Dysfunction of Complementarity Determining Region 1 Encoded by T Cell Receptor Beta Variable Gene Is Potentially Associated with African Swine Fever Virus Infection in Pigs
Abstract
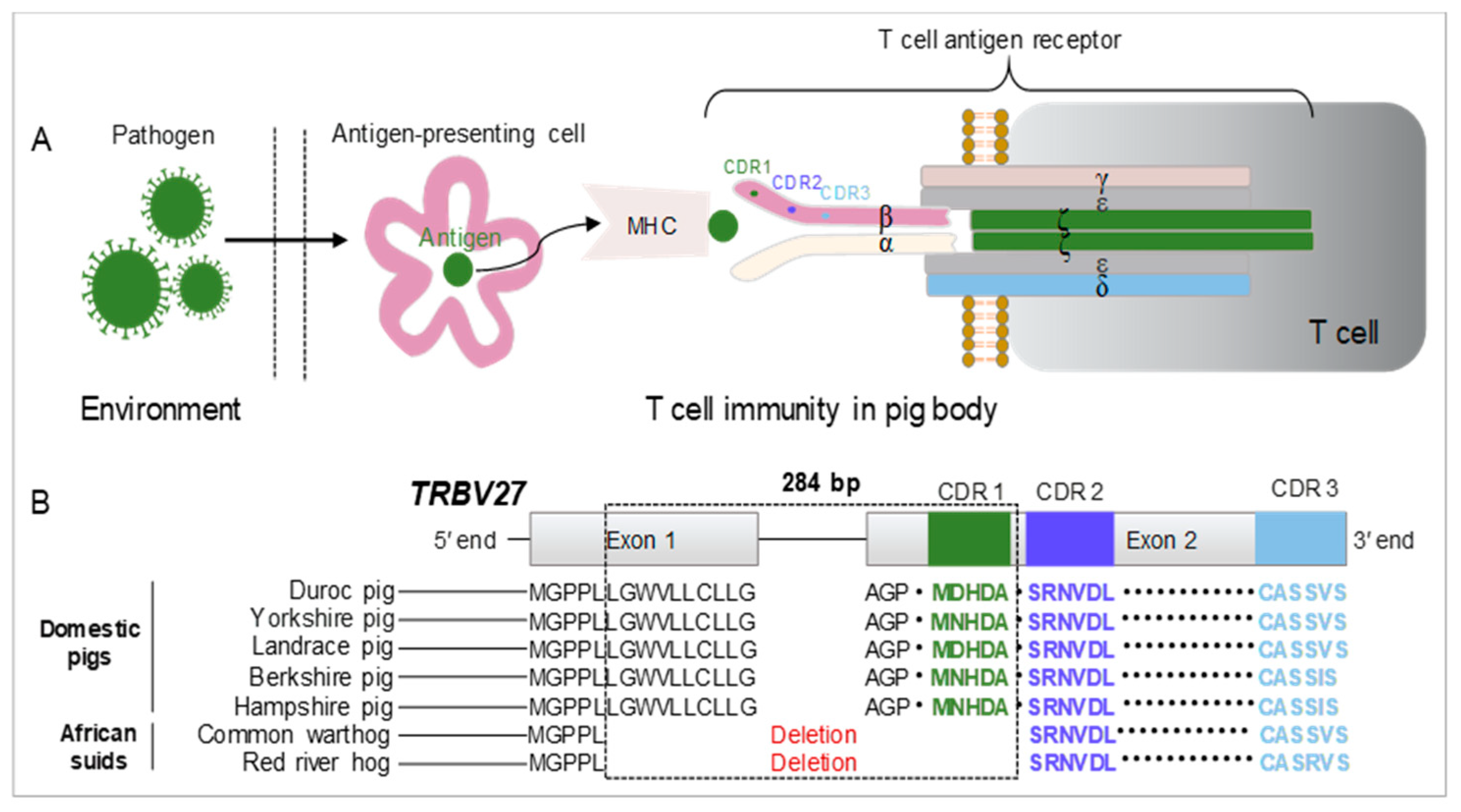
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Construction of TRBV-Edited Pigs
2.3. Live African Swine Fever Virus
2.4. Detection of African Swine Fever Virus
2.5. Animal Infection Experiments
2.6. Monitoring of Infection Intensity and Disease Process
2.7. Transcriptome Analysis of the TRBV Gene Family
2.8. Statistical Analysis
3. Results
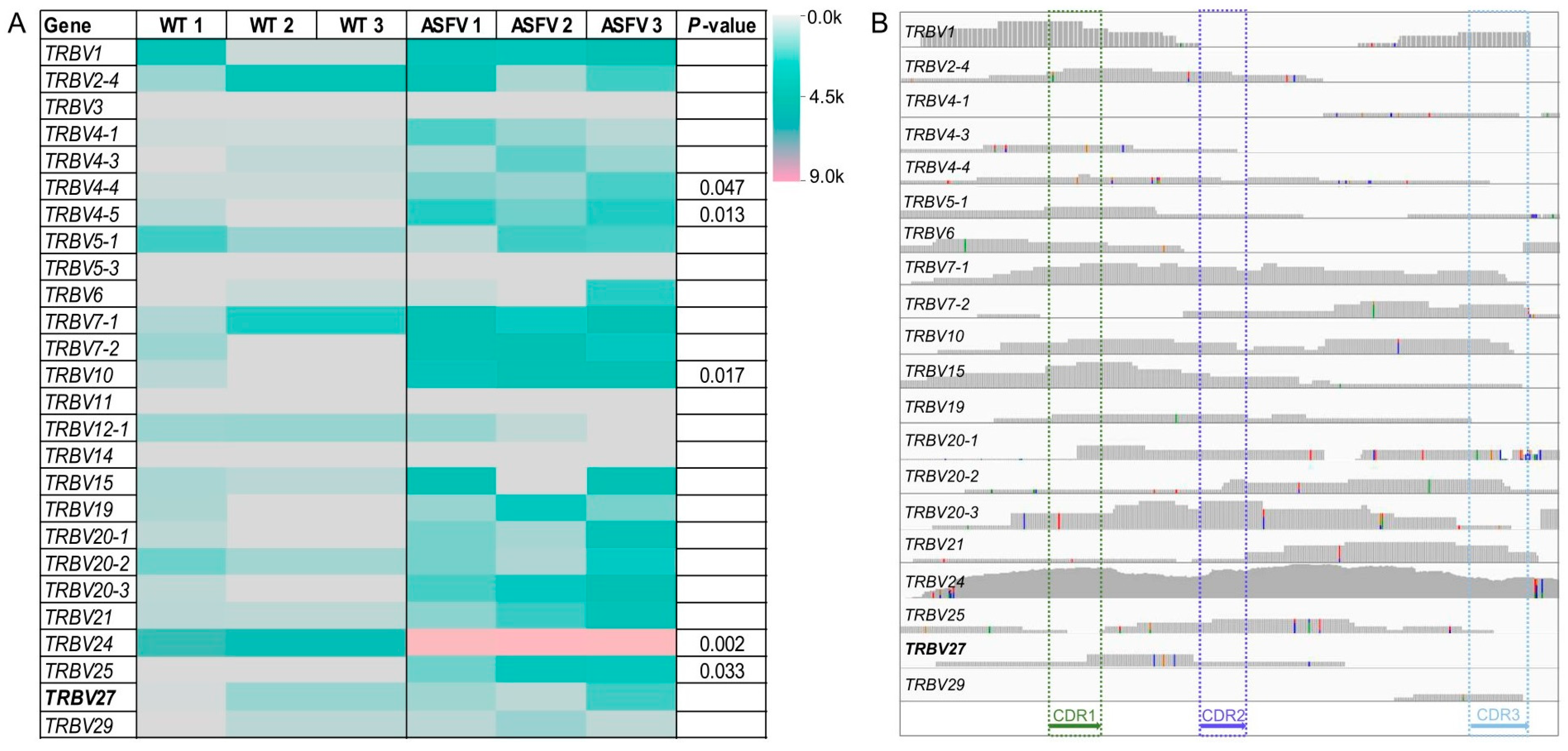
3.1. Expression Profile of the TRBV Gene Family in ASFV-Infected Pigs
3.2. Generation of TRBV27-Edited Pigs
3.3. Replication of ASFV in TRBV27-Edited and Wild-Type Pigs
3.4. Differences in Clinical Signs and Organ Lesions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Galindo, I.; Alonso, C. African swine fever virus: A review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef]
- Wang, F.X.; Zhang, H.; Hou, L.N.; Yang, C.; Wen, Y.J. Advance of African swine fever virus in recent years. Res. Vet. Sci. 2021, 136, 535–539. [Google Scholar] [CrossRef]
- Sanchez-Cordon, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Zeng, D.X.; Qian, B.X.; Li, Y.F.; Zong, K.; Peng, W.Q.; Liao, K.; Yu, X.F.; Sun, J.J.; Lv, X.Y.; Ding, L.; et al. Prospects for the application of infectious virus detection technology based on propidium monoazide in African swine fever management. Front. Microbiol. 2022, 13, 1025758. [Google Scholar] [CrossRef]
- Dixon, L.K.; Sun, H.; Roberts, H. African swine fever. Antivir. Res. 2019, 165, 34–41. [Google Scholar] [CrossRef]
- Wu, K.K.; Liu, J.M.; Wang, L.X.; Fan, S.Q.; Li, Z.Y.; Li, Y.W.; Yi, L.; Ding, H.X.; Zhao, M.Q.; Chen, J.D. Current state of global African swine fever vaccine development under the prevalence and transmission of ASF in China. Vaccines 2020, 8, 531. [Google Scholar] [CrossRef]
- Penrith, M.L. African swine fever. Onderstepoort J. Vet. Res. 2009, 76, 91–95. [Google Scholar] [CrossRef]
- Jo, E.K. Interplay between host and pathogen: Immune defense and beyond. Exp. Mol. Med. 2019, 51, 1–3. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Ai, Q.Y.; Huang, S.L.; Ou, Y.T.; Gao, Y.Z.; Tong, T.Z.; Fan, H.Y. Immune escape mechanism and vaccine research progress of African swine fever virus. Vaccines 2022, 10, 344. [Google Scholar] [CrossRef]
- Munoz-Moreno, R.; Galindo, I.; Cuesta-Geijo, M.A.; Barrado-Gil, L.; Alonso, C. Host cell targets for African swine fever virus. Virus Res. 2015, 209, 118–127. [Google Scholar] [CrossRef]
- Galindo, I.; Cuesta-Geijo, M.A.; Hlavova, K.; Munoz-Moreno, R.; Barrado-Gil, L.; Dominguez, J.; Alonso, C. African swine fever virus infects macrophages, the natural host cells, via clathrin- and cholesterol-dependent endocytosis. Virus Res. 2015, 200, 45–55. [Google Scholar] [CrossRef]
- Franzoni, G.; Graham, S.P.; Giudici, S.D.; Bonelli, P.; Pilo, G.; Anfossi, A.G.; Pittau, M.; Nicolussi, P.S.; Laddomada, A.; Oggiano, A. Characterization of the interaction of African swine fever virus with monocytes and derived macrophage subsets. Vet. Microbiol. 2017, 198, 88–98. [Google Scholar] [CrossRef]
- Lithgow, P.; Takamatsu, H.; Werling, D.; Dixon, L.; Chapman, D. Correlation of cell surface marker expression with African swine fever virus infection. Vet. Microbiol. 2014, 168, 413–419. [Google Scholar] [CrossRef]
- Sanchez-Torres, C.; Gomez-Puertas, P.; Gomez-del-Moral, M.; Alonso, F.; Escribano, J.M.; Ezquerra, A.; Dominguez, J. Expression of porcine CD163 on monocytes/macrophages correlates with permissiveness to African swine fever infection. Arch. Virol. 2003, 148, 2307–2323. [Google Scholar] [CrossRef]
- Van Gorp, H.; Van Breedam, W.; Van Doorsselaere, J.; Delputte, P.L.; Nauwynck, H.J. Identification of the CD163 protein domains involved in infection of the porcine reproductive and respiratory syndrome virus. J. Virol. 2010, 84, 3101–3105. [Google Scholar] [CrossRef]
- Popescu, L.; Gaudreault, N.N.; Whitworth, K.M.; Murgia, M.V.; Nietfeld, J.C.; Mileham, A.; Samuel, M.; Wells, K.D.; Prather, R.S.; Rowland, R.R.R. Genetically edited pigs lacking CD163 show no resistance following infection with the African swine fever virus isolate, Georgia 2007/1. Virology 2017, 501, 102–106. [Google Scholar] [CrossRef]
- Sun, L.; Su, Y.; Jiao, A.; Wang, X.; Zhang, B. T cells in health and disease. Signal Transduct. Target. Ther. 2023, 8, 235. [Google Scholar]
- Davis, M.M. T cell receptor gene diversity and selection. Annu. Rev. Biochem. 1990, 59, 475–496. [Google Scholar] [CrossRef]
- Eguchi-Ogawa, T.; Toki, D.; Uenishi, H. Genomic structure of the whole D-J-C clusters and the upstream region coding V segments of the TRB locus in pig. Dev. Comp. Immunol. 2009, 33, 1111–1119. [Google Scholar] [CrossRef]
- Irvine, D.J.; Purbhoo, M.A.; Krogsgaard, M.; Davis, M.M. Direct observation of ligand recognition by T cells. Nature 2002, 419, 845–849. [Google Scholar] [CrossRef]
- Demuth, J.P.; Hahn, M.W. The life and death of gene families. Bioessays 2009, 31, 29–39. [Google Scholar] [CrossRef]
- Olinski, R.P.; Lundin, L.G.; Hallbook, F. Genome duplication-driven evolution of gene families: Insights from the formation of the insulin family. Ann. N. Y. Acad. Sci. 2005, 1040, 426–428. [Google Scholar] [CrossRef]
- Massari, S.; Bellini, M.; Ciccarese, S.; Antonacci, R. Overview of the germline and expressed repertoires of the TRB genes in Sus scrofa. Front. Immunol. 2018, 9, 2526. [Google Scholar] [CrossRef]
- Xie, H.B.; Yan, C.; Adeola, A.C.; Wang, K.; Huang, C.P.; Xu, M.M.; Qiu, Q.; Yin, X.; Fan, C.Y.; Ma, Y.F.; et al. African suid genomes provide insights into the local adaptation to diverse African environments. Mol. Biol. Evol. 2022, 39, msac256. [Google Scholar] [CrossRef]
- Yang, L.H.; Church, G.; Zhao, H.Y.; Huang, L.S.; Gao, Y.B.; Wei, H.J.; Yang, G. Porcine germline genome engineering. Proc. Natl. Acad. Sci. USA 2021, 118, e2004836117. [Google Scholar] [CrossRef]
- Niu, D.; Wei, H.J.; Lin, L.; George, H.; Wang, T.; Lee, H.; Zhao, H.Y.; Wang, Y.; Kan, Y.N.; Shrock, E.; et al. Inactivation of porcine endogenous retrovirus in pigs using CRISPR-Cas9. Science 2017, 357, 1303–1307. [Google Scholar] [CrossRef]
- Yu, H.H.; Zhao, H.; Qing, Y.B.; Pan, W.R.; Jia, B.Y.; Zhao, H.Y.; Huang, X.X.; Wei, H.J. Porcine zygote injection with Cas9/sgRNA results in DMD-modified pig with muscle dystrophy. Int. J. Mol. Sci. 2016, 17, 1668. [Google Scholar] [CrossRef]
- Cheng, W.M.; Zhao, H.; Yu, H.H.; Xin, J.G.; Wang, J.; Zeng, L.Y.; Yuan, Z.M.; Qing, Y.B.; Li, H.H.; Jia, B.Y.; et al. Efficient generation of GGTA1-null Diannan miniature pigs using TALENs combined with somatic cell nuclear transfer. Reprod. Biol. Endocrin. 2016, 14, 77. [Google Scholar] [CrossRef]
- Shen, Y.F.; Xu, K.X.; Yuan, Z.M.; Guo, J.X.; Zhao, H.; Zhang, X.Z.; Zhao, L.; Qing, Y.B.; Li, H.H.; Pan, W.R.; et al. Efficient generation of P53 biallelic knockout miniature pigs via TALENs and somatic cell nuclear transfer. J. Transl. Med. 2017, 15, 224. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Lee, K.; Benne, J.A.; Beaton, B.P.; Spate, L.D.; Murphy, S.L.; Samuel, M.S.; Mao, J.; O’Gorman, C.; Walters, E.M.; et al. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol. Reprod. 2014, 91, 78. [Google Scholar] [CrossRef]
- Zhang, J.; Khazalwa, E.M.; Abkallo, H.M.; Zhou, Y.; Nie, X.; Ruan, J.; Zhao, C.; Wang, J.; Xu, J.; Li, X.; et al. The advancements, challenges, and future implications of the CRISPR/Cas9 system in swine research. J. Genet. Genom. 2021, 48, 347–360. [Google Scholar] [CrossRef]
- Wei, H.; Qing, Y.; Pan, W.; Zhao, H.; Li, H.; Cheng, W.; Zhao, L.; Xu, C.; Li, H.; Li, S.; et al. Comparison of the efficiency of Banna miniature inbred pig somatic cell nuclear transfer among different donor cells. PLoS ONE 2013, 8, e57728. [Google Scholar] [CrossRef]
- Zhao, D.; Liu, R.; Zhang, X.; Li, F.; Wang, J.; Zhang, J.; Liu, X.; Wang, L.; Zhang, J.; Wu, X.; et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes Infect. 2019, 8, 438–447. [Google Scholar] [CrossRef]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.; Drew, T.W. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef]
- Galindo-Cardiel, I.; Ballester, M.; Solanes, D.; Nofrarias, M.; Lopez-Soria, S.; Argilaguet, J.M.; Lacasta, A.; Accensi, F.; Rodriguez, F.; Segales, J. Standardization of pathological investigations in the framework of experimental ASFV infections. Virus Res. 2013, 173, 180–190. [Google Scholar] [CrossRef]
- Sun, H.L.; Niu, Q.L.; Yang, J.F.; Zhao, Y.R.; Tian, Z.C.; Fan, J.; Zhang, Z.H.; Wang, Y.W.; Geng, S.X.; Zhang, Y.L.; et al. Transcriptome profiling reveals features of immune response and metabolism of acutely infected, dead and asymptomatic infection of African swine fever virus in pigs. Front. Immunol. 2021, 12, 808545. [Google Scholar] [CrossRef]
- Babraham Bioinformatics: Fastqc: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 April 2024).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; No. LBNL-7065E; Ernest Orlando Lawrence Berkeley National Laboratory: Berkeley, CA, USA, 2014. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Jori, F.; Bastos, A.D. Role of wild suids in the epidemiology of African swine fever. EcoHealth 2009, 6, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Dayhoff, M.O. The origin and evolution of protein superfamilies. Fed. Proc. 1976, 35, 2132–2138. [Google Scholar] [PubMed]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Xing, H.; Liu, K.; Fan, N.; Xu, K.; Zhao, H.; Jiao, D.; Wei, T.; Cheng, W.; Guo, J.; et al. Dysfunction of Complementarity Determining Region 1 Encoded by T Cell Receptor Beta Variable Gene Is Potentially Associated with African Swine Fever Virus Infection in Pigs. Microorganisms 2024, 12, 1113. https://doi.org/10.3390/microorganisms12061113
Li J, Xing H, Liu K, Fan N, Xu K, Zhao H, Jiao D, Wei T, Cheng W, Guo J, et al. Dysfunction of Complementarity Determining Region 1 Encoded by T Cell Receptor Beta Variable Gene Is Potentially Associated with African Swine Fever Virus Infection in Pigs. Microorganisms. 2024; 12(6):1113. https://doi.org/10.3390/microorganisms12061113
Chicago/Turabian StyleLi, Jiayu, Huiyan Xing, Kai Liu, Ninglin Fan, Kaixiang Xu, Heng Zhao, Deling Jiao, Taiyun Wei, Wenjie Cheng, Jianxiong Guo, and et al. 2024. "Dysfunction of Complementarity Determining Region 1 Encoded by T Cell Receptor Beta Variable Gene Is Potentially Associated with African Swine Fever Virus Infection in Pigs" Microorganisms 12, no. 6: 1113. https://doi.org/10.3390/microorganisms12061113
APA StyleLi, J., Xing, H., Liu, K., Fan, N., Xu, K., Zhao, H., Jiao, D., Wei, T., Cheng, W., Guo, J., Zhang, X., Zhu, F., Bu, Z., Zhao, D., Wang, W., & Wei, H.-J. (2024). Dysfunction of Complementarity Determining Region 1 Encoded by T Cell Receptor Beta Variable Gene Is Potentially Associated with African Swine Fever Virus Infection in Pigs. Microorganisms, 12(6), 1113. https://doi.org/10.3390/microorganisms12061113






