Targeting Transcriptional Regulators Affecting Acarbose Biosynthesis in Actinoplanes sp. SE50/110 Using CRISPRi Silencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Media, and Reagents
2.2. Construction of CRISPRi Vector Systems
2.3. Conjugation and Plasmid Curing in Actinoplanes
2.4. Cultivation of Actinoplanes
2.5. Acarbose Quantification
2.6. RNA Isolation and Quantification of mRNA Levels of Targeted Genes
3. Results
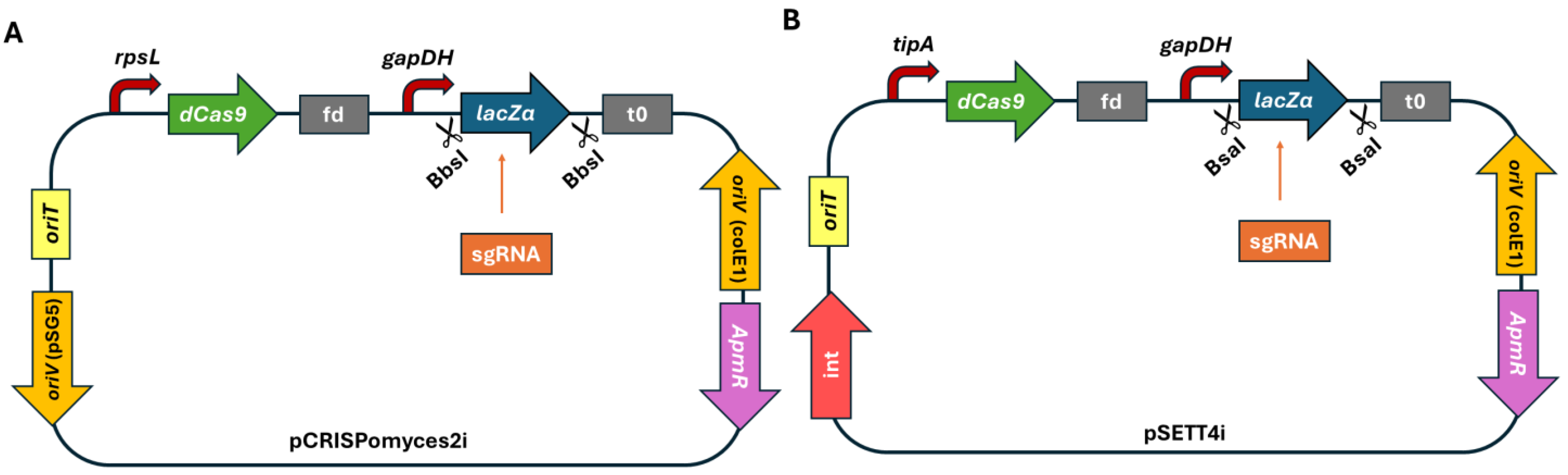
3.1. Design of Two Types of CRISPRi Vectors as a Basis for Silencing Experiments in Actinoplanes sp. SE50/110
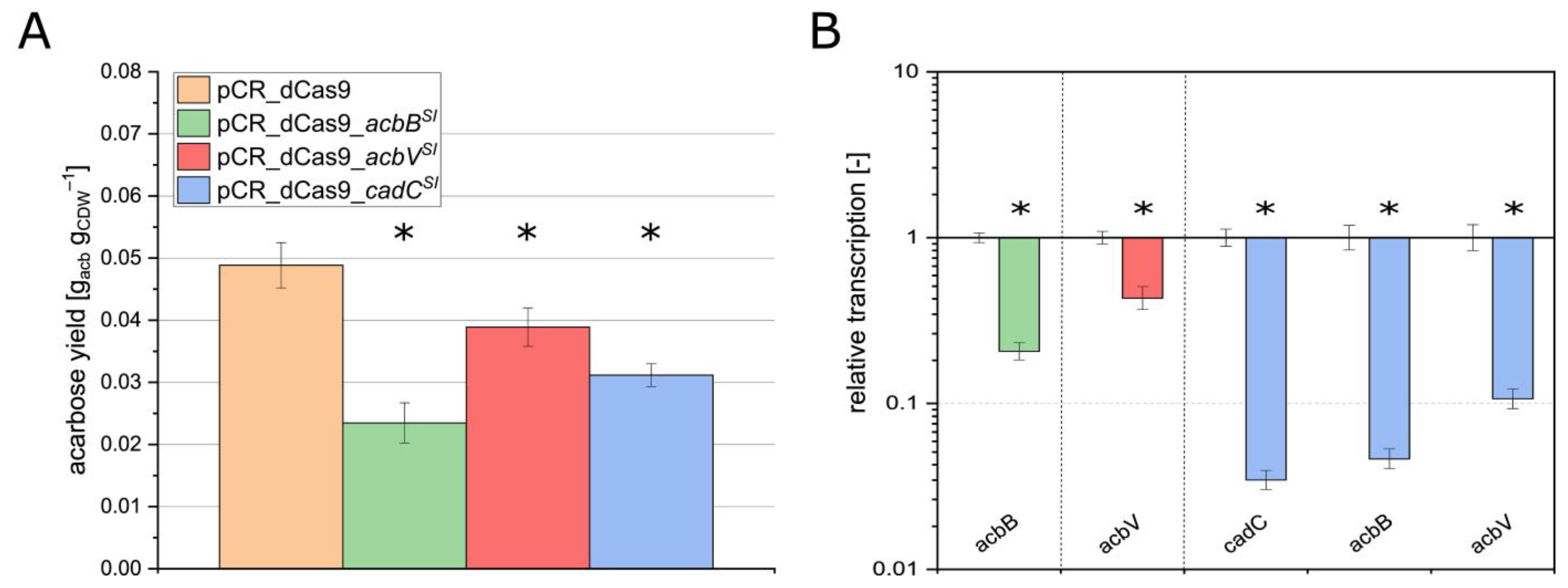
3.2. Application of pCR_dCas9 for Reversible Silencing of the Genes acbB, acbV, and cadC
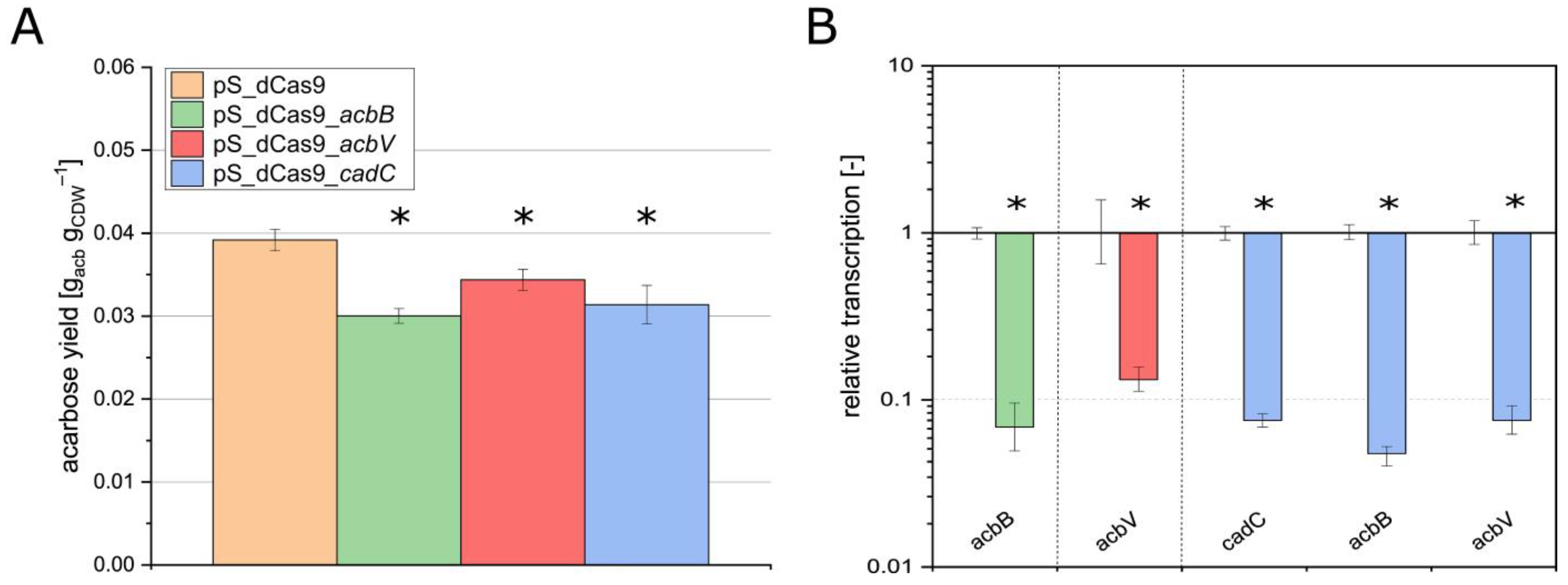
3.3. Application of pSETT4i as a Rapid Screening Vector for Silencing of the Genes acbB, acbV, and cadC
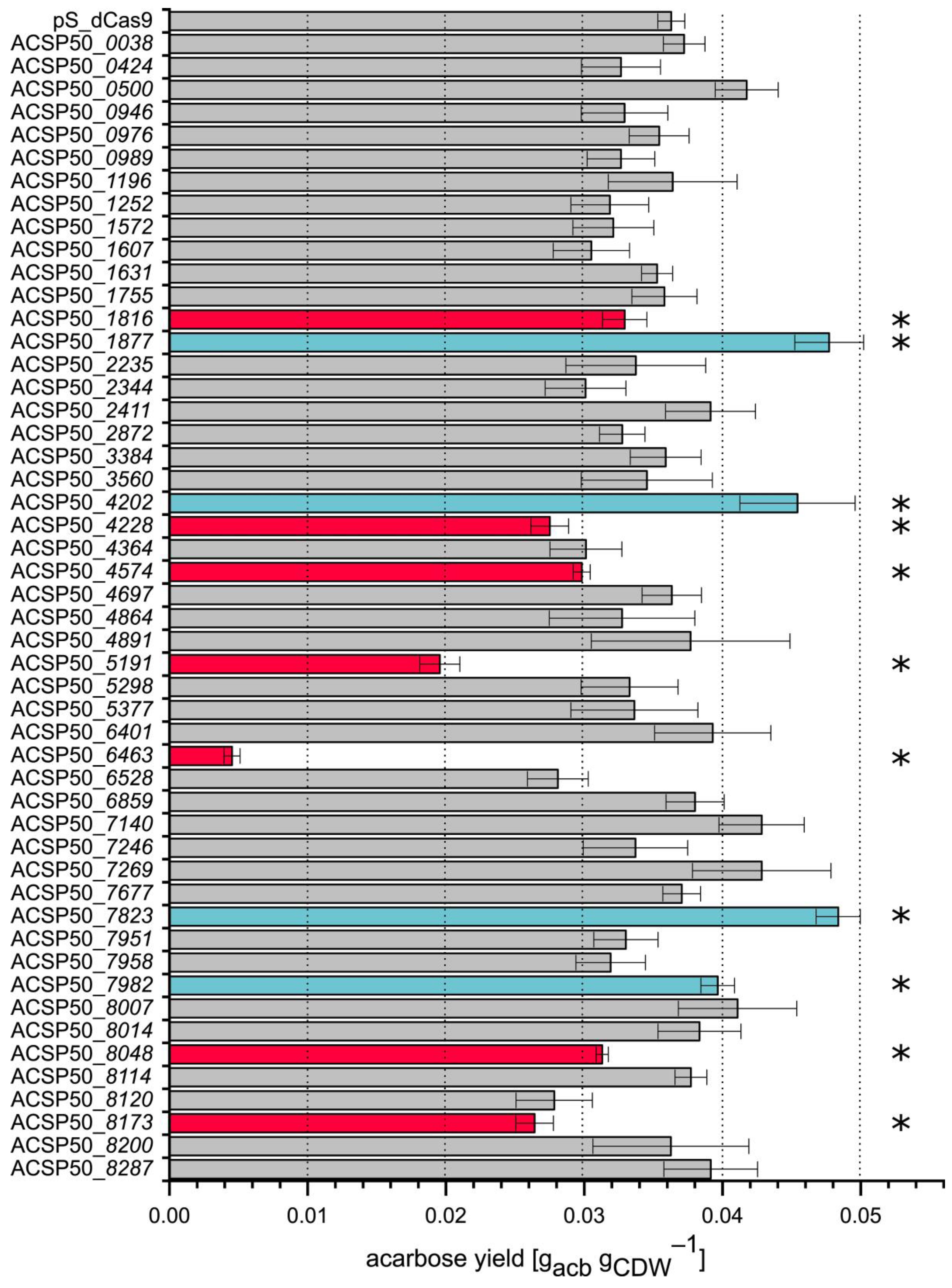
3.4. Identification of Eleven Regulators Influencing Acarbose Biosynthesis Through Screening of CRISPRi Strain Library
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- de Simeis, D.; Serra, S. Actinomycetes: A Never-Ending Source of Bioactive Compounds-An Overview on Antibiotics Production. Antibiotics 2021, 10, 483. [Google Scholar] [CrossRef]
- Takahashi, Y.; Nakashima, T. Actinomycetes, an Inexhaustible Source of Naturally Occurring Antibiotics. Antibiotics 2018, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Vobis, G.; Schäfer, J.; Kämpfer, P. Actinoplanes. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; Wiley: Hoboken, NJ, USA, 2015; pp. 1–41. ISBN 9781118960608. [Google Scholar]
- Parra, J.; Beaton, A.; Seipke, R.F.; Wilkinson, B.; Hutchings, M.I.; Duncan, K.R. Antibiotics from rare actinomycetes, beyond the genus Streptomyces. Curr. Opin. Microbiol. 2023, 76, 102385. [Google Scholar] [CrossRef] [PubMed]
- Debono, M.; Merkel, K.E.; Molloy, R.M.; Barnhart, M.; Presti, E.; Hunt, A.H.; Hamill, R.L. Actaplanin, new glycopeptide antibiotics produced by Actinoplanes missouriensis. The isolation and preliminary chemical characterization of actaplanin. J. Antibiot. 1984, 37, 85–95. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, M.; González, I.; Parish, C.A.; Onishi, R.; Tormo, J.R.; Martín, J.; Peláez, F.; Zink, D.; El Aouad, N.; Reyes, F.; et al. Production of Ramoplanin and Ramoplanin Analogs by Actinomycetes. Front. Microbiol. 2017, 8, 343. [Google Scholar] [CrossRef] [PubMed]
- Sosio, M.; Kloosterman, H.; Bianchi, A.; de Vreugd, P.; Dijkhuizen, L.; Donadio, S. Organization of the teicoplanin gene cluster in Actinoplanes teichomyceticus. Microbiology 2004, 150, 95–102. [Google Scholar] [CrossRef]
- Truscheit, E.; Frommer, W.; Junge, B.; Müller, L.; Schmidt, D.D.; Wingender, W. Chemistry and Biochemistry of Microbial α-Glucosidase Inhibitors. Angew. Chem. Int. Ed. Engl. 1981, 20, 744–761. [Google Scholar] [CrossRef]
- Wehmeier, U.F.; Piepersberg, W. Biotechnology and molecular biology of the α-glucosidase inhibitor acarbose. Appl. Microbiol. Biotechnol. 2004, 63, 613–625. [Google Scholar] [CrossRef]
- Derosa, G.; Maffioli, P. α-Glucosidase inhibitors and their use in clinical practice. Arch. Med. Sci. 2012, 8, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Tsunoda, T.; Samadi, A.; Burade, S.; Mahmud, T. Complete biosynthetic pathway to the antidiabetic drug acarbose. Nat. Commun. 2022, 13, 3455. [Google Scholar] [CrossRef]
- Schwientek, P.; Szczepanowski, R.; Rückert, C.; Kalinowski, J.; Klein, A.; Selber, K.; Wehmeier, U.F.; Stoye, J.; Pühler, A. The complete genome sequence of the acarbose producer Actinoplanes sp. SE50/110. BMC Genom. 2012, 13, 112. [Google Scholar] [CrossRef] [PubMed]
- Droste, J.; Ortseifen, V.; Schaffert, L.; Persicke, M.; Schneiker-Bekel, S.; Pühler, A.; Kalinowski, J. The expression of the acarbose biosynthesis gene cluster in Actinoplanes sp. SE50/110 is dependent on the growth phase. BMC Genom. 2020, 21, 818. [Google Scholar] [CrossRef] [PubMed]
- Schwientek, P.; Wendler, S.; Neshat, A.; Eirich, C.; Rückert, C.; Klein, A.; Wehmeier, U.F.; Kalinowski, J.; Stoye, J.; Pühler, A. Comparative RNA-sequencing of the acarbose producer Actinoplanes sp. SE50/110 cultivated in different growth media. J. Biotechnol. 2013, 167, 166–177. [Google Scholar] [CrossRef]
- Wendler, S.; Hürtgen, D.; Kalinowski, J.; Klein, A.; Niehaus, K.; Schulte, F.; Schwientek, P.; Wehlmann, H.; Wehmeier, U.F.; Pühler, A. The cytosolic and extracellular proteomes of Actinoplanes sp. SE50/110 led to the identification of gene products involved in acarbose metabolism. J. Biotechnol. 2013, 167, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Wendler, S.; Ortseifen, V.; Persicke, M.; Klein, A.; Neshat, A.; Niehaus, K.; Schneiker-Bekel, S.; Walter, F.; Wehmeier, U.F.; Kalinowski, J.; et al. Carbon source dependent biosynthesis of acarviose metabolites in Actinoplanes sp. SE50/110. J. Biotechnol. 2014, 191, 113–120. [Google Scholar] [CrossRef]
- Wendler, S.; Otto, A.; Ortseifen, V.; Bonn, F.; Neshat, A.; Schneiker-Bekel, S.; Walter, F.; Wolf, T.; Zemke, T.; Wehmeier, U.F.; et al. Comprehensive proteome analysis of Actinoplanes sp. SE50/110 highlighting the location of proteins encoded by the acarbose and the pyochelin biosynthesis gene cluster. J. Proteom. 2015, 125, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wendler, S.; Otto, A.; Ortseifen, V.; Bonn, F.; Neshat, A.; Schneiker-Bekel, S.; Wolf, T.; Zemke, T.; Wehmeier, U.F.; Hecker, M.; et al. Comparative proteome analysis of Actinoplanes sp. SE50/110 grown with maltose or glucose shows minor differences for acarbose biosynthesis proteins but major differences for saccharide transporters. J. Proteom. 2016, 131, 140–148. [Google Scholar] [CrossRef]
- Rockser, Y.; Wehmeier, U.F. The gac-gene cluster for the production of acarbose from Streptomyces glaucescens GLA.O: Identification, isolation and characterization. J. Biotechnol. 2009, 140, 114–123. [Google Scholar] [CrossRef]
- Guo, X.; Geng, P.; Bai, F.; Bai, G.; Sun, T.; Li, X.; Shi, L.; Zhong, Q. Draft genome sequence of Streptomyces coelicoflavus ZG0656 reveals the putative biosynthetic gene cluster of acarviostatin family α-amylase inhibitors. Lett. Appl. Microbiol. 2012, 55, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Droste, J.; Gren, T.; Ortseifen, V.; Schneiker-Bekel, S.; Zemke, T.; Pühler, A.; Kalinowski, J. The MalR type regulator AcrC is a transcriptional repressor of acarbose biosynthetic genes in Actinoplanes sp. SE50/110. BMC Genom. 2017, 18, 562. [Google Scholar] [CrossRef]
- Gren, T. Development and Application of Genetic Engineering Methods for Actinoplanes sp. SE50/110. Ph.D. Dissertation, Universität Bielefeld, Bielefeld, Germany, 2017. [Google Scholar]
- Zhao, Q.; Luo, Y.; Zhang, X.; Kang, Q.; Zhang, D.; Zhang, L.; Bai, L.; Deng, Z. A severe leakage of intermediates to shunt products in acarbose biosynthesis. Nat. Commun. 2020, 11, 1468. [Google Scholar] [CrossRef]
- Schaffert, L. Studies on the Acarviosyl-Maltose Metabolism in Actinoplanes sp. SE50/110 by Gene Deletion and Overexpression. Ph.D. Dissertation, Bielefeld University, Bielefeld, Germany, 2019. [Google Scholar]
- Gren, T.; Ortseifen, V.; Wibberg, D.; Schneiker-Bekel, S.; Bednarz, H.; Niehaus, K.; Zemke, T.; Persicke, M.; Pühler, A.; Kalinowski, J. Genetic engineering in Actinoplanes sp. SE50/110—Development of an intergeneric conjugation system for the introduction of actinophage-based integrative vectors. J. Biotechnol. 2016, 232, 79–88. [Google Scholar] [CrossRef]
- Schaffert, L.; Jacob, L.; Schneiker-Bekel, S.; Persicke, M.; März, C.; Rückert, C.; Pühler, A.; Kalinowski, J. pSETT4, an Improved φC31-Based Integrative Vector System for Actinoplanes sp. SE50/110. Microbiol. Resour. Announc. 2020, 9, e00596-20. [Google Scholar] [CrossRef] [PubMed]
- Schaffert, L.; März, C.; Burkhardt, L.; Droste, J.; Brandt, D.; Busche, T.; Rosen, W.; Schneiker-Bekel, S.; Persicke, M.; Pühler, A.; et al. Evaluation of vector systems and promoters for overexpression of the acarbose biosynthesis gene acbC in Actinoplanes sp. SE50/110. Microb. Cell Fact. 2019, 18, 114. [Google Scholar] [CrossRef]
- Zhao, Q.; Xie, H.; Peng, Y.; Wang, X.; Bai, L. Improving acarbose production and eliminating the by-product component C with an efficient genetic manipulation system of Actinoplanes sp. SE50/110. Synth. Syst. Biotechnol. 2017, 2, 302–309. [Google Scholar] [CrossRef] [PubMed]
- Gust, B.; Kieser, T.; Chater, K.F. REDIRECT Technology: PCR-Targeting System in Streptomyces coelicolor; The John Innes Centre: Norwich, UK, 2002. [Google Scholar]
- Wolf, T.; Gren, T.; Thieme, E.; Wibberg, D.; Zemke, T.; Pühler, A.; Kalinowski, J. Targeted genome editing in the rare actinomycete Actinoplanes sp. SE50/110 by using the CRISPR/Cas9 System. J. Biotechnol. 2016, 231, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Luzhetskyy, A.; Fedoryshyn, M.; Gromyko, O.; Ostash, B.; Rebets, Y.; Bechthold, A.; Fedorenko, V. IncP plasmids are most effective in mediating conjugation between Escherichia coli and Streptomycetes. Russ. J. Genet. 2006, 42, 476–481. [Google Scholar] [CrossRef]
- Horbal, L.; Zaburannyy, N.; Ostash, B.; Shulga, S.; Fedorenko, V. Manipulating the regulatory genes for teicoplanin production in Actinoplanes teichomyceticus. World J. Microbiol. Biotechnol. 2012, 28, 2095–2100. [Google Scholar] [CrossRef] [PubMed]
- Voeykova, T.; Emelyanova, L.; Tabakov, V.; Mkrtumyan, N. Transfer of plasmid pTO1 from Escherichia coli to various representatives of the order Actinomycetales by intergeneric conjugation. FEMS Microbiol. Lett. 1998, 162, 47–52. [Google Scholar] [CrossRef]
- Cobb, R.E.; Wang, Y.; Zhao, H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth. Biol. 2015, 4, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef]
- Arroyo-Olarte, R.D.; Bravo Rodríguez, R.; Morales-Ríos, E. Genome Editing in Bacteria: CRISPR-Cas and Beyond. Microorganisms 2021, 9, 844. [Google Scholar] [CrossRef] [PubMed]
- Barman, A.; Deb, B.; Chakraborty, S. A glance at genome editing with CRISPR-Cas9 technology. Curr. Genet. 2020, 66, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, W.; Shao, S.; Wang, Q. Gene Silencing Through CRISPR Interference in Bacteria: Current Advances and Future Prospects. Front. Microbiol. 2021, 12, 635227. [Google Scholar] [CrossRef]
- Schultenkämper, K.; Brito, L.F.; Wendisch, V.F. Impact of CRISPR interference on strain development in biotechnology. Biotechnol. Appl. Biochem. 2020, 67, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Grant, S.G.; Jessee, J.; Bloom, F.R.; Hanahan, D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA 1990, 87, 4645–4649. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics: John Innes Foundation; Norwich Research Park: Colney, UK, 2000; pp. 44–61. [Google Scholar]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Blin, K.; Pedersen, L.E.; Weber, T.; Lee, S.Y. CRISPy-web: An online resource to design sgRNAs for CRISPR applications. Synth. Syst. Biotechnol. 2016, 1, 118–121. [Google Scholar] [CrossRef] [PubMed]
- Muth, G.; Nußbaumer, B.; Wohlleben, W.; Pühler, A. A vector system with temperature-sensitive replication for gene disruption and mutational cloning in Streptomycetes. Mol. Gen. Genet. 1989, 219, 341–348. [Google Scholar] [CrossRef]
- Engler, C.; Marillonnet, S. Golden Gate cloning. Methods Mol. Biol. 2014, 1116, 119–131. [Google Scholar] [CrossRef]
- Rey, D.A.; Pühler, A.; Kalinowski, J. The putative transcriptional repressor McbR, member of the TetR-family, is involved in the regulation of the metabolic network directing the synthesis of sulfur containing amino acids in Corynebacterium glutamicum. J. Biotechnol. 2003, 103, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Wang, X. Method for Enhancing Positive Regulatory Protein Gene Expression to Increase Acarbose Fermentation Level. CN112592878A, 25 December 2020. [Google Scholar]
- Rudolph, M.M.; Vockenhuber, M.-P.; Suess, B. Synthetic riboswitches for the conditional control of gene expression in Streptomyces coelicolor. Microbiology 2013, 159, 1416–1422. [Google Scholar] [CrossRef] [PubMed]
- Uguru, G.C.; Mondhe, M.; Goh, S.; Hesketh, A.; Bibb, M.J.; Good, L.; Stach, J.E.M. Synthetic RNA Silencing of Actinorhodin Biosynthesis in Streptomyces coelicolor A3(2). PLoS ONE 2013, 8, e67509. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.L.; Heelan, W.J.; Ward, R.D.; Peters, J.M. CRISPRi functional genomics in bacteria and its application to medical and industrial research. Microbiol. Mol. Biol. Rev. 2024, 88, e0017022. [Google Scholar] [CrossRef]
- Schaffert, L.; Schneiker-Bekel, S.; Dymek, S.; Droste, J.; Persicke, M.; Busche, T.; Brandt, D.; Pühler, A.; Kalinowski, J. Essentiality of the Maltase AmlE in Maltose Utilization and Its Transcriptional Regulation by the Repressor AmlR in the Acarbose-Producing Bacterium Actinoplanes sp. SE50/110. Front. Microbiol. 2019, 10, 2448. [Google Scholar] [CrossRef]
- Zhang, D.; Zhao, Q.; Jiang, M.; Kang, Q.; Bai, L. Biosynthetic pathway of deoxyaminosugar moiety in acarbose from Actinoplanes sp. SE50/110. Acta Microbiol. Sin. 2020, 60, 118–134. (In Chinese) [Google Scholar] [CrossRef]
- Göttl, V.L.; Schmitt, I.; Braun, K.; Peters-Wendisch, P.; Wendisch, V.F.; Henke, N.A. CRISPRi-Library-Guided Target Identification for Engineering Carotenoid Production by Corynebacterium glutamicum. Microorganisms 2021, 9, 670. [Google Scholar] [CrossRef] [PubMed]
- Ameruoso, A.; Villegas Kcam, M.C.; Cohen, K.P.; Chappell, J. Activating natural product synthesis using CRISPR interference and activation systems in Streptomyces. Nucleic Acids Res. 2022, 50, 7751–7760. [Google Scholar] [CrossRef]
- Geissler, A.S.; Fehler, A.O.; Poulsen, L.D.; González-Tortuero, E.; Kallehauge, T.B.; Alkan, F.; Anthon, C.; Seemann, S.E.; Rasmussen, M.D.; Breüner, A.; et al. CRISPRi screen for enhancing heterologous α-amylase yield in Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2023, 50, kuac028. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Cao, Y.; Wang, Y.; Sun, L.; Song, H. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis. Microb. Cell Fact. 2019, 18, 90. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-H.; Shen, C.R.; Li, H.; Sung, L.-Y.; Wu, M.-Y.; Hu, Y.-C. CRISPR interference (CRISPRi) for gene regulation and succinate production in cyanobacterium S. elongatus PCC 7942. Microb. Cell Fact. 2016, 15, 196. [Google Scholar] [CrossRef]
- Craney, A.; Ahmed, S.; Nodwell, J. Towards a new science of secondary metabolism. J. Antibiot. 2013, 66, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, L.; Busche, T.; Bondzio, L.; Hütten, A.; Niehaus, K.; Schneiker-Bekel, S.; Pühler, A.; Kalinowski, J. Sigma Factor Engineering in Actinoplanes sp. SE50/110: Expression of the Alternative Sigma Factor Gene ACSP50_0507 (σHAs) Enhances Acarbose Yield and Alters Cell Morphology. Microorganisms 2024, 12, 1241. [Google Scholar] [CrossRef] [PubMed]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef]
- Mao, X.-M.; Luo, S.; Li, Y.-Q. Negative regulation of daptomycin production by DepR2, an ArsR-family transcriptional factor. J. Ind. Microbiol. Biotechnol. 2017, 44, 1653–1658. [Google Scholar] [CrossRef]
- Chen, H.; Wang, J.; Cui, J.; Wang, C.; Liang, S.; Liu, H.; Wen, J. Negative regulation of bleomycins biosynthesis by ArsR/SmtB family repressor BlmR in Streptomyces verticillus. Appl. Microbiol. Biotechnol. 2019, 103, 6629–6644. [Google Scholar] [CrossRef] [PubMed]
- Bibb, M.J. Regulation of secondary metabolism in Streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Horinouchi, S. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front Biosci 2002, 7, A897. [Google Scholar]
- Ryu, Y.-G.; Kim, E.-S.; Kim, D.-W.; Kim, S.-K.; Lee, K.-J. Differential stringent responses of Streptomyces coelicolor M600 to starvation of specific nutrients. J. Microbiol. Biotechnol. 2007, 17, 305–312. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Schwientek, P. Genomics and Transcriptomics of the Industiral Acarbose Produces Actinoplanes sp. SE50/110. Ph. D. Dissertation, Bielefeld University, Bielefeld, Germany, 2012. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dymek, S.; Jacob, L.; Pühler, A.; Kalinowski, J. Targeting Transcriptional Regulators Affecting Acarbose Biosynthesis in Actinoplanes sp. SE50/110 Using CRISPRi Silencing. Microorganisms 2025, 13, 1. https://doi.org/10.3390/microorganisms13010001
Dymek S, Jacob L, Pühler A, Kalinowski J. Targeting Transcriptional Regulators Affecting Acarbose Biosynthesis in Actinoplanes sp. SE50/110 Using CRISPRi Silencing. Microorganisms. 2025; 13(1):1. https://doi.org/10.3390/microorganisms13010001
Chicago/Turabian StyleDymek, Saskia, Lucas Jacob, Alfred Pühler, and Jörn Kalinowski. 2025. "Targeting Transcriptional Regulators Affecting Acarbose Biosynthesis in Actinoplanes sp. SE50/110 Using CRISPRi Silencing" Microorganisms 13, no. 1: 1. https://doi.org/10.3390/microorganisms13010001
APA StyleDymek, S., Jacob, L., Pühler, A., & Kalinowski, J. (2025). Targeting Transcriptional Regulators Affecting Acarbose Biosynthesis in Actinoplanes sp. SE50/110 Using CRISPRi Silencing. Microorganisms, 13(1), 1. https://doi.org/10.3390/microorganisms13010001





