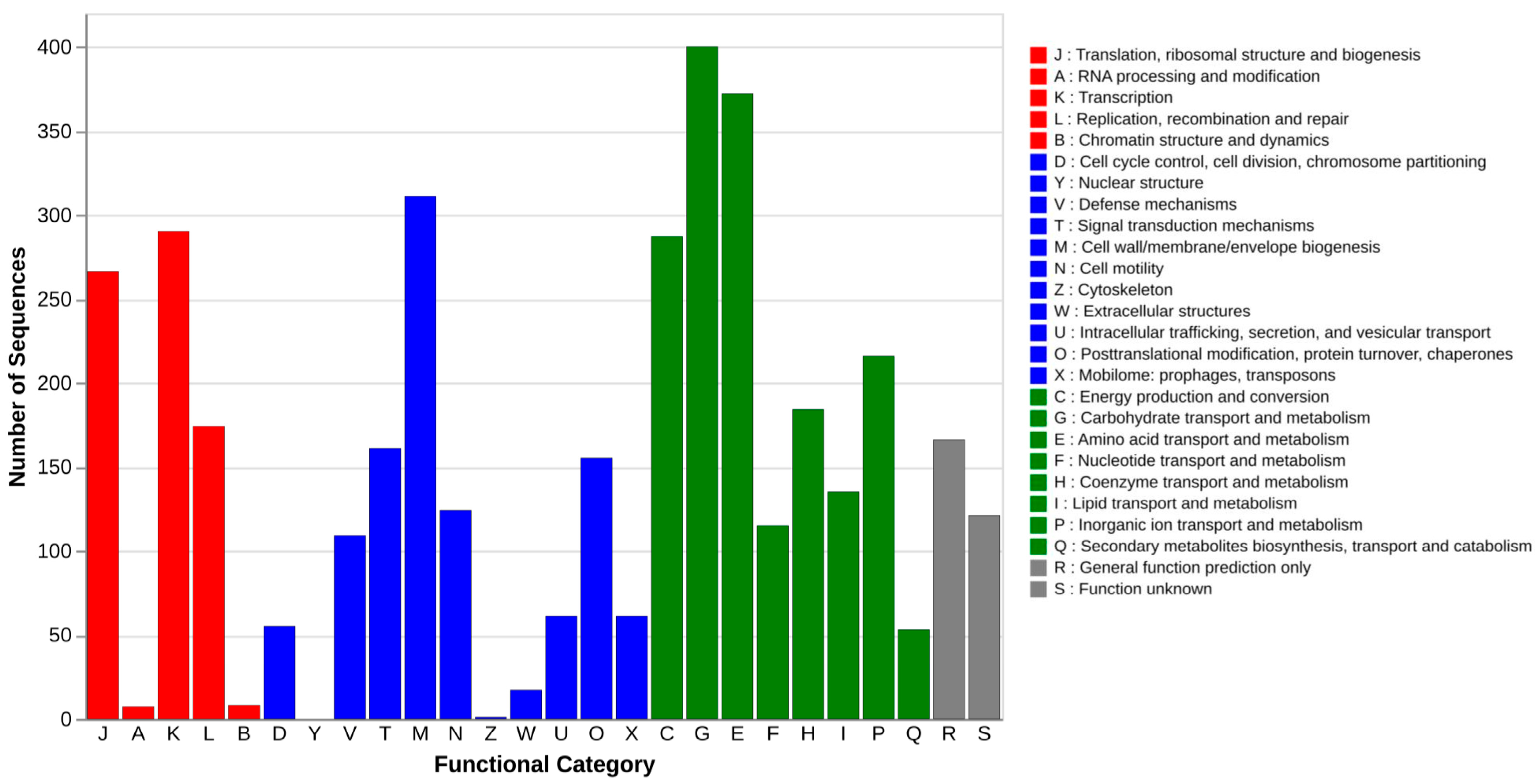
3.5.1. General Genomic Profiling Based on COG Classification
3.5.2. Biological Pathways Profiling with KEGG
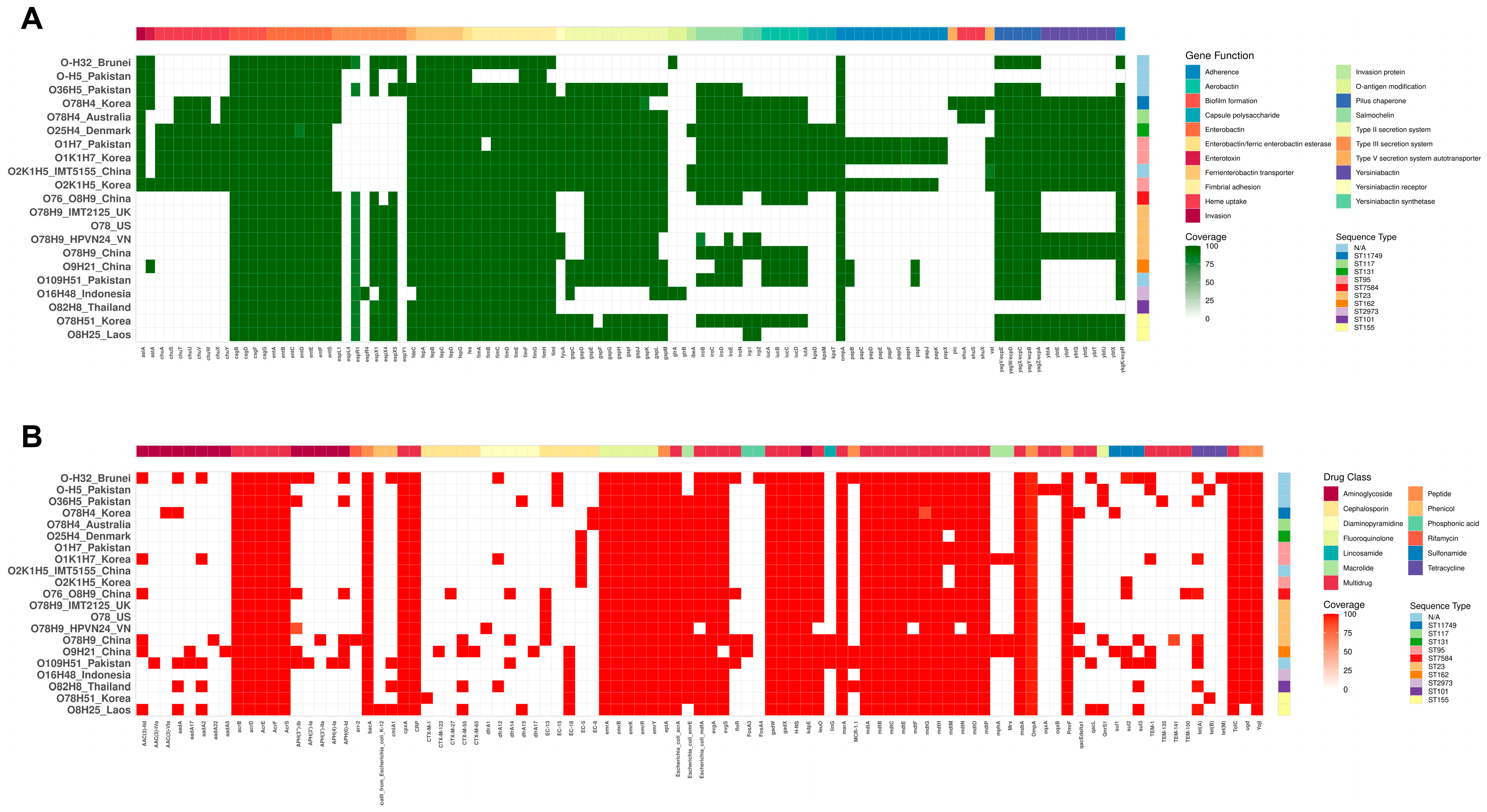
3.6. Virulence Genes
3.7. Antibiotic Resistance Genes
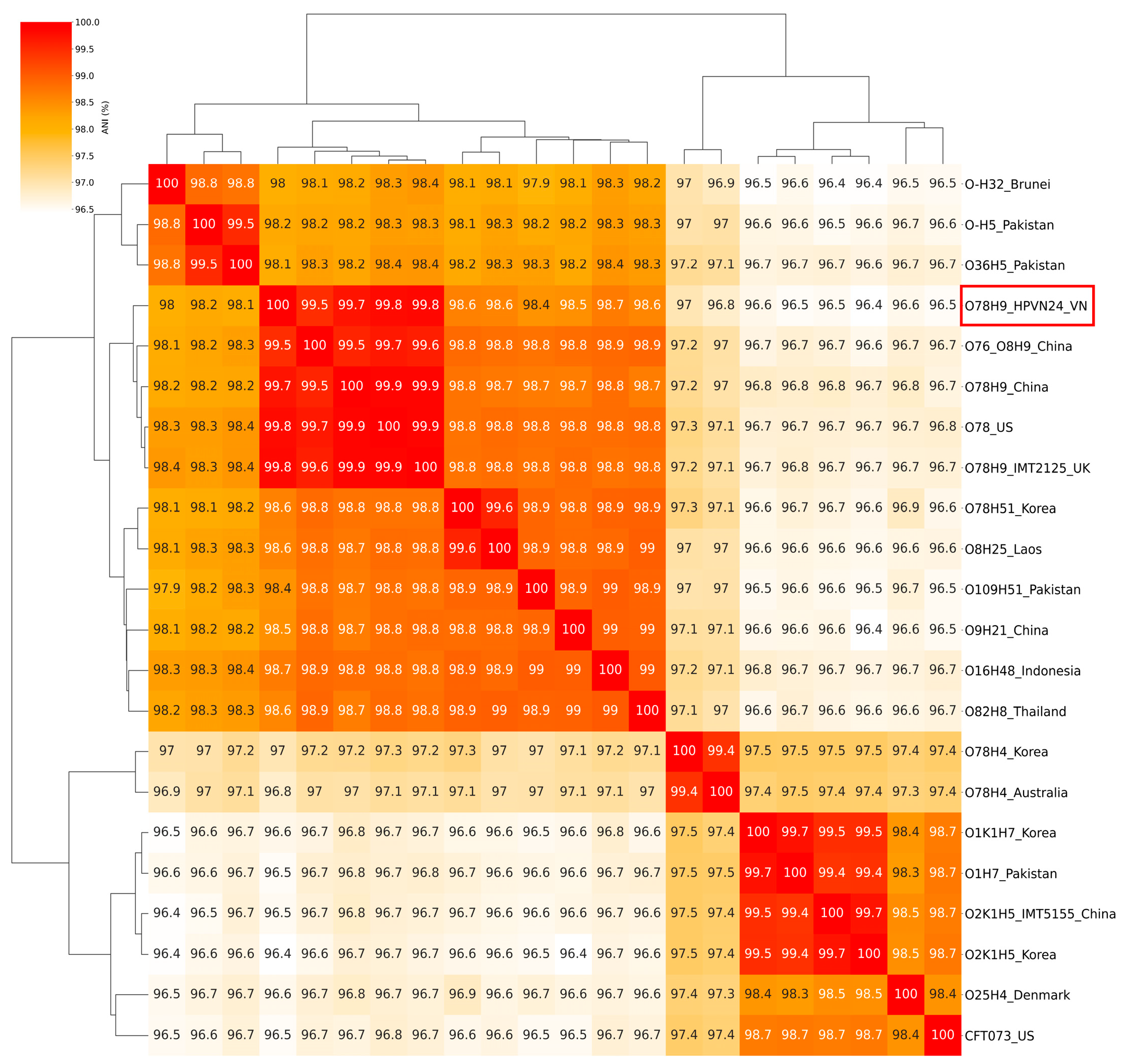
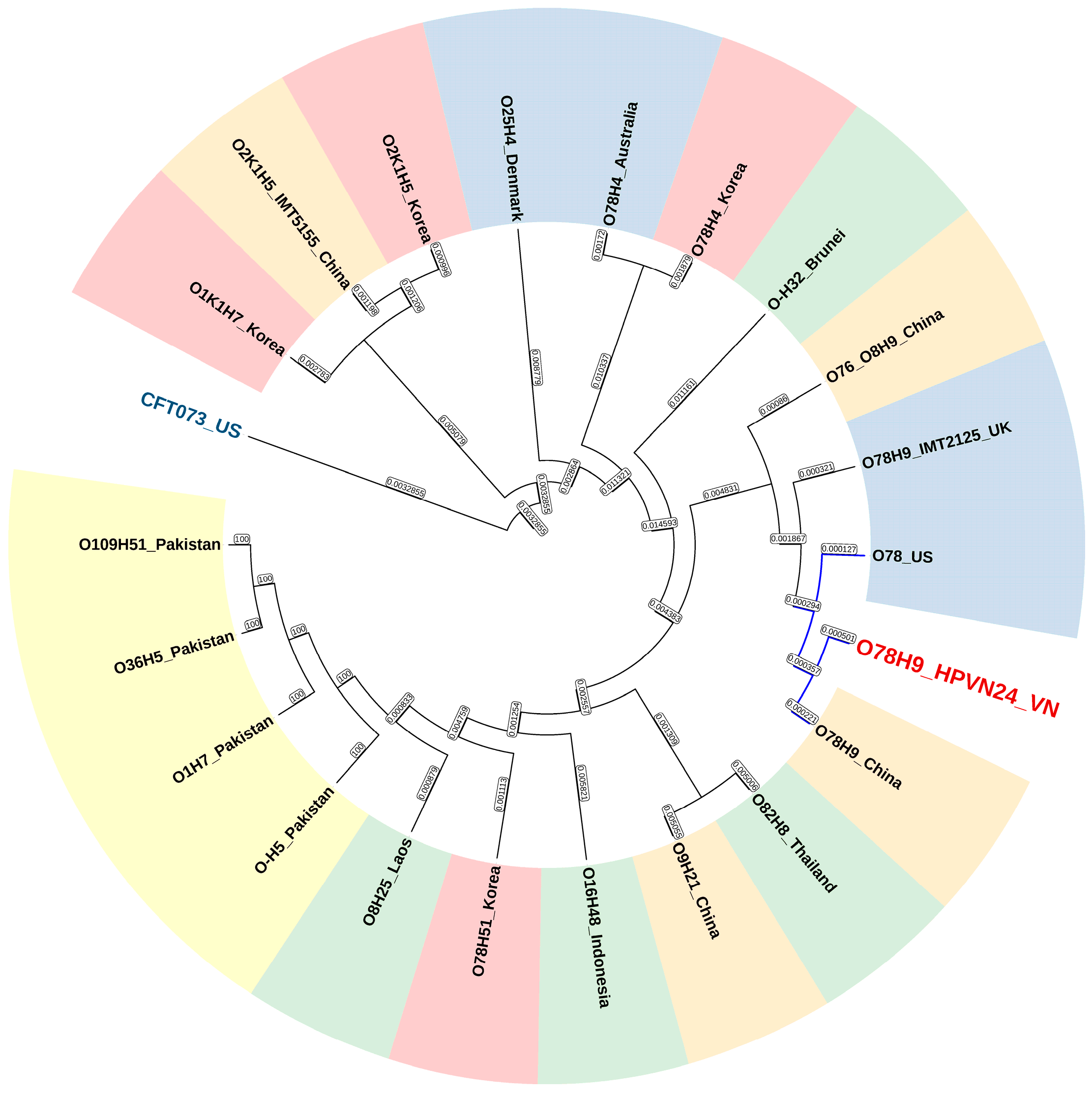
3.8. Genomic Comparative and Phylogenetic Analyses
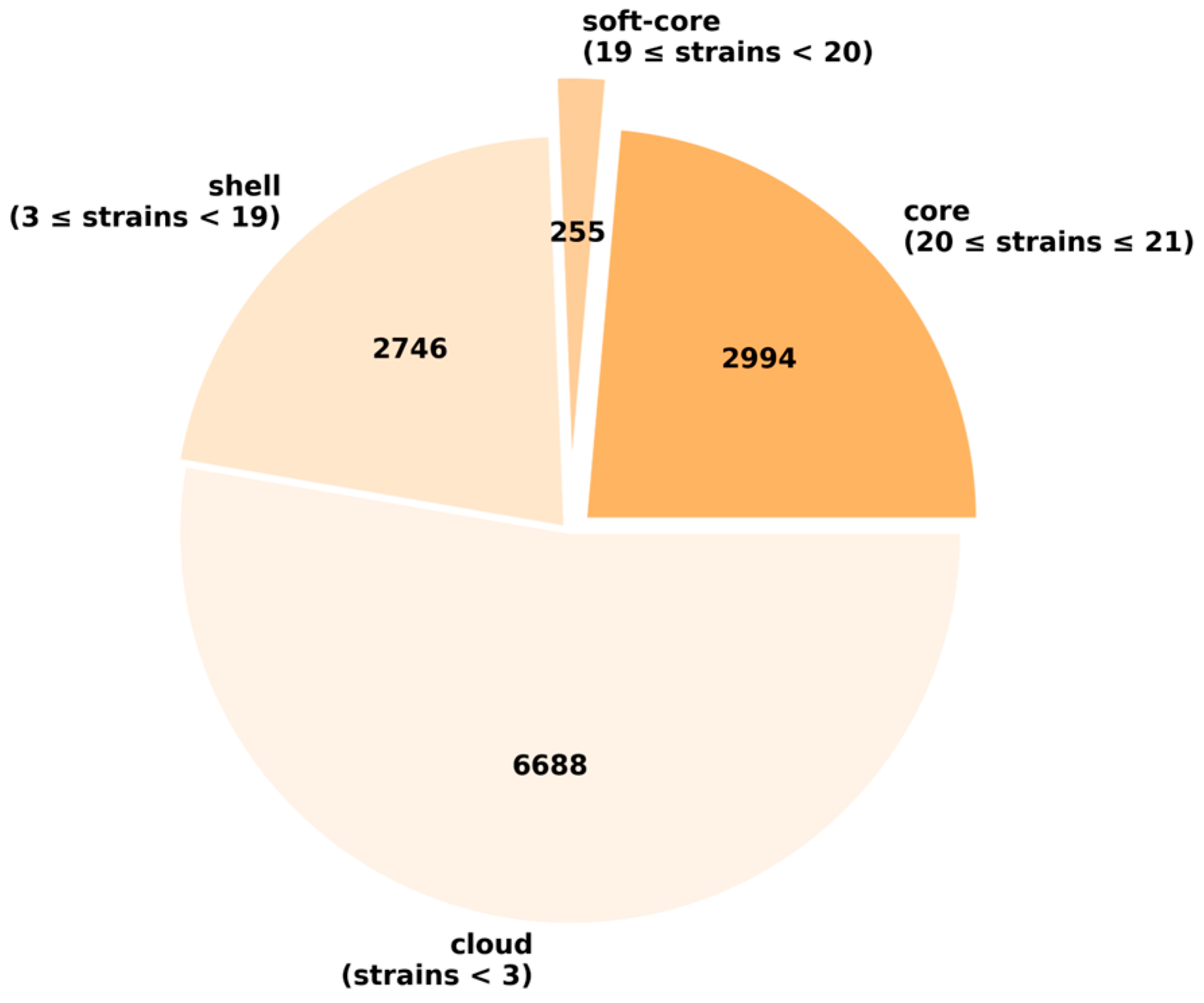
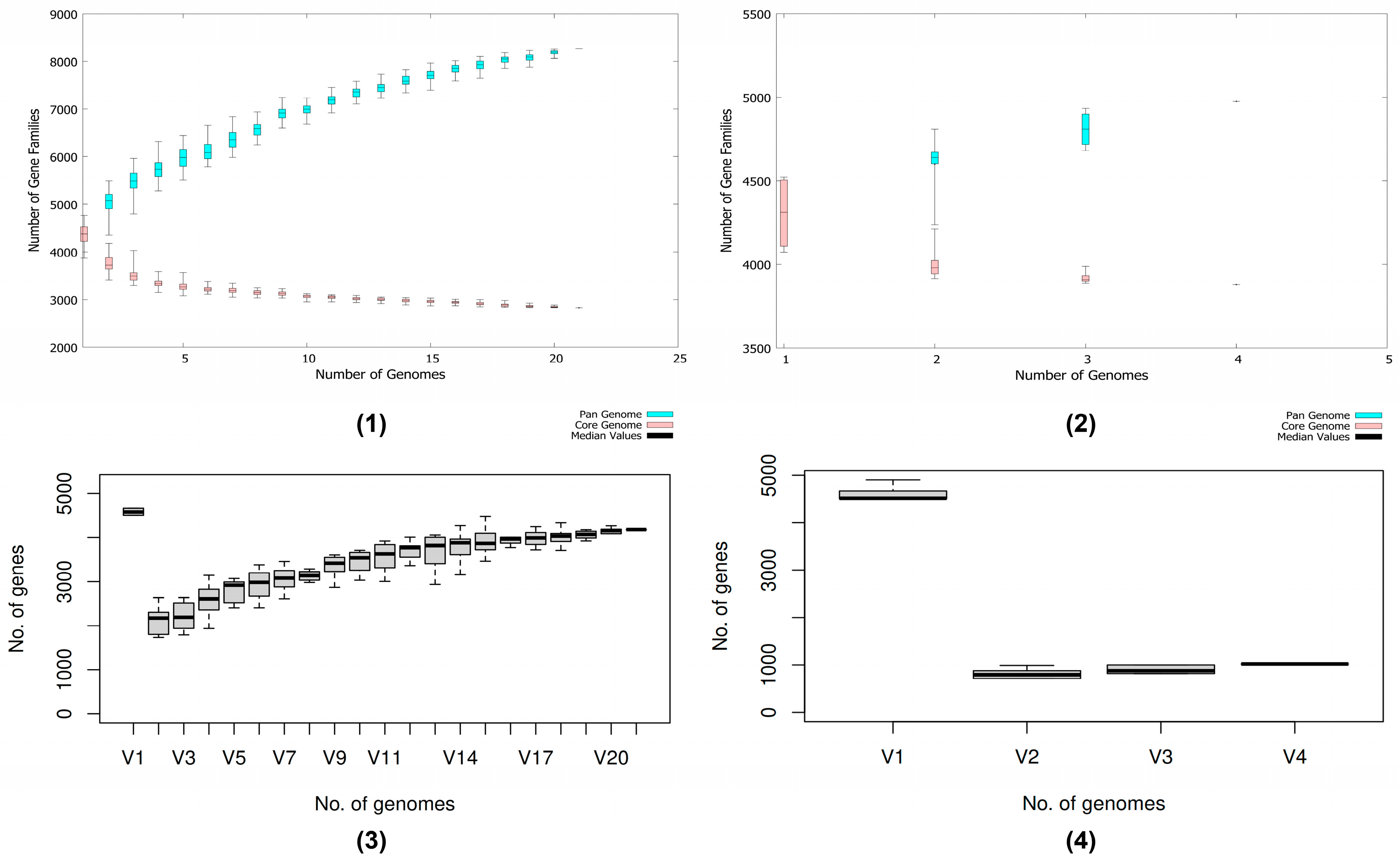
3.9. Pan-Genome and Genetic Profiles Comparison
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| UV | Ultraviolet |
| CLSI | Clinical and Laboratory Standards Institute |
| EUCAST | European Committee on Antimicrobial Susceptibility Testing |
| GC | Guanine–Cytosine |
| E-test | Epsilometer test |
| SNP | Single-nucleotide polymorphism |
References
- FAOSTAT. Food Balances: World Food Supply Quantity. Available online: https://www.fao.org/faostat/en/#data/FBS (accessed on 16 June 2025).
- FAOSTAT. Crops and Livestock Products: World Production Quantity. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 16 June 2025).
- Renub Research. Global Chicken Market Size, Share & Forecast 2025–2033. Available online: https://www.renub.com/ (accessed on 16 June 2025).
- National Statistics Office. Agriculture, Forestry and Fishery. Available online: https://www.nso.gov.vn/en/agriculture-forestry-and-fishery/ (accessed on 16 June 2025).
- One Health Poultry Hub. Poultry in Vietnam. Available online: https://www.onehealthpoultry.org/where-we-work/vietnam/poultry-in-vietnam/ (accessed on 16 June 2025).
- Guabiraba, R.; Schouler, C. Avian colibacillosis: Still many black holes. FEMS Microbiol. Lett. 2015, 362, fnv118. [Google Scholar] [CrossRef]
- Dho-Moulin, M.; Morris Fairbrother, J. Avian pathogenic Escherichia coli (APEC). Vet. Res. 1999, 30, 299–316. [Google Scholar]
- Ghunaim, H.; Abu-Madi, M.A.; Kariyawasam, S. Advances in vaccination against avian pathogenic Escherichia coli respiratory disease: Potentials and limitations. Vet. Microbiol. 2014, 172, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Dziva, F.; Stevens, M.P. Colibacillosis in poultry: Unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathol. 2008, 37, 355–366. [Google Scholar] [CrossRef]
- Huja, S.; Oren, Y.; Trost, E.; Brzuszkiewicz, E.; Biran, D.; Blom, J.; Goesmann, A.; Gottschalk, G.; Hacker, J.; Ron, E.Z.; et al. Genomic Avenue to Avian Colisepticemia. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Thuan, N.K.; Tam, N.T.; Huyen Trang, C.T.; Khanh, N.P.; Bich, T.N.; Taniguchi, T.; Hayashidani, H.; Lien Khai, L.T. Prevalence and Genetic Relationship of Predominant Escherichia coli Serotypes Isolated from Poultry, Wild Animals, and Environment in the Mekong Delta, Vietnam. Vet. Med. Int. 2021, 2021, 6504648. [Google Scholar] [CrossRef]
- Nawaz, S.; Wang, Z.; Zhang, Y.; Jia, Y.; Jiang, W.; Chen, Z.; Yin, H.; Huang, C.; Han, X. Avian pathogenic Escherichia coli (APEC): Current insights and future challenges. Poult. Sci. 2024, 103, 104359. [Google Scholar] [CrossRef]
- Janßen, T.; Schwarz, C.; Preikschat, P.; Voss, M.; Philipp, H.-C.; Wieler, L.H. Virulence-associated genes in avian pathogenic Escherichia coli (APEC) isolated from internal organs of poultry having died from colibacillosis. Int. J. Med. Microbiol. 2001, 291, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.M.; Barbieri, N.L.; de Oliveira, A.L.; Willis, D.; Nolan, L.K.; Logue, C.M. Characterizing avian pathogenic Escherichia coli (APEC) from colibacillosis cases, 2018. PeerJ 2021, 9, e11025. [Google Scholar] [CrossRef]
- Chauvin, C.; Clement, C.; Bruneau, M.; Pommeret, D. Time-patterns of antibiotic exposure in poultry production—A Markov chains exploratory study of nature and consequences. Prev. Vet. Med. 2007, 80, 230–240. [Google Scholar] [CrossRef]
- Cloud, S.S.; Rosenberger, J.K.; Fries, P.A.; Wilson, R.A.; Odor, E.M. In Vitro and In Vivo Characterization of Avian Escherichia coli. I. Serotypes, Metabolic Activity, and Antibiotic Sensitivity. Avian Dis. 1985, 29, 1084–1093. [Google Scholar] [CrossRef]
- Gregersen, R.H.; Christensen, H.; Ewers, C.; Bisgaard, M. Impact of Escherichia coli vaccine on parent stock mortality, first week mortality of broilers and population diversity of E. coli in vaccinated flocks. Avian Pathol. 2010, 39, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Solà-Ginés, M.; Cameron-Veas, K.; Badiola, I.; Dolz, R.; Majó, N.; Dahbi, G.; Viso, S.; Mora, A.; Blanco, J.; Piedra-Carrasco, N.; et al. Diversity of Multi-Drug Resistant Avian Pathogenic Escherichia coli (APEC) Causing Outbreaks of Colibacillosis in Broilers during 2012 in Spain. PLoS ONE 2015, 10, e0143191. [Google Scholar] [CrossRef]
- Luu, Q.H.; Nguyen, T.L.A.; Pham, T.N.; Vo, N.G.; Padungtod, P. Antimicrobial use in household, semi-industrialized, and industrialized pig and poultry farms in Viet Nam. Prev. Vet. Med. 2021, 189, 105292. [Google Scholar] [CrossRef]
- Rahman, M.R.T.; Fliss, I.; Biron, E. Insights in the Development and Uses of Alternatives to Antibiotic Growth Promoters in Poultry and Swine Production. Antibiotics 2022, 11, 766. [Google Scholar] [CrossRef]
- Ho, T.V.T.; Doan, T.L.A.; Le, V.D. Escherichia coli infection in ducks in the Mekong Delta: Bacterial isolation, serogroup distribution and antibiotic resistance. CTU J. Innov. Sustain. Dev. 2019, 11, 24–29. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.K.; Virdi, J.S. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 2015. [Google Scholar] [CrossRef] [PubMed]
- Turista, D.D.R.; Puspitasari, E. The Growth of Staphylococcus aureus in the blood agar plate media of sheep blood and human blood groups A, B, AB, and O. J. Teknol. Lab. 2019, 8, 1–7. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- de Sena Brandine, G.; Smith, A. Falco: High-speed FastQC emulation for quality control of sequencing data [version 2; peer review: 2 approved]. F1000Research 2021, 8, 1874. [Google Scholar] [CrossRef]
- Chen, S. Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef] [PubMed]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Alonge, M.; Lebeigle, L.; Kirsche, M.; Jenike, K.; Ou, S.; Aganezov, S.; Wang, X.; Lippman, Z.B.; Schatz, M.C.; Soyk, S. Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 2022, 23, 258. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Hirsch, P.; Gurevich, A. Webquast: Online evaluation of genome assemblies. Nucleic Acids Res. 2023, 51, W601–W606. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- The Galaxy Community. The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 2024, 52, W83–W94. [Google Scholar] [CrossRef]
- Bartual, S.G.; Seifert, H.; Hippler, C.; Luzon, M.A.D.; Wisplinghoff, H.; Rodríguez-Valera, F. Development of a Multilocus Sequence Typing Scheme for Characterization of Clinical Isolates of Acinetobacter baumannii. J. Clin. Microbiol. 2005, 43, 4382–4390, Erratum in J. Clin. Microbiol. 2007, 45. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus Sequence Typing of Total-Genome-Sequenced Bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef]
- Wirth, T.; Falush, D.; Lan, R.; Colles, F.; Mensa, P.; Wieler, L.H.; Karch, H.; Reeves, P.R.; Maiden, M.C.J.; Ochman, H.; et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006, 60, 1136–1151. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy In Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Roer, L.; Johannesen, T.B.; Hansen, F.; Stegger, M.; Tchesnokova, V.; Sokurenko, E.; Garibay, N.; Allesøe, R.; Thomsen, M.C.F.; Lund, O.; et al. CHTyper, a Web Tool for Subtyping of Extraintestinal Pathogenic Escherichia coli Based on the fumC and fimH Alleles. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2022, 51, D690–D699. [Google Scholar] [CrossRef]
- Shimoyama, Y. COGclassifier: A Tool for Classifying Prokaryote Protein Sequences into COG Functional Category, 2022. Available online: https://github.com/moshi4/COGclassifier (accessed on 26 February 2025).
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Popov, I. KEGGaNOG: Tool for Mining KEGG Pathways Completeness Data from eggNOG-Mapper Annotations, 2024. Available online: https://github.com/iliapopov17/KEGGaNOG (accessed on 17 July 2025).
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-Time Whole-Genome Sequencing for Routine Typing, Surveillance, and Outbreak Detection of Verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Tetzschner, A.M.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate: Mass Screening of Contigs for Antimicrobial Resistance or Virulence Genes, 2020. Available online: https://github.com/tseemann/abricate (accessed on 19 May 2025).
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2021, 50, D912–D917. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Shimoyama, Y. ANIclustermap: A Tool for Drawing ANI Clustermap Between All-Vs-All Microbial Genomes, 2022. Available online: https://github.com/moshi4/ANIclustermap (accessed on 26 February 2025).
- Ha, E.-J.; Hong, S.-M.; Kim, S.-J.; Ahn, S.-M.; Kim, H.-W.; Choi, K.-S.; Kwon, H.-J. Tracing the Evolutionary Pathways of Serogroup O78 Avian Pathogenic Escherichia coli. Antibiotics 2023, 12, 1714. [Google Scholar] [CrossRef]
- Zhu Ge, X.; Jiang, J.; Pan, Z.; Hu, L.; Wang, S.; Wang, H.; Leung, F.C.; Dai, J.; Fan, H. Comparative Genomic Analysis Shows That Avian Pathogenic Escherichia coli Isolate IMT5155 (O2:K1:H5; ST Complex 95, ST140) Shares Close Relationship with ST95 APEC O1:K1 and Human ExPEC O18:K1 Strains. PLoS ONE 2014, 9, e112048. [Google Scholar] [CrossRef]
- Zhang, J.; Cherbuin, J.D.R.; Khan, M.; Niaz, H.; Adnan, F. Complete genome sequences of three avian pathogenic Escherichia coli strains isolated from colibacillosis-affected poultry in Pakistan. Microbiol. Resour. Announc. 2025, 14, e00307–e00325. [Google Scholar] [CrossRef]
- Yang, Q.E.; Tansawai, U.; Andrey, D.O.; Wang, S.; Wang, Y.; Sands, K.; Kiddee, A.; Assawatheptawee, K.; Bunchu, N.; Hassan, B.; et al. Environmental dissemination of mcr-1 positive Enterobacteriaceae by Chrysomya spp. (common blowfly): An increasing public health risk. Environ. Int. 2019, 122, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Moser, A.I.; Kuenzli, E.; Campos-Madueno, E.I.; Büdel, T.; Rattanavong, S.; Vongsouvath, M.; Hatz, C.; Endimiani, A. Antimicrobial-Resistant Escherichia coli Strains and Their Plasmids in People, Poultry, and Chicken Meat in Laos. Front. Microbiol. 2021, 12, 2021. [Google Scholar] [CrossRef]
- Dziva, F.; Hauser, H.; Connor, T.R.; Diemen, P.M.v.; Prescott, G.; Langridge, G.C.; Eckert, S.; Chaudhuri, R.R.; Ewers, C.; Mellata, M.; et al. Sequencing and Functional Annotation of Avian Pathogenic Escherichia coli Serogroup O78 Strains Reveal the Evolution of E. coli Lineages Pathogenic for Poultry via Distinct Mechanisms. Infect. Immun. 2013, 81, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Mangiamele, P.; Nicholson, B.; Wannemuehler, Y.; Seemann, T.; Logue, C.M.; Li, G.; Tivendale, K.A.; Nolan, L.K. Complete Genome Sequence of the Avian Pathogenic Escherichia coli Strain APEC O78. Genome Announc. 2013, 1. [Google Scholar] [CrossRef] [PubMed]
- Clairfeuille, T.; Buchholz, K.R.; Li, Q.; Verschueren, E.; Liu, P.; Sangaraju, D.; Park, S.; Noland, C.L.; Storek, K.M.; Nickerson, N.N.; et al. Structure of the essential inner membrane lipopolysaccharide–PbgA complex. Nature 2020, 584, 479–483. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Katz, L. Lskatz/Lskscripts/ConvertAlignment.pl, 2018. Available online: https://github.com/lskatz/lskScripts (accessed on 13 June 2025).
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.M.; Gupta, V.K.; Dutta, C. BPGA- an ultra-fast pan-genome analysis pipeline. Sci. Rep. 2016, 6, 24373. [Google Scholar] [CrossRef]
- Bagel, S.; Hüllen, V.; Wiedemann, B.; Heisig, P. Impact of gyrA and parC Mutations on Quinolone Resistance, Doubling Time, and Supercoiling Degree of Escherichia coli. Antimicrob. Agents Chemother. 1999, 43, 868–875. [Google Scholar] [CrossRef]
- Oethinger, M.; Podglajen, I.; Kern, W.V.; Levy, S.B. Overexpression of the marA or soxS Regulatory Gene in Clinical Topoisomerase Mutants of Escherichia coli. Antimicrob. Agents Chemother. 1998, 42, 2089–2094. [Google Scholar] [CrossRef]
- Zayed, A.A.-F.; Essam, T.M.; Hashem, A.-G.M.; El-Tayeb, O.M. ‘Supermutators’ found amongst highly levofloxacin-resistant E. coli isolates: A rapid protocol for the detection of mutation sites. Emerg. Microbes Infect. 2015, 4, 1–8. [Google Scholar] [CrossRef]
- Watts, A.; Wigley, P. Avian Pathogenic Escherichia coli: An Overview of Infection Biology, Antimicrobial Resistance and Vaccination. Antibiotics 2024, 13, 809. [Google Scholar] [CrossRef]
- Yousef, H.M.Y.; Hashad, M.E.; Osman, K.M.; Alatfeehy, N.M.; Hassan, W.M.M.; Elebeedy, L.A.; Salem, H.M.; Shami, A.; Al-Saeed, F.A.; El-Saadony, M.T.; et al. Surveillance of Escherichia coli in different types of chicken and duck hatcheries: One health outlook. Poult. Sci. 2023, 102, 103108. [Google Scholar] [CrossRef]
- Subedi, M.; Luitel, H.; Devkota, B.; Bhattarai, R.K.; Phuyal, S.; Panthi, P.; Shrestha, A.; Chaudhary, D.K. Antibiotic resistance pattern and virulence genes content in avian pathogenic Escherichia coli (APEC) from broiler chickens in Chitwan, Nepal. BMC Vet. Res. 2018, 14, 113. [Google Scholar] [CrossRef]
- Zhao, S.; Maurer, J.J.; Hubert, S.; De Villena, J.F.; McDermott, P.F.; Meng, J.; Ayers, S.; English, L.; White, D.G. Antimicrobial susceptibility and molecular characterization of avian pathogenic Escherichia coli isolates. Vet. Microbiol. 2005, 107, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Dheilly, A.; Le Devendec, L.; Mourand, G.; Jouy, E.; Kempf, I. Antimicrobial resistance selection in avian pathogenic E. coli during treatment. Vet. Microbiol. 2013, 166, 655–658. [Google Scholar] [CrossRef]
- Li, L.; Kromann, S.; Olsen, J.E.; Svenningsen, S.W.; Olsen, R.H. Insight into synergetic mechanisms of tetracycline and the selective serotonin reuptake inhibitor, sertraline, in a tetracycline-resistant strain of Escherichia coli. J. Antibiot. 2017, 70, 944–953. [Google Scholar] [CrossRef]
- Babai, R.; Blum-Oehler, G.; Stern, B.E.; Hacker, J.; Ron, E.Z. Virulence patterns from septicemic Escherichia coli O78 strains. FEMS Microbiol. Lett. 2006, 149, 99–105. [Google Scholar] [CrossRef][Green Version]
- Kathayat, D.; Lokesh, D.; Ranjit, S.; Rajashekara, G. Avian Pathogenic Escherichia coli (APEC): An Overview of Virulence and Pathogenesis Factors, Zoonotic Potential, and Control Strategies. Pathogens 2021, 10, 467. [Google Scholar] [CrossRef]
- Hu, J.; Afayibo, D.J.A.; Zhang, B.; Zhu, H.; Yao, L.; Guo, W.; Wang, X.; Wang, Z.; Wang, D.; Peng, H.; et al. Characteristics, pathogenic mechanism, zoonotic potential, drug resistance, and prevention of avian pathogenic Escherichia coli (APEC). Front. Microbiol. 2022, 13, 2022. [Google Scholar] [CrossRef]
- Nawaz, S.; Shoaib, M.; Huang, C.; Jiang, W.; Bao, Y.; Wu, X.; Nie, L.; Fan, W.; Wang, Z.; Chen, Z.; et al. Molecular Characterization, Antibiotic Resistance, and Biofilm Formation of Escherichia coli Isolated from Commercial Broilers from Four Chinese Provinces. Microorganisms 2025, 13, 1017. [Google Scholar] [CrossRef]
- Ovi, F.; Zhang, L.; Nabors, H.; Jia, L.; Adhikari, P. A compilation of virulence-associated genes that are frequently reported in avian pathogenic Escherichia coli (APEC) compared to other E. coli. J. Appl. Microbiol. 2023, 134, lxad014. [Google Scholar] [CrossRef]
- Arné, P.; Marc, D.; Brée, A.; Schouler, C.; Dho-Moulin, M. Increased Tracheal Colonization in Chickens without Impairing Pathogenic Properties of Avian Pathogenic Escherichia coli MT78 with a fimH Deletion. Avian Dis. 2000, 44, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Mathers, A.J.; Peirano, G.; Pitout, J.D.D. The Role of Epidemic Resistance Plasmids and International High-Risk Clones in the Spread of Multidrug-Resistant Enterobacteriaceae. Clin. Microbiol. Rev. 2015, 28, 565–591. [Google Scholar] [CrossRef] [PubMed]
- Bélanger, L.; Garenaux, A.; Harel, J.; Boulianne, M.; Nadeau, E.; Dozois, C.M. Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli. FEMS Immunol. Med. Microbiol. 2011, 62, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Franchini, A.G.; Ihssen, J.; Egli, T. Effect of Global Regulators RpoS and Cyclic-AMP/CRP on the Catabolome and Transcriptome of Escherichia coli K12 during Carbon- and Energy-Limited Growth. PLoS ONE 2015, 10, e0133793. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hunt, S.; Green, J.; Artymiuk, P.J. Hemolysin E (HlyE, ClyA, SheA) and Related Toxins. In Proteins Membrane Binding and Pore Formation; Anderluh, G., Lakey, J., Eds.; Springer: New York, NY, USA, 2010; pp. 116–126. [Google Scholar]
- Ahsan, C.R.; Shamma, F.; Ahsan, N.; Islam, M.J. Environmental factors regulate the hlye gene expression in both S. typhi and E. coli in a similar way to display haemolytic activity. Bangladesh Med. Res. Counc. Bull. 2017, 42, 33–38. [Google Scholar] [CrossRef][Green Version]
- Murase, K.; Ooka, T.; Iguchi, A.; Ogura, Y.; Nakayama, K.; Asadulghani, M.; Islam, M.R.; Hiyoshi, H.; Kodama, T.; Beutin, L.; et al. Haemolysin E- and enterohaemolysin-derived haemolytic activity of O55/O157 strains and other Escherichia coli lineages. Microbiology 2012, 158, 746–758. [Google Scholar] [CrossRef][Green Version]
- Murase, K. Cytolysin A (ClyA): A Bacterial Virulence Factor with Potential Applications in Nanopore Technology, Vaccine Development, and Tumor Therapy. Toxins 2022, 14, 78. [Google Scholar] [CrossRef]
- Hu, J.; Lv, X.; Niu, X.; Yu, F.; Zuo, J.; Bao, Y.; Yin, H.; Huang, C.; Nawaz, S.; Zhou, W.; et al. Effect of nutritional and environmental conditions on biofilm formation of avian pathogenic Escherichia coli. J. Appl. Microbiol. 2022, 132, 4236–4251. [Google Scholar] [CrossRef]
- Sande, C.; Whitfield, C. Capsules and Extracellular Polysaccharides in Escherichia coli and Salmonella. EcoSal Plus 2021, 9, Eesp-0033-2020, Erratum in EcoSal Plus 2022, 10. [Google Scholar] [CrossRef]
- Usman, S.; Anjum, A.; Usman, M.; Imran, M.S.; Ali, M.; Moustafa, M.; Rehman, M.S.; Hussain, T.; Sarwar, F.; Azad, A.; et al. Antibiotic resistance pattern and pathological features of avian pathogenic Escherichia coli O78:K80 in chickens. Braz. J. Biol. 2024, 84, e257179. [Google Scholar] [CrossRef]
- Piddock, L.J.V. Multidrug-resistance efflux pumps? Not just for resistance. Nat. Rev. Microbiol. 2006, 4, 629–636. [Google Scholar] [CrossRef]
- Dulanto Chiang, A.; Dekker, J.P. Efflux pump-mediated resistance to new beta lactam antibiotics in multidrug-resistant gram-negative bacteria. Commun. Med. 2024, 4, 170. [Google Scholar] [CrossRef]
- Afridi, O.K.; Ali, J.; Chang, J.H. Next-Generation Sequencing Based Gut Resistome Profiling of Broiler Chickens Infected with Multidrug-Resistant Escherichia coli. Animals 2020, 10, 2350. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 1996, 178, 5853–5859. [Google Scholar] [CrossRef]
- Zgurskaya, H.I.; Nikaido, H. Multidrug resistance mechanisms: Drug efflux across two membranes. Mol. Microbiol. 2000, 37, 219–225. [Google Scholar] [CrossRef]
- Greene, G.; Koolman, L.; Whyte, P.; Burgess, C.; Lynch, H.; Coffey, A.; Lucey, B.; O’Connor, L.; Bolton, D. Effect of Doxycycline Use in the Early Broiler Production Cycle on the Microbiome. Front. Microbiol. 2022, 13, 2022. [Google Scholar] [CrossRef]
- Devi, B.T.; Aditya, V.; Kini, S.; Kumar, B.K.; Deekshit, V.K. Synergistic effects of antibiotics and efflux pump inhibitors on multidrug-resistant Escherichia coli and Klebsiella pneumoniae. J. Appl. Microbiol. 2025, 136, lxaf169. [Google Scholar] [CrossRef]
- Jahantigh, M.; Samadi, K.; Dizaji, R.E.; Salari, S. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Vet. Res. 2020, 16, 267. [Google Scholar] [CrossRef]
- Møller, T.S.B.; Overgaard, M.; Nielsen, S.S.; Bortolaia, V.; Sommer, M.O.A.; Guardabassi, L.; Olsen, J.E. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli. BMC Microbiol. 2016, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Goudarztalejerdi, A.; Mohammadzadeh, A.; Niazi, K.; Mohammad Mirzaei, M. High Prevalence of Multidrug Resistance and Biofilm-Formation Ability Among Avian Escherichia coli Isolated from Broilers in Iran. Microb. Drug Resist. 2022, 28, 244–254. [Google Scholar] [CrossRef]
- Krucinska, J.; Lombardo, M.N.; Erlandsen, H.; Estrada, A.; Si, D.; Viswanathan, K.; Wright, D.L. Structure-guided functional studies of plasmid-encoded dihydrofolate reductases reveal a common mechanism of trimethoprim resistance in Gram-negative pathogens. Commun. Biol. 2022, 5, 459. [Google Scholar] [CrossRef]
- Mammeri, H.; Poirel, L.; Fortineau, N.; Nordmann, P. Naturally Occurring Extended-Spectrum Cephalosporinases in Escherichia coli. Antimicrob. Agents Chemother. 2006, 50, 2573–2576. [Google Scholar] [CrossRef]
- Hainrichson, M.; Yaniv, O.; Cherniavsky, M.; Nudelman, I.; Shallom-Shezifi, D.; Yaron, S.; Baasov, T. Overexpression and Initial Characterization of the Chromosomal Aminoglycoside 3′-O-Phosphotransferase APH(3′)-IIb from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2007, 51, 774–776. [Google Scholar] [CrossRef]
- Xu, J.; Sangthong, R.; McNeil, E.; Tang, R.; Chongsuvivatwong, V. Antibiotic use in chicken farms in northwestern China. Antimicrob. Resist. Infect. Control 2020, 9, 10. [Google Scholar] [CrossRef]
- Bâtie, C.; Tran Minh, H.; Thi Vu, V.A.; Thuy Luong, D.; Thi Pham, T.; Fortané, N.; Pham Duc, P.; Goutard, F.L. Reducing antimicrobial use in chicken production in Vietnam: Exploring the systemic dimension of change. PLoS ONE 2023, 18, e0290296. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Q.; Ji, Z.; Andom, O.; Wang, X.; Guo, X.; Li, Z. Current Status and Spatiotemporal Evolution of Antibiotic Residues in Livestock and Poultry Manure in China. Agriculture 2023, 13, 1877. [Google Scholar] [CrossRef]
- Singer, R.S.; Schrag, N.F.D.; Ricke, I.; Apley, M.D. Antimicrobial usage in broiler chicken production in the United States, 2013–2021. Front. Vet. Sci. 2023, 10, 2023. [Google Scholar] [CrossRef]
- The British Poultry Council. “The Key to Unlocking Continuous Improvement”—BPC Antibiotic Stewardship Report 2023. Available online: https://britishpoultry.org.uk/the-key-to-unlocking-continuous-improvement-bpc-antibiotic-stewardship-report-2023/ (accessed on 16 September 2025).
- Flo, T.H.; Smith, K.D.; Sato, S.; Rodriguez, D.J.; Holmes, M.A.; Strong, R.K.; Akira, S.; Aderem, A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 2004, 432, 917–921. [Google Scholar] [CrossRef]
- Caza, M.; Lépine, F.; Milot, S.; Dozois, C.M. Specific Roles of the iroBCDEN Genes in Virulence of an Avian Pathogenic Escherichia coli O78 Strain and in Production of Salmochelins. Infect. Immun. 2008, 76, 3539–3549. [Google Scholar] [CrossRef]

| Categories | Pathways | Completeness Score | Pathways | Completeness Score |
|---|---|---|---|---|
| Amino acid metabolism | Alanine | 1.0 | Lysine | 1.0 |
| Arginine | 1.0 | Methionine | 1.0 | |
| Asparagine | 1.0 | Phenylalanine | 1.0 | |
| Aspartate | 1.0 | Proline | 1.0 | |
| Cysteine | 1.0 | Serine | 1.0 | |
| Glutamate | 1.0 | Formaldehyde assimilation | 0.4 | |
| Glycine | 1.0 | Threonine | 1.0 | |
| Histidine | 1.0 | Tryptophan | 1.0 | |
| Isoleucine | 1.0 | Tyrosine | 1.0 | |
| Leucine | 1.0 | Valine | 1.0 | |
| Arsenic reduction | Arsenic reduction | 0.75 | ||
| Bacterial secretion systems | Sec-SRP | 1.0 | Type III secretion | 0.47 |
| Twin Arginine targetting | 1.0 | Type IV secretion | 0.75 | |
| Type I secretion | 0.66 | Type VI secretion | 0.11 | |
| Type II secretion | 0.92 | |||
| Biofilm formation | PGS Synthesis protein | 1.0 | Colanic acid and Biofilm transcription regulator | 1.0 |
| Biofilm regulator BSS | 1.0 | Curli fimbriae biosynthesis | 0.99 | |
| Colanic acid and Biofilm protein A | 1.0 | |||
| Carbohydrate metabolism | Alpha-amylase | 1.0 | Mixed acid: Formate | 1.0 |
| Entner–Doudoroff | 1.0 | Mixed acid: Formate to CO2 & H2 | 0.5 | |
| Glycolysis | 0.89 | Mixed acid: PEP to Succinate via OAA, malate and fumarate | 0.88 | |
| Glyoxylate shunt | 1.0 | Polyhydroxybutyrate synthesis | 0.17 | |
| Mixed acid: Acetate | 1.0 | Starch/glycogen degradation | 1.0 | |
| Mixed acid: Ethanol, Acetate to Acetylaldehyde | 1.0 | Starch/glycogen synthesis | 0.99 | |
| Mixed acid: Ethanol, Acetyl-CoA to Acetylaldehyde (reversible) | 1.0 | TCA Cycle | 0.88 | |
| Carbon degradation | Beta-glucosidase | 1.0 | Diacetylchitobiose deacetylase | 1.0 |
| Bifunctional chitinase/ lysozyme | 1.0 | Naphthalene degradation to salicylate | 0.17 | |
| Chitinase | 1.0 | Pullulanase | 1.0 | |
| D-galacturonate isomerase | 1.0 | beta-N-acetylhexosaminidase | 1.0 | |
| Carbon fixation | 3-Hydroxypropionate Bicycle | 0.24 | Gluconeogenesis | 0.89 |
| 4-Hydroxybutyrate/ 3-hydroxypropionate | 0.2 | |||
| Cell mobility | Adhesion | 1.0 | Flagellum | 1.0 |
| Chemotaxis | 0.75 | |||
| Genetic competence | Competence-related core components | 0.14 | ||
| Hydrogen redox | Hydrogen:quinone oxidoreductase | 1.0 | NiFe hydrogenase Hyd-l | 0.99 |
| Metal transporters | Cobalt transporter CorA | 1.0 | Ferrous iron transporter FeoB | 1.0 |
| Copper transporter CopA | 1.0 | Nickel ABC-type substrate-binding NikA | 1.0 | |
| Fe-Mn transporter MntH | 1.0 | |||
| Miscellaneous | Anaplerotic genes | 0.75 | Staphyloaxanthin biosynthesis | 0.17 |
| Nitrogen metabolism | DRNA | 1.0 | Nitrite oxidation | 1.0 |
| Dissimilatory nitrate reduction | 1.0 | |||
| Oxidative phosphorylation | Cytochrome bd complex | 1.0 | F-type ATPase | 1.0 |
| Cytochrome o ubiquinol oxidase | 1.0 | NADH-quinone oxidoreductase | 0.84 | |
| Sulfur metabolism | DMSO reductase | 0.99 | Thiosulfate/polysulfide reductase | 0.66 |
| Sulfur assimilation | 1.0 | |||
| Transporters | Bidirectional polyphosphate | 1.0 | Transporter: phosphate | 1.0 |
| C-P lyase cleavage PhnJ | 1.0 | Transporter: phosphonate | 0.99 | |
| CP-lyase complex | 1.0 | Transporter: thiamin | 0.99 | |
| CP-lyase operon | 0.99 | transporter: vitamin B12 | 0.99 | |
| Vitamin biosynthesis | Cobalamin biosynthesis | 0.62 | Retinal biosynthesis | 0.25 |
| MEP-DOXP pathway | 0.99 | Riboflavin biosynthesis | 1.0 | |
| Mevalonate pathway | 0.2 | Thiamin biosynthesis | 0.91 |
| Resistance Mechanisms | Antibiotic Resistance Ontology (ARO) Category | Resistance Genes | Involved Antimicrobial Classes |
|---|---|---|---|
| Antibiotic target replacements | Perfect | dfrA1 | diaminopyrimidine antibiotic |
| Antibiotic target alteration | PmrF, bacA | peptide antibiotic | |
| Reduced permeability to antibiotic | marA | nitroimidazole, macrolide, fluoroquinolone, aminoglycoside, carbapenem, cephalosporin, glycylcycline, penicillin beta-lactam, tetracycline, peptide, aminocoumarin, rifamycin, phenicol, phosphonic acid, disinfecting agents and antiseptics | |
| Antibiotic efflux | msbA, acrA, acrB, marA, mdtE, AcrS, AcrE, evgA, H-NS, emrB, emrR, mdtH, mdtG, cpxA, qacEDelta1 | ||
| Reduced permeability to antibiotic | Strict | SoxS | fluoroquinolone, monobactam, carbapenem, cephalosporin, glycylcycline, penicillin beta-lactam, tetracycline, rifamycin, phenicol, disinfecting agents and antiseptics |
| Antibiotic inactivation | EC-13 | cephalosporin | |
| Antibiotic efflux | TolC, mdtC, mdtB, mdtA, Yojl, AcrF, acrD, emrY, emrK, evgS, mdtP, mdtO, mdtM, kdpE, emrA, KpnF, KpnE, CRP, gadX, rsmA, baeR, acrR, marR, SoxS, SoxR | macrolide, fluoroquinolone, aminoglycoside, nucleoside, carbapenem, cephalosporin, glycylcycline, lincosamide, penicillin beta-lactam, tetracycline, glycopeptide, peptide, aminocoumarin, rifamycin, phenicol, phosphonic acid, diaminopyrimidine, disinfecting agents and antiseptics | |
| Antibiotic target alteration | ArnT, eptA, vanG, ugd, acrR, marR, soxS, soxR | peptide, glycopeptide, fluoroquinolone, cephalosporin, glycylcycline, penicillin beta-lactam, tetracycline, rifamycin, phenicol, disinfecting agents and antiseptics |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoang, M.D.; Lanh, P.T.; Hien, V.T.; Kao, C.-Y.; Quyen, D.V. Comprehensive Genomic and Phenotypic Characterization of Escherichia coli O78:H9 Strain HPVN24 Isolated from Diarrheic Poultry in Vietnam. Microorganisms 2025, 13, 2265. https://doi.org/10.3390/microorganisms13102265
Hoang MD, Lanh PT, Hien VT, Kao C-Y, Quyen DV. Comprehensive Genomic and Phenotypic Characterization of Escherichia coli O78:H9 Strain HPVN24 Isolated from Diarrheic Poultry in Vietnam. Microorganisms. 2025; 13(10):2265. https://doi.org/10.3390/microorganisms13102265
Chicago/Turabian StyleHoang, Minh Duc, Pham Thi Lanh, Vu Thi Hien, Cheng-Yen Kao, and Dong Van Quyen. 2025. "Comprehensive Genomic and Phenotypic Characterization of Escherichia coli O78:H9 Strain HPVN24 Isolated from Diarrheic Poultry in Vietnam" Microorganisms 13, no. 10: 2265. https://doi.org/10.3390/microorganisms13102265
APA StyleHoang, M. D., Lanh, P. T., Hien, V. T., Kao, C.-Y., & Quyen, D. V. (2025). Comprehensive Genomic and Phenotypic Characterization of Escherichia coli O78:H9 Strain HPVN24 Isolated from Diarrheic Poultry in Vietnam. Microorganisms, 13(10), 2265. https://doi.org/10.3390/microorganisms13102265







