Assessment of the Safety and Probiotic Properties of Enterococcus faecium B13 Isolated from Fermented Chili
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Whole-Genome Sequencing
2.2.1. Genomic DNA Extraction
2.2.2. Illumina and PacBio Sequencing
2.2.3. Average Nucleotide Identity
2.2.4. Identification of Virulence Factors and Antibiotic Resistance Genes
2.3. Phenotypic Safety and Probiotic Characteristics Assessment
2.3.1. Acid and Bile Salt Tolerance
2.3.2. Cell Surface Hydrophobicity
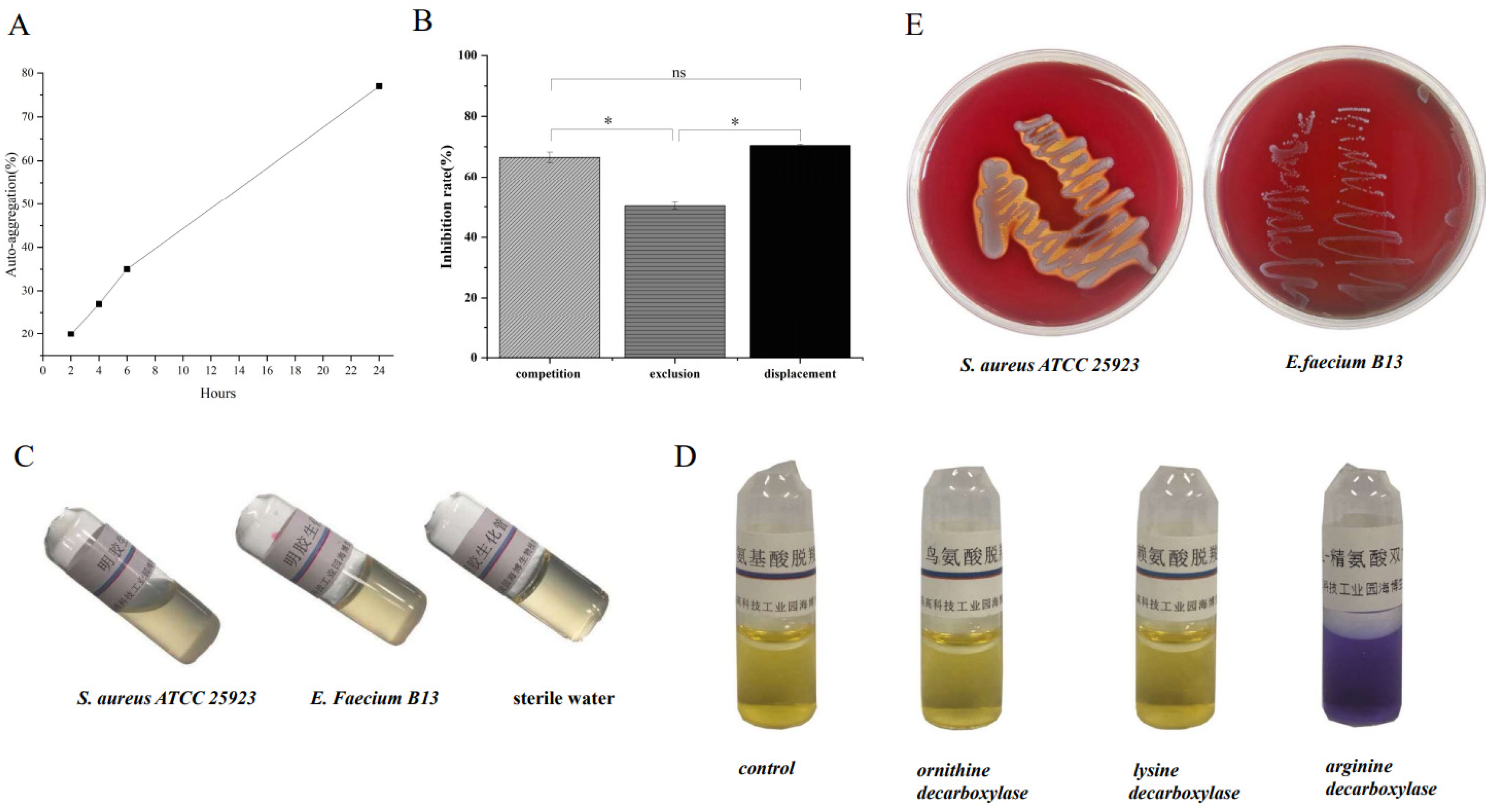
2.3.3. Auto-Aggregation
2.3.4. Antioxidant Activity and Total Antioxidant Capacity
2.3.5. Bile Salt Hydrolase (BSH) Activity
2.3.6. Gelatinase Activity
2.3.7. Decarboxylase Activity
2.3.8. Indole Experiment
2.3.9. Hemolysin Activity
2.3.10. Antibiotic Susceptibility
2.3.11. Cell Adhesion In Vitro
2.3.12. Inhibitive Pathogen Adhesion
2.4. Effects In Vivo
2.4.1. Animals and Bacteria
2.4.2. Intestinal Distribution of E. faecium B13
2.4.3. Animals Experiment
2.4.4. Metagenome Sequencing of Colon Microbiota
2.5. Statistical Analysis
3. Results and Discussion
3.1. The Whole Genome Sequence Analysis of the E. faecium B13
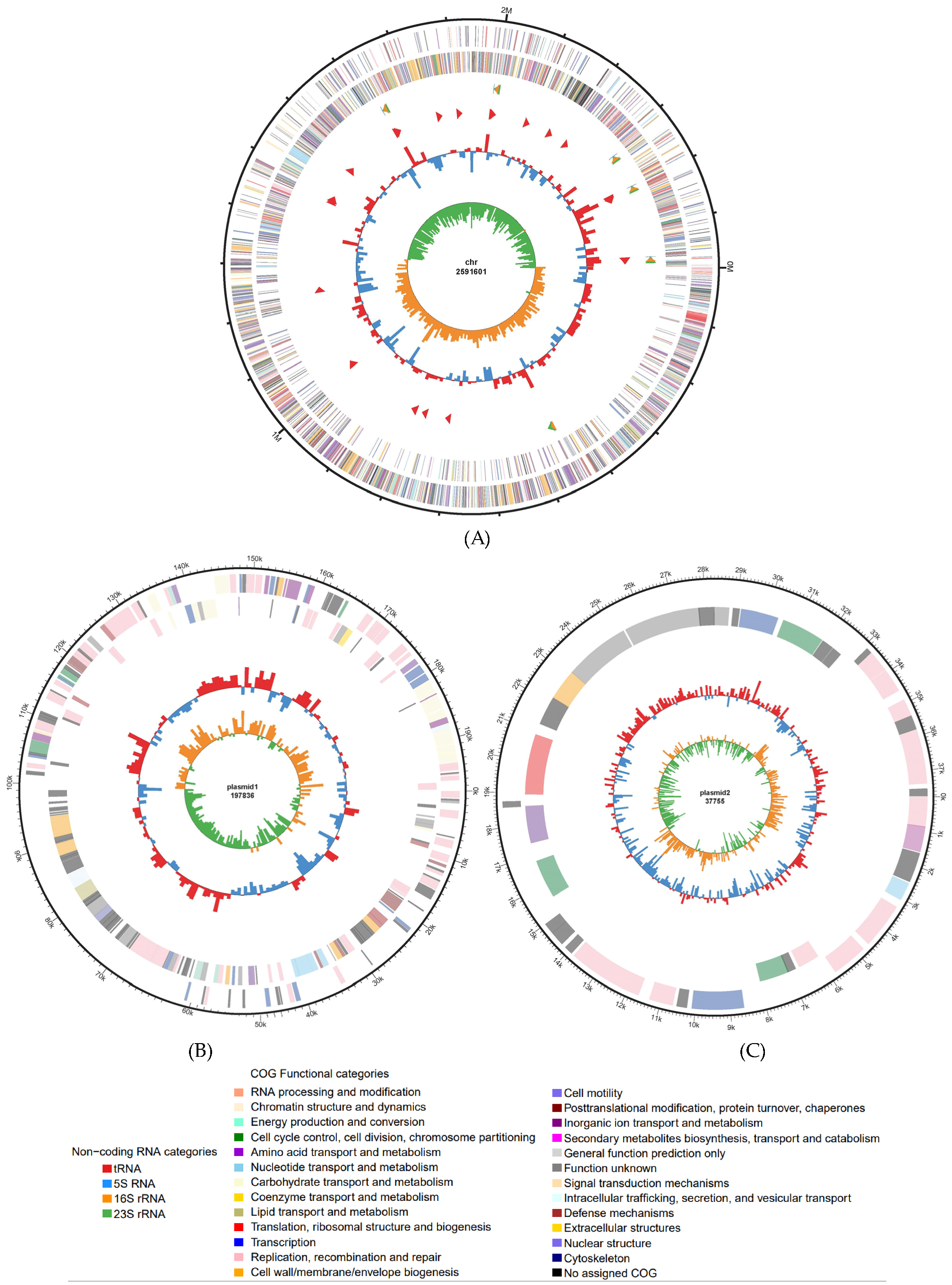
3.1.1. General Genome Features
3.1.2. Functional Annotation
3.1.3. Average Nucleotide Identity
3.1.4. Virulence Genes
3.1.5. Antibiotic Resistance Genes
3.2. Probiotic Properties Assessment
3.2.1. Acid and Bile Salt Tolerance
3.2.2. Antioxidant Activity In Vitro
3.2.3. Cell Adhesion
3.2.4. Hydrophobicity and Auto-Aggregation
3.3. Safety Assessment In Vitro
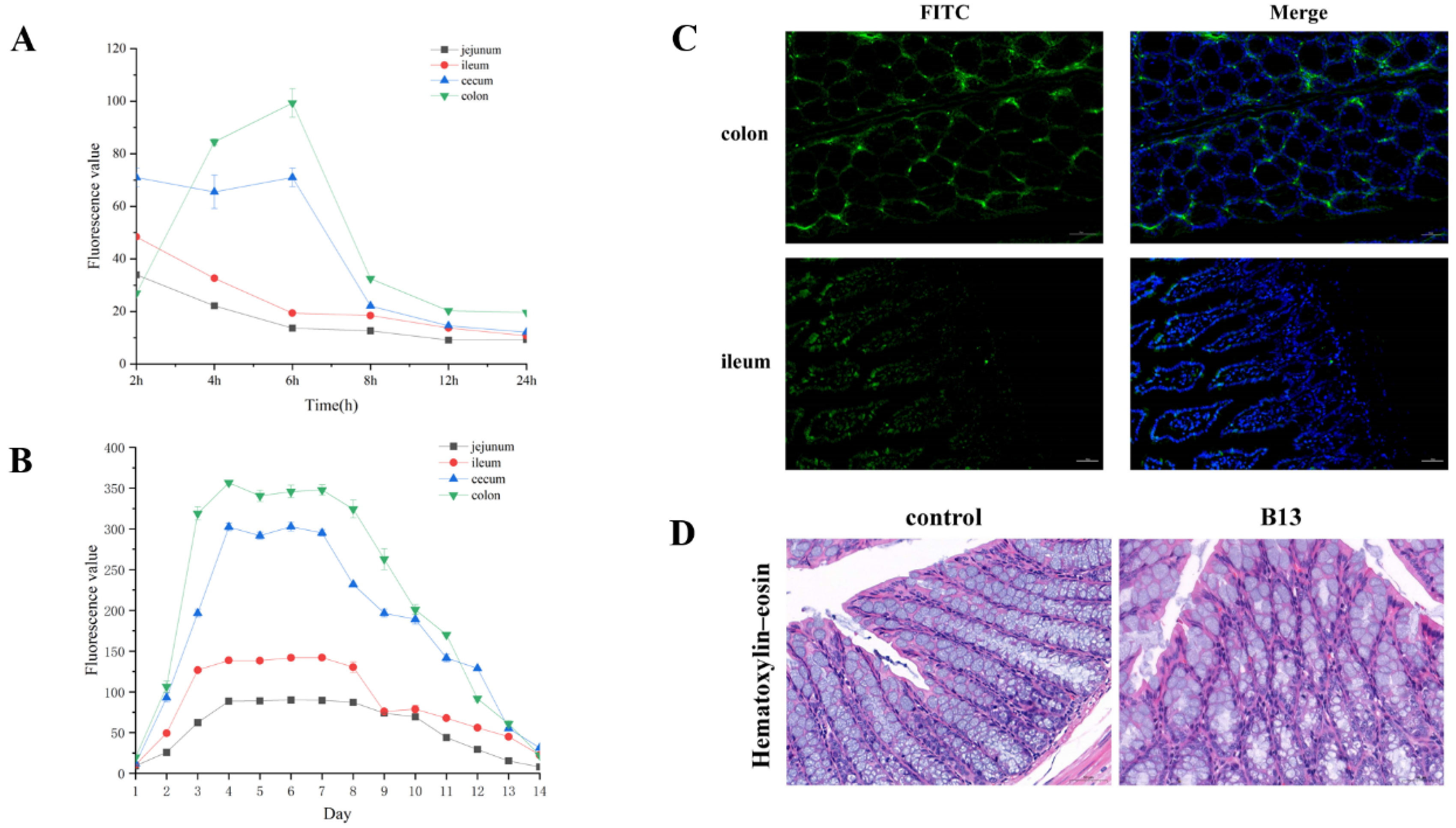
3.4. Intestinal Localization of E. faecium B13
3.5. Animal Experiment
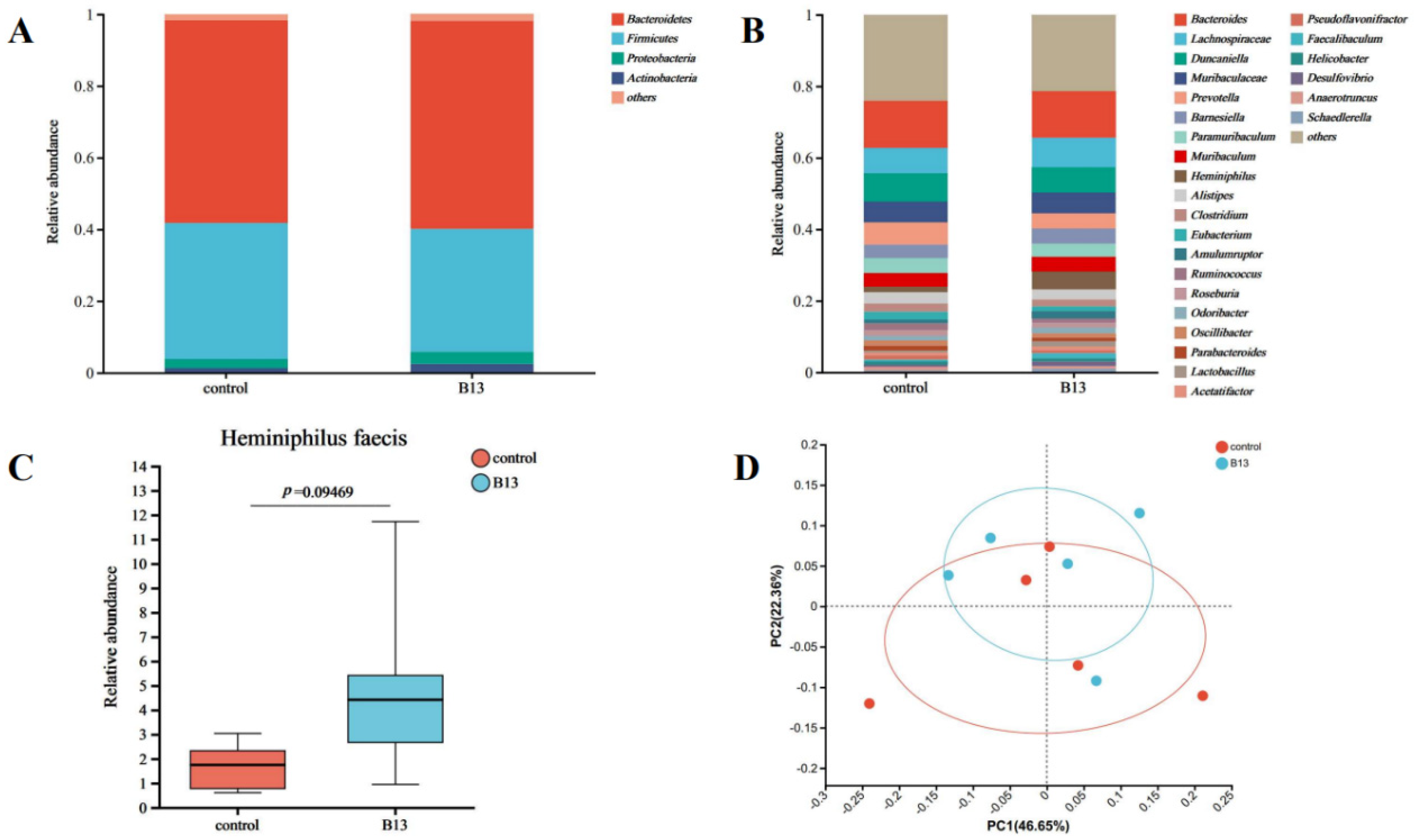
3.6. Metagenome Sequencing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yoon, S.; Lee, Y.J. Molecular Characteristics of Enterococcus faecalis and Enterococcus faecium from Bulk Tank Milk in Korea. Animals 2021, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Koo, M. Occurrence, Antimicrobial Resistance and Molecular Diversity of Enterococcus faecium in Processed Pork Meat Products in Korea. Foods 2020, 9, 1283. [Google Scholar] [CrossRef] [PubMed]
- Amaral, D.M.F.; Silva, L.F.; Casarotti, S.N.; Nascimento, L.C.S.; Penna, A.L.B. Enterococcus faecium and Enterococcus durans isolated from cheese: Survival in the presence of medications under simulated gastrointestinal conditions and adhesion properties. J. Dairy Sci. 2017, 100, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Žugić Petrović, T.D.; Ilić, P.D.; Grujović, M.; Mladenović, K.G.; Kocić-Tanackov, S.D.; Čomić, L.R. Assessment of safety aspect and probiotic potential of autochthonous Enterococcus faecium strains isolated from spontaneous fermented sausage. Biotechnol. Lett. 2020, 42, 1513–1525. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Choi, S.I.; Jeong, Y.; Kang, C.H. Evaluation of Safety and Probiotic Potential of Enterococcus faecalis MG5206 and Enterococcus faecium MG5232 Isolated from Kimchi, a Korean Fermented Cabbage. Microorganisms 2022, 10, 2070. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Valenzuela, A.; Lavilla Lerma, L.; Benomar, N.; Gálvez, A.; Pérez Pulido, R.; Abriouel, H. Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne Pathog. Dis. 2013, 10, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Villaluenga, C.; Torino, M.I.; Martín, V.; Arroyo, R.; Garcia-Mora, P.; Estrella Pedrola, I.; Vidal-Valverde, C.; Rodriguez, J.M.; Frias, J. Multifunctional properties of soy milk fermented by Enterococcus faecium strains isolated from raw soy milk. J. Agric. Food Chem. 2012, 60, 10235–10244. [Google Scholar] [CrossRef] [PubMed]
- Quintela-Baluja, M.; Jobling, K.; Graham, D.W.; Tabraiz, S.; Shamurad, B.; Alnakip, M.; Böhme, K.; Barros-Velázquez, J.; Carrera, M.; Calo-Mata, P. Rapid Proteomic Characterization of Bacteriocin-Producing Enterococcus faecium Strains from Foodstuffs. Int. J. Mol. Sci. 2022, 23, 13830. [Google Scholar] [CrossRef] [PubMed]
- Centeno, J.A.; Lorenzo, J.M.; Carballo, J. Effects of autochthonous Kluyveromyces lactis and commercial Enterococcus faecium adjunct cultures on the volatile profile and the sensory characteristics of short-ripened acid-curd Cebreiro cheese. Food Microbiol. 2022, 108, 104101. [Google Scholar] [CrossRef]
- Hassanzadazar, H.; Ehsani, A.; Mardani, K. Antibacterial activity of Enterococcus faecium derived from Koopeh cheese against Listeria monocytogenes in probiotic ultra-filtrated cheese. Vet. Res. Forum 2014, 5, 169–175. [Google Scholar]
- Tsanasidou, C.; Asimakoula, S.; Sameli, N.; Fanitsios, C.; Vandera, E.; Bosnea, L.; Koukkou, A.I.; Samelis, J. Safety Evaluation, Biogenic Amine Formation, and Enzymatic Activity Profiles of Autochthonous Enterocin-Producing Greek Cheese Isolates of the Enterococcus faecium/durans Group. Microorganisms 2021, 9, 777. [Google Scholar] [CrossRef] [PubMed]
- Yerlikaya, O.; Akbulut, N. Potential use of probiotic Enterococcus faecium and Enterococcus durans strains in Izmir Tulum cheese as adjunct culture. J. Food Sci. Technol. 2019, 56, 2175–2185. [Google Scholar] [CrossRef] [PubMed]
- Kern, M.; Aschenbach, J.R.; Tedin, K.; Pieper, R.; Loss, H.; Lodemann, U. Characterization of Inflammasome Components in Pig Intestine and Analysis of the Influence of Probiotic Enterococcus faecium during an Escherichia coli Challenge. Immunol. Investig. 2017, 46, 742–757. [Google Scholar] [CrossRef] [PubMed]
- Tarasova, E.; Yermolenko, E.; Donets, V.; Sundukova, Z.; Bochkareva, A.; Borshev, I.; Suvorova, M.; Ilyasov, I.; Simanenkov, V.; Suvorov, A.N. The influence of probiotic Enterococcus faecium strain L5 on the microbiota and cytokines expression in rats with dysbiosis induced by antibiotics. Benef. Microbes 2010, 1, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Benmouna, Z.; Dalache, F.; Zadi-Karam, H.; Karam, N.E.; Vuotto, C. Ability of Three Lactic Acid Bacteria to Grow in Sessile Mode and to Inhibit Biofilm Formation of Pathogenic Bacteria. Adv. Exp. Med. Biol. 2020, 1282, 105–114. [Google Scholar] [PubMed]
- Griffin, M.E.; Espinosa, J.; Becker, J.L.; Luo, J.D.; Carroll, T.S.; Jha, J.K.; Fanger, G.R.; Hang, H.C. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 2021, 373, 1040–1046. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chai, W.; Burwinkel, M.; Twardziok, S.; Wrede, P.; Palissa, C.; Esch, B.; Schmidt, M.F. Inhibitory influence of Enterococcus faecium on the propagation of swine influenza A virus in vitro. PLoS ONE 2013, 8, e53043. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.S.; Molina, M.A.; Di Sciullo, P.; Azpiroz, M.B.; Leocata Nieto, F.; Sterín-Speziale, N.B.; Mongini, C.; Manghi, M.A. Beneficial activity of Enterococcus faecalis CECT7121 in the anti-lymphoma protective response. J. Appl. Microbiol. 2010, 109, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Thirabunyanon, M.; Hongwittayakorn, P. Potential probiotic lactic acid bacteria of human origin induce antiproliferation of colon cancer cells via synergic actions in adhesion to cancer cells and short-chain fatty acid bioproduction. Appl. Biochem. Biotechnol. 2013, 169, 511–525. [Google Scholar] [CrossRef]
- Guo, L.; Li, T.; Tang, Y.; Yang, L.; Huo, G. Probiotic properties of Enterococcus strains isolated from traditional naturally fermented cream in China. Microb. Biotechnol. 2016, 9, 737–745. [Google Scholar] [CrossRef]
- Molina, M.A.; Díaz, A.M.; Hesse, C.; Ginter, W.; Gentilini, M.V.; Nuñez, G.G.; Canellada, A.M.; Sparwasser, T.; Berod, L.; Castro, M.S.; et al. Immunostimulatory Effects Triggered by Enterococcus faecalis CECT7121 Probiotic Strain Involve Activation of Dendritic Cells and Interferon-Gamma Production. PLoS ONE 2015, 10, e0127262. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.; Arini, A.; Aeschbacher, M.; Coppolecchia, R.; Pot, B. Enterococcus faecium SF68 as a model for efficacy and safety evaluation of pharmaceutical probiotics. Benef. Microbes 2018, 9, 375–388. [Google Scholar] [CrossRef]
- Arai, E.N.; Yoneda, S.; Yoneda, N.; Ito, M.; Tsuda, S.; Shiozaki, A.; Nohira, T.; Hyodo, H.; Kumazawa, K.; Suzuki, T.; et al. Probiotics including Clostridium butyricum, Enterococcus faecium, and Bacillus subtilis may prevent recurrent spontaneous preterm delivery. J. Obstet. Gynaecol. Res. 2022, 48, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Livermore, D.M. Infections caused by Gram-positive bacteria: A review of the global challenge. J. Infect. 2009, 59 (Suppl. S1), S4–S16. [Google Scholar] [CrossRef] [PubMed]
- Golob, M.; Pate, M.; Kušar, D.; Dermota, U.; Avberšek, J.; Papić, B.; Zdovc, I. Antimicrobial Resistance and Virulence Genes in Enterococcus faecium and Enterococcus faecalis from Humans and Retail Red Meat. Biomed. Res. Int. 2019, 2019, 2815279. [Google Scholar] [CrossRef] [PubMed]
- Willems, R.J.; van Schaik, W. Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol. 2009, 4, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Mirzaii, M.; Alebouyeh, M.; Sohrabi, M.B.; Eslami, P.; Fazli, M.; Ebrahimi, M.; HajiAsgarli, P.; Rashidan, M. Antibiotic resistance assessment and multi-drug efflux pumps of Enterococcus faecium isolated from clinical specimens. J. Infect. Dev. Ctries. 2023, 17, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Zouain, M.G.; Araj, G.F. Antimicrobial resistance of Enterococci in Lebanon. Int. J. Antimicrob. Agents 2001, 17, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Oh, T.; Baek, S.Y. Multidrug Resistance, Biofilm Formation, and Virulence of Escherichia coli Isolates from Commercial Meat and Vegetable Products. Foodborne Pathog. Dis. 2018, 15, 782–789. [Google Scholar] [CrossRef]
- Yoon, Y.K.; Park, G.C.; An, H.; Chun, B.C.; Sohn, J.W.; Kim, M.J. Trends of Antibiotic Consumption in Korea According to National Reimbursement Data (2008–2012): A Population-Based Epidemiologic Study. Medicine 2015, 94, e2100. [Google Scholar] [CrossRef]
- Ding, S.; Guo, C.-H.; Zhang, Z.-F.; Bai, X.; Wei, J.; Zhang, M.; Luo, F. Effects of Enterococcus faecium B13 Yielding Bacteriocins on Growth Performance, Nutrient Digestibility, Serum Immune Indexes and Fecal Microbiota of Weaned Piglets. Acta Vet. Zootech. Sin. 2017, 48, 1902–1911. [Google Scholar]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, J.; Yu, J.; Yao, Z.; Sun, L.; Shen, Y.; Jin, Q. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Hamada, T.; Sameshima, Y.; Honda, K.; Omasa, T.; Kato, J.; Ohtake, H. A comparison of various methods to predict bacterial predilection for organic solvents used as reaction media. J. Biosci. Bioeng. 2008, 106, 357–362. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Rubayet Ul Alam, A.S.M.; Jahid, I.K. Characterization and evaluation of lactic acid bacteria from indigenous raw milk for potential probiotic properties. J. Dairy Sci. 2020, 103, 1223–1237. [Google Scholar] [CrossRef]
- Wu, S.; Chen, Y.; Chen, Z.; Zhou, Q.; Wei, F.; Li, P.; Gu, Q. Antioxidant properties and molecular mechanisms of Lactiplantibacillus plantarum ZJ316: A potential probiotic resource. LWT 2023, 187, 115269. [Google Scholar] [CrossRef]
- Al Atya, A.K.; Drider-Hadiouche, K.; Ravallec, R.; Silvain, A.; Vachee, A.; Drider, D. Probiotic potential of Enterococcus faecalis strains isolated from meconium. Front. Microbiol. 2015, 6, 227. [Google Scholar] [CrossRef]
- Gök, Ş.M.; Türk Dağı, H.; Kara, F.; Arslan, U.; Fındık, D. Investigation of Antibiotic Resistance and Virulence Factors of Enterococcus faecium and Enterococcus faecalis Strains Isolated from Clinical Samples. Mikrobiyol. Bul. 2020, 54, 26–39. [Google Scholar] [CrossRef]
- Zommiti, M.; Cambronel, M.; Maillot, O.; Barreau, M.; Sebei, K.; Feuilloley, M.; Ferchichi, M.; Connil, N. Evaluation of Probiotic Properties and Safety of Enterococcus faecium Isolated From Artisanal Tunisian Meat “Dried Ossban”. Front. Microbiol. 2018, 9, 1685. [Google Scholar] [CrossRef]
- Choeisoongnern, T.; Sirilun, S.; Waditee-Sirisattha, R.; Pintha, K.; Peerajan, S.; Chaiyasut, C. Potential Probiotic Enterococcus faecium OV3-6 and Its Bioactive Peptide as Alternative Bio-Preservation. Foods 2021, 10, 2264. [Google Scholar] [CrossRef] [PubMed]
- Gerkins, C.; Hajjar, R.; Oliero, M.; Santos, M.M. Assessment of Gut Barrier Integrity in Mice Using Fluorescein-Isothiocyanate-Labeled Dextran. J. Vis. Exp. 2022, 189, e64710. [Google Scholar]
- GB 14925–2010; Laboratory Animal Requirements of Environment and Housing Facilities. Administration of Quality Supervision, Inspection and Quarantine of People’s Republic of China and Standardization Administration of China: Beijing, China, 2010.
- Gilmore, M.S.; Salamzade, R.; Selleck, E.; Bryan, N.; Mello, S.S.; Manson, A.L.; Earl, A.M. Genes Contributing to the Unique Biology and Intrinsic Antibiotic Resistance of Enterococcus faecalis. mBio 2020, 11, e02962-20. [Google Scholar] [CrossRef] [PubMed]
- Ghattargi, V.C.; Nimonkar, Y.S.; Burse, S.A.; Davray, D.; Kumbhare, S.V.; Shetty, S.A.; Gaikwad, M.A.; Suryavanshi, M.V.; Doijad, S.P.; Utage, B.; et al. Genomic and physiological analyses of an indigenous strain, Enterococcus faecium 17OM39. Funct. Integr. Genom. 2018, 18, 385–399. [Google Scholar] [CrossRef]
- Banik, A.; Anjum, H.; Habib, H.; Abony, M.; Begum, A.; Ahmed, Z. Characterization of lactic acid bacteria isolated from street pickles of Dhaka, Bangladesh. Heliyon 2023, 9, e17508. [Google Scholar] [CrossRef]
- Shridhar, P.B.; Amachawadi, R.G.; Tokach, M.; Patel, I.; Gangiredla, J.; Mammel, M.; Nagaraja, T.G. Whole genome sequence analyses-based assessment of virulence potential and antimicrobial susceptibilities and resistance of Enterococcus faecium strains isolated from commercial swine and cattle probiotic products. J. Anim. Sci. 2022, 100, skac030. [Google Scholar] [CrossRef] [PubMed]
- Alfano, J.R.; Collmer, A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 2004, 42, 385–414. [Google Scholar] [CrossRef]
- Bourgogne, A.; Hilsenbeck, S.G.; Dunny, G.M.; Murray, B.E. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: The Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J. Bacteriol. 2006, 188, 2875–2884. [Google Scholar] [CrossRef]
- Nogawa, H.; Kuwae, A.; Matsuzawa, T.; Abe, A. The Type III Secreted Protein BopD in Bordetella bronchiseptica is Complexed with BopB for Pore Formation on the Host Plasma Membrane. J. Bacteriol. 2004, 186, 3806–3813. [Google Scholar] [CrossRef] [PubMed]
- Fatoba, D.O.; Amoako, D.G.; Akebe, A.L.K.; Ismail, A.; Essack, S.Y. Genomic analysis of antibiotic-resistant Enterococcus spp. reveals novel enterococci strains and the spread of plasmid-borne Tet(M), Tet(L) and Erm(B) genes from chicken litter to agricultural soil in South Africa. J. Environ. Manag. 2022, 302 Pt B, 114101. [Google Scholar] [CrossRef]
- Thumu, S.C.R.; Halami, P.M. Conjugal transfer of erm(B) and multiple tet genes from Lactobacillus spp. to bacterial pathogens in animal gut, in vitro and during food fermentation. Food Res. Int. 2019, 116, 1066–1075. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Q.; Wang, X.M.; Li, H.; Shang, Y.H.; Pan, Y.S.; Wu, C.M.; Wang, Y.; Du, X.D.; Shen, J.Z. Novel lnu(G) gene conferring resistance to lincomycin by nucleotidylation, located on Tn6260 from Enterococcus faecalis E531. J. Antimicrob. Chemother. 2017, 72, 993–997. [Google Scholar] [PubMed]
- Wei, J.; Ding, S.; Zhang, M.; Luo, M.; Zhou, S.; Luo, F. Optimization of Fermentation Conditions for Bacteriocin Production from Enterococcus faecium B13. Food Res. Dev. 2020, 41, 194–199. [Google Scholar]
- Zimmermann, R.A.; Moellering, R.C., Jr.; Weinberg, A.N. Mechanism of resistance to antibiotic synergism in enterococci. J. Bacteriol. 1971, 105, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Boehr, D.D.; Daigle, D.M.; Wright, G.D. Domain-domain interactions in the aminoglycoside antibiotic resistance enzyme AAC(6′)-APH(2″). Biochemistry 2004, 43, 9846–9855. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.H.; Sabo, J.L.; Rice-Trujillo, C.; Hernandez, J.; McDermott, P.F. Whole-genome sequencing based characterization of antimicrobial resistance in Enterococcus. Pathog. Dis. 2018, 76, fty018. [Google Scholar] [CrossRef] [PubMed]
- Ilavenil, S.; Vijayakumar, M.; Kim, D.H.; Valan Arasu, M.; Park, H.S.; Ravikumar, S.; Choi, K.C. Assessment of probiotic, antifungal and cholesterol lowering properties of Pediococcus pentosaceus KCC-23 isolated from Italian ryegrass. J. Sci. Food Agric. 2016, 96, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Abbasiliasi, S.; Tan, J.S.; Ibrahim, T.A.; Ramanan, R.N.; Vakhshiteh, F.; Mustafa, S.; Ling, T.C.; Rahim, R.A.; Ariff, A.B. Isolation of Pediococcus acidilactici Kp10 with ability to secrete bacteriocin-like inhibitory substance from milk products for applications in food industry. BMC Microbiol. 2012, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Noohi, N.; Ebrahimipour, G.; Rohani, M.; Talebi, M.; Pourshafie, M.R. Evaluation of potential probiotic characteristics and antibacterial effects of strains of Pediococcus species isolated from broiler chickens. Br. Poult. Sci. 2016, 57, 317–323. [Google Scholar] [CrossRef]
- Zommiti, M.; Bouffartigues, E.; Maillot, O.; Barreau, M.; Szunerits, S.; Sebei, K.; Feuilloley, M.; Connil, N.; Ferchichi, M. In vitro Assessment of the Probiotic Properties and Bacteriocinogenic Potential of Pediococcus pentosaceus MZF16 Isolated from Artisanal Tunisian Meat “Dried Ossban”. Front. Microbiol. 2018, 9, 2607. [Google Scholar] [CrossRef]
- Zhou, Y.; Gong, W.; Xu, C.; Zhu, Z.; Peng, Y.; Xie, C. Probiotic assessment and antioxidant characterization of Lactobacillus plantarum GXL94 isolated from fermented chili. Front. Microbiol. 2022, 13, 997940. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Q.; Gao, N.; Wang, Z.; Li, F.; Li, J.; Shan, A. Exopolysaccharides produced by Lactobacillus rhamnosus GG alleviate hydrogen peroxide-induced intestinal oxidative damage and apoptosis through the Keap1/Nrf2 and Bax/Bcl-2 pathways in vitro. Food Funct. 2021, 12, 9632–9641. [Google Scholar] [CrossRef] [PubMed]
- Mu, G.; Li, H.; Tuo, Y.; Gao, Y.; Zhang, Y. Antioxidative effect of Lactobacillus plantarum Y44 on 2,2′-azobis(2-methylpropionamidine) dihydrochloride (ABAP)-damaged Caco-2 cells. J. Dairy Sci. 2019, 102, 6863–6875. [Google Scholar] [CrossRef] [PubMed]
- Del Re, B.; Sgorbati, B.; Miglioli, M.; Palenzona, D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett. Appl. Microbiol. 2000, 31, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Rickard, A.H.; Gilbert, P.; High, N.J.; Kolenbrander, P.E.; Handley, P.S. Bacterial coaggregation: An integral process in the development of multi-species biofilms. Trends Microbiol. 2003, 11, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Han, S.H.; Kim, Y.G.; Jeong, Y.; Paek, N.S. Antibacterial Activity and Probiotic Properties of Lactic Acid Bacteria Isolated from Traditional Fermented Foods. Korean Soc. Biotechnol. Bioeng. J. 2017, 32, 199–205. [Google Scholar]
- Zhang, C.; Ma, K.; Nie, K.; Deng, M.; Luo, W.; Wu, X.; Huang, Y.; Wang, X. Assessment of the safety and probiotic properties of Roseburia intestinalis: A potential “Next Generation Probiotic”. Front. Microbiol. 2022, 13, 973046. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, K.M.; Vieira, A.D.; Salles, H.O.; Oliveira Jda, S.; Rocha, C.R.; Borges Mde, F.; Bruno, L.M.; Franco, B.D.; Todorov, S.D. Safety, beneficial and technological properties of Enterococcus faecium isolated from Brazilian cheeses. Braz. J. Microbiol. 2015, 46, 237–249. [Google Scholar] [CrossRef]
- Favaro, L.; Basaglia, M.; Casella, S.; Hue, I.; Dousset, X.; Gombossy de Melo Franco, B.D.; Todorov, S.D. Bacteriocinogenic potential and safety evaluation of non-starter Enterococcus faecium strains isolated from home made white brine cheese. Food Microbiol. 2014, 38, 228–239. [Google Scholar] [CrossRef]
- Shukla, S.; Park, H.K.; Kim, J.K.; Kim, M. Determination of biogenic amines in Korean traditional fermented soybean paste (Doenjang). Food Chem. Toxicol. 2010, 48, 1191–1195. [Google Scholar] [CrossRef]
- Del Rio, B.; Fernandez, M.; Redruello, B.; Ladero, V.; Alvarez, M.A. New insights into the toxicological effects of dietary biogenic amines. Food Chem. 2024, 435, 137558. [Google Scholar] [CrossRef] [PubMed]
- Tsigkrimani, M.; Panagiotarea, K.; Paramithiotis, S.; Bosnea, L.; Pappa, E.; Drosinos, E.H.; Skandamis, P.N.; Mataragas, M. Microbial Ecology of Sheep Milk, Artisanal Feta, and Kefalograviera Cheeses. Part II: Technological, Safety, and Probiotic Attributes of Lactic Acid Bacteria Isolates. Foods 2022, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Sun, J.; Guo, Z.; Guo, H.; Ren, F. Safety evaluation of Lactobacillus paracasei subsp. paracasei LC-01, a probiotic bacterium. J. Microbiol. 2013, 51, 633–638. [Google Scholar] [CrossRef]
- Stasiak, M.; Maćkiw, E.; Kowalska, J.; Kucharek, K.; Postupolski, J. Silent Genes: Antimicrobial Resistance and Antibiotic Production. Pol. J. Microbiol. 2021, 70, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Lanz, R.; Kuhnert, P.; Boerlin, P. Antimicrobial resistance and resistance gene determinants in clinical Escherichia coli from different animal species in Switzerland. Vet. Microbiol. 2003, 91, 73–84. [Google Scholar] [CrossRef]
- Lipszyc, A.; Szuplewska, M.; Bartosik, D. How Do Transposable Elements Activate Expression of Transcriptionally Silent Antibiotic Resistance Genes? Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Surujon, D.; Ortiz-Marquez, J.C.; Huo, W.; Isberg, R.R.; Bento, J.; van Opijnen, T. Entropy of a bacterial stress response is a generalizable predictor for fitness and antibiotic sensitivity. Nat. Commun. 2020, 11, 4365. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Shen, Y.; Xin, J.; Xu, X.; Ding, Q.; Chen, W.; Wang, J.; Lv, Y.; Wei, X.; Wei, Y.; et al. Cryptotanshinone alleviates radiation-induced lung fibrosis via modulation of gut microbiota and bile acid metabolism. Phytother. Res. 2023, 37, 4557–4571. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Y.; Liu, Z.; Feng, X.; Zhou, R.; He, Y.; Zhou, H.; Peng, H.; Huang, Y. Augmented temperature fluctuation aggravates muscular atrophy through the gut microbiota. Nat. Commun. 2023, 14, 3494. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, Y.; Huang, M.; Chen, J.; Zhang, Z.; Li, J.; Yang, R.; Liu, Y.; Cai, S. Fuzhengjiedu formula exerts protective effect against LPS-induced acute lung injury via gut-lung axis. Phytomedicine 2023, 123, 155190. [Google Scholar] [CrossRef]
- Park, J.K.; Chang, D.H.; Rhee, M.S.; Jeong, H.; Song, J.; Ku, B.J.; Kim, S.B.; Lee, M.; Kim, B.C. Heminiphilus faecis gen. nov., sp. nov., a member of the family Muribaculaceae, isolated from mouse faeces and emended description of the genus Muribaculum. Antonie Leeuwenhoek 2021, 114, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Bäuerl, C.; Collado, M.C.; Diaz Cuevas, A.; Viña, J.; Pérez Martínez, G. Shifts in gut microbiota composition in an APP/PSS1 transgenic mouse model of Alzheimer’s disease during lifespan. Lett. Appl. Microbiol. 2018, 66, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.J.; Miller, R.A.; Ericsson, A.C.; Harrison, D.C.; Strong, R.; Schmidt, T.M. Changes in the gut microbiome and fermentation products concurrent with enhanced longevity in acarbose-treated mice. BMC Microbiol. 2019, 19, 130. [Google Scholar] [CrossRef] [PubMed]
- McNamara, M.P.; Singleton, J.M.; Cadney, M.D.; Ruegger, P.M.; Borneman, J.; Garland, T. Early-life effects of juvenile Western diet and exercise on adult gut microbiome composition in mice. J. Exp. Biol. 2021, 224, jeb239699. [Google Scholar] [CrossRef] [PubMed]
- Dowden, R.A.; McGuinness, L.R.; Wisniewski, P.J.; Campbell, S.C.; Guers, J.J.; Oydanich, M.; Vatner, S.F.; Häggblom, M.M.; Kerkhof, L.J. Host genotype and exercise exhibit species-level selection for members of the gut bacterial communities in the mouse digestive system. Sci. Rep. 2020, 10, 8984. [Google Scholar] [CrossRef]
- Zou, X.; Pan, L.; Xu, M.; Wang, X.; Wang, Q.; Han, Y. Probiotic potential of Lactobacillus sakei L-7 in regulating gut microbiota and metabolism. Microbiol. Res. 2023, 274, 127438. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Ro, K.S.; Jiang, C.; Yin, D.; Zhao, L.; Zhang, D.; Du, L.; Xie, J. The anti-hyperuricemic and gut microbiota regulatory effects of a novel purine assimilatory strain, Lactiplantibacillus plantarum X7022. Eur. J. Nutr. 2023, 63, 697–711. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.; Zhang, H.; Zhu, L.; Deehan, E.C.; Fu, J.; Wang, Y.; Jin, M. Auricularia auricula polysaccharides attenuate obesity in mice through gut commensal Papillibacter cinnamivorans. J. Adv. Res. 2023, 52, 203–218. [Google Scholar] [CrossRef]
- Zheng, Y.; Yu, Z.; Zhang, W.; Sun, T. Lactobacillus rhamnosus Probio-M9 Improves the Quality of Life in Stressed Adults by Gut Microbiota. Foods 2021, 10, 2384. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, J.; Jiang, S.; Xu, M.; Ma, T.; Sun, Z.; Zhang, J. Lactobacillus fermentum HNU312 alleviated oxidative damage and behavioural abnormalities during brain development in early life induced by chronic lead exposure. Ecotoxicol. Environ. Saf. 2023, 251, 114543. [Google Scholar] [CrossRef] [PubMed]
| Virulence genes | Product | Function | Location | Identify(%) | VFid | GeneID |
|---|---|---|---|---|---|---|
| Adherence | ||||||
| pilB | pilB-type pili | Involved in type IV pili biosynthesis | chr | 99.2 | VFG042991 | B13000560 |
| pilF | minor pilin subunit | Involved in type IV pili biosynthesis | plasmid1 | 99.1 | VFG042984 | B13002625 |
| pilE | cell wall-associated LPXTG-like protein | Involved in type IV pili biosynthesis | plasmid1 | 99.2 | VFG042986 | B13002627 |
| pilA | pilA-type pili | Involved in type IV pili biosynthesis | plasmid1 | 98.3 | VFG042988 | B13002629 |
| Biofilm formation | ||||||
| sgrA | cell wall anchored protein SgrA | Adherence to cell wall | chr | 79.6 | VFG043511 | B13001199 |
| bopD | Sugar binding transcriptional regulator | biofilm on plastic surfaces | chr | 86.3 | VFG002197 | B13000384 |
| Immune Evasion | ||||||
| hasC | Hyaluronic acid HA capsule | Evasion of host immune system | chr | 73.3 | VFG005865 | B13001994 |
| pathogenicity | ||||||
| sagA | Streptolysin S core peptide | hemolytic activity | chr | 95.4 | VFG043441 | B13002370 |
| Location | Identify(%) | Gene family | Product | Drug Class | Resistance Mechanism | Origin Species |
|---|---|---|---|---|---|---|
| plasmid2 | 99.75 | tet(L) | tetracycline efflux MFS transporter Tet(L) | tetracycline | antibiotic efflux | Geobacillus stearothermophilus |
| chr | 98.901 | AAC(6′)-Ii | aminoglycoside N-acetyltransferase AAC(6′)-Ii | aminoglycoside | antibiotic inactivation | Enterococcus faecium |
| plasmid1 | 99.625 | lnu(G) | lincosamide nucleotidyltransferase Lnu(G) | lincosamide | antibiotic inactivation | Enterococcus faecalis |
| plasmid1 | 100 | ANT(6)-Ia | aminoglycoside nucleotidyltransferase ANT(6)-Ia | aminoglycoside | antibiotic inactivation | Exiguobacterium |
| chr | 97.531 | liaF | three-component signaling pathway regulator LiaF | lipopeptide | antibiotic target alteration | Enterococcus faecium |
| chr | 98.941 | cls | cardiolipin synthase Cls | lipopeptide | antibiotic target alteration | Enterococcus faecium |
| chr | 99.524 | liaR | response regulator transcription factor LiaR | lipopeptide | antibiotic target alteration | Enterococcus faecium |
| chr | 100 | liaS | sensor histidine kinase LiaS | lipopeptide | antibiotic target alteration | Enterococcus faecium |
| plasmid2 | 95.149 | tet(M) | tetracycline resistance ribosomal protection protein Tet(M) | tetracycline | antibiotic target protection | Staphylococcus aureus |
| chr | 97.154 | msrC | ABC-F type ribosomal protection protein Msr | macrolide/streptogramin | antibiotic target protection | Enterococcus faecium |
| chr | 99 | eatA | ABC-F type ribosomal protection protein Eat(A) | pleuromutilin | antibiotic target protection | Enterococcus faecium |
| pH | Viable Count (log10 CFU/mL) | Survival Rate (%) |
|---|---|---|
| 1.0 | - | 0 |
| 2.0 | - | 0 |
| 3.0 | 6.74 ± 0.03 | 97.97 |
| 4.0 | 6.95 ± 0.04 | 101 |
| Times of Exposure (h) | Viable Count (log10 CFU/mL) | Survival Rate (%) |
|---|---|---|
| 1 | 7.80 | 95% |
| 2 | 7.61 | 92.69% |
| 3 | 7.45 | 90.62% |
| Subjects | CFS | Vitamin C |
|---|---|---|
| DPPH radical (%) | 32.83 ± 0.37 | 88.39 ± 1.64 |
| total antioxidant capacity (U/mL) | 19.28 ± 3.14 | 37.45 ± 1.35 |
| Antibiotic | ZOI (mm) | Antibiotic Susceptibility | Antibiotic | ZOI (mm) | Antibiotic Susceptibility |
|---|---|---|---|---|---|
| Nitrofurantoin | 21.76 ± 0.93 | S | Teikoplanin | 22.4 ± 0.6 | S |
| Zyvox | 28.9 ± 1.3 | S | Gentamicin | 20.8 ± 0.4 | S |
| Erythromycin | 12.0 ± 0.6 | R | Doxycycline | 8.0 ± 0.1 | R |
| Vancomycin | 25.8 ± 0.5 | S | Minocycline | 8.9 ± 0.1 | R |
| Rifampicin | 11.1 ± 0.6 | R | Gatifloxacin | 25.1 ± 0.5 | S |
| Tetracycline | 5.2 ± 0.1 | R | Norfloxacin | 22.0 ± 0.1 | S |
| Ampicillin | 25.3 ± 0.6 | S | Penicillin | 17.7 ± 1.4 | S |
| Ciprofloxacin | 24.8 ± 0.4 | S | Levofloxacin | 21.6 ± 0.4 | S |
| Chloramphenicol | 25.9 ± 1.8 | S |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Chen, C.; Fu, Z.; Wang, S.; Luo, F. Assessment of the Safety and Probiotic Properties of Enterococcus faecium B13 Isolated from Fermented Chili. Microorganisms 2024, 12, 994. https://doi.org/10.3390/microorganisms12050994
Xiao J, Chen C, Fu Z, Wang S, Luo F. Assessment of the Safety and Probiotic Properties of Enterococcus faecium B13 Isolated from Fermented Chili. Microorganisms. 2024; 12(5):994. https://doi.org/10.3390/microorganisms12050994
Chicago/Turabian StyleXiao, Jingmin, Cai Chen, Zhuxian Fu, Shumin Wang, and Fan Luo. 2024. "Assessment of the Safety and Probiotic Properties of Enterococcus faecium B13 Isolated from Fermented Chili" Microorganisms 12, no. 5: 994. https://doi.org/10.3390/microorganisms12050994
APA StyleXiao, J., Chen, C., Fu, Z., Wang, S., & Luo, F. (2024). Assessment of the Safety and Probiotic Properties of Enterococcus faecium B13 Isolated from Fermented Chili. Microorganisms, 12(5), 994. https://doi.org/10.3390/microorganisms12050994





