Identification of the Microbiota in Coconut Water, Kefir, Coconut Water Kefir and Coconut Water Kefir-Fermented Sourdough Using Culture-Dependent Techniques and Illumina–MiSeq Sequencing
Abstract
1. Introduction
2. Materials and methods
2.1. pH, D-Lactic Acid, L-Lactic Acid and Total Titratable Acidity
2.2. Phenotypic Characterisation and Identification of Species
2.2.1. Samples
2.2.2. Methods of Analysis
2.2.3. Preparation of Samples
2.3. Identification Using Sanger Sequencing Method
2.4. MiSeq High-Throughput Sequencing Methods (Illumina Sequencing)
2.5. Bioinformatics Analysis
3. Results and Discussion
3.1. pH, D-Lactic Acid, L-Lactic Acid and Total Titratable Acidity Determination
3.2. Identification
3.3. Relative Abundance of Microorganisms Using Miseq High-Throughput Sequencing Method
4. General Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Salvadori, M.; Bertoldi, D.; Molinari, F.; Nobile, M. Exploring the Microbial Diversity of Sourdough: An Ancient Fermentation Method. J. Food Sci. 2023, 68, 421–435. [Google Scholar]
- Bonatsou, S.; Gialitaki, M.; Mallouchos, A.; Tassou, C. Utilizing Sourdough as a Substrate for Fruit-Based Kefir: Implications on Nutritional Composition and Probiotic Diversity. Food Microbiol. 2024, 82, 78–89. [Google Scholar]
- Huys, G.; Daniel, H.-M.; De Vuyst, L. Taxonomy and Biodiversity of Sourdough Yeasts and Lactic Acid Bacteria Handbook on Sourdough Biotechnology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 105–154. [Google Scholar]
- Vrancken, G.; De Vuyst, L.; Van der Meulen, R.; Huys, G.; Vandamme, P.; Daniel, H.-M. Yeast species composition differs between artisan bakery and spontaneous laboratory sourdoughs. FEMS Yeast Res. 2010, 10, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Daniel, H.-M.; Moons, M.-C.; Huret, S.; Vrancken, G.; De Vuyst, L. Wickerhamomyces anomalus in the sourdough microbial ecosystem. Antonie Leeuwenhoek 2011, 99, 63–73. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Vermeulen, N.; Vogel, R.F. Carbohydrate, peptide and lipid metabolism of lactic acid bacteria in sourdough. Food Microbiol. 2007, 24, 128–138. [Google Scholar] [CrossRef]
- Gobbetti, M.; De Angelis, M.; Corsetti, A.; Di Cagno, R. Biochemistry and physiology of sourdough lactic acid bacteria. Trends Food Sci. Technol. 2005, 16, 57–69. [Google Scholar] [CrossRef]
- Rollan, G.; Lorca, G.; de Valdez, G.F. Arginine catabolism and acid tolerance response in Lactobacillus reuteri isolated from sourdough. Food Microbiol. 2003, 20, 313–319. [Google Scholar] [CrossRef]
- Araujo Filho, A.A.L.; Sousa PHM de Vieira, I.G.P.; Fernandes, V.B.; Cunha, F.E.T.; Magalhaes, F.E.A.; da Silva, L.M.R. Kombucha and kefir fermentation dynamics on cashew nut beverage (Anacardium occidentale L.). Int. J. Gastron. Food Sci. 2023, 33, 100778. [Google Scholar] [CrossRef]
- Yan, R.; Zeng, X.; Shen, J.; Wu, Z.; Guo, Y.; Du, Q.; Tu, M.; Pan, D. New clues for postbiotics to improve host health: A review from the perspective of function and mechanisms. J. Sci. Food Agric. 2024; early view. [Google Scholar] [CrossRef]
- Limbad, M.; Gutierrez Maddox, N.; Hamid, N.; Kantono, K. Sensory and physicochemical characterization of sourdough bread prepared with a coconut water kefir starter. Foods 2020, 9, 1165. [Google Scholar] [CrossRef]
- Limbad, M.J.; Gutierrez-Maddox, N.; Hamid, N. Coconut water: An essential health drink in both natural and fermented forms. In Fruit and Pomace Extracts: Biological Activity, Potential Applications and Beneficial Health Effects; Nova Science Publishers Inc.: New York, NY, USA, 2015; pp. 145–156. [Google Scholar]
- Rosa, D.D.; Dias, M.M.; Grześkowiak, Ł.M.; Reis, S.A.; Conceição, L.L.; Maria do Carmo, G.P. Milk kefir: Nutritional, microbiological and health benefits. Nutr. Res. Rev. 2017, 30, 82–96. [Google Scholar] [CrossRef]
- Marsh, A.J.; O’Sullivan, O.; Hill, C.; Ross, R.P.; Cotter, P.D. Sequence-based analysis of the microbial composition of water kefir from multiple sources. FEMS Microbiol. Lett. 2013, 348, 79–85. [Google Scholar] [CrossRef]
- Dallas, D.C.; Citerne, F.; Tian, T.; Silva, V.L.M.; Kalanetra, K.M.; Frese, S.A.; Robinson, R.C.; Mills, D.A.; Barile, D. Peptidomic analysis reveals proteolytic activity of kefir microorganisms on bovine milk proteins. Food Chem. 2016, 197, 273–284. [Google Scholar] [CrossRef]
- Walsh, S.D.; De Clercq, B.; Molcho, M.; Harel-Fisch, Y.; Davison, C.M.; Madsen, K.R.; Stevens, G.W.J.M. The Relationship Between Immigrant School Composition, Classmate Support and Involvement in Physical Fighting and Bullying among Adolescent Immigrants and Non-immigrants in 11 Countries. J. Youth Adolesc. 2016, 45, 1–16. [Google Scholar] [CrossRef]
- Dertli, E.; Çon, A.H. Microbial diversity of traditional kefir grains and their role on kefir aroma. LWT—Food Sci. Technol. 2017, 85, 151–157. [Google Scholar] [CrossRef]
- Gao, X.; Li, B. Chemical and microbiological characteristics of kefir grains and their fermented dairy products: A review. Cogent Food Agric. 2016, 2, 1272152. [Google Scholar] [CrossRef]
- Garofalo, C.; Bancalari, E.; Milanović, V.; Cardinali, F.; Osimani, A.; Sardaro, M.L.S.; Bottari, B.; Bernini, V.; Aquilanti, L.; Clementi, F.; et al. Study of the bacterial diversity of foods: PCR-DGGE versus LH-PCR. Int. J. Food Microbiol. 2017, 242, 24–36. [Google Scholar] [CrossRef]
- Leite, A.M.O.; Mayo, B.; Rachid, C.T.C.C.; Peixoto, R.S.; Silva, J.T.; Paschoalin, V.M.F.; Delgado, S. Assessment of the microbial diversity of Brazilian kefir grains by PCR-DGGE and pyrosequencing analysis. Food Microbiol. 2012, 31, 215–221. [Google Scholar] [CrossRef]
- Yang, Y.; Shevchenko, A.; Knaust, A.; Abuduresule, I.; Li, W.; Hu, X.; Wang, C.; Shevchenko, A. Proteomics evidence for kefir dairy in Early Bronze Age China. J. Archaeol. Sci. 2014, 45, 178–186. [Google Scholar] [CrossRef]
- Zhang, T.; Hong, S.; Zhang, J.-R.; Liu, P.-H.; Li, S.; Wen, Z.; Xiao, J.; Zhang, G.; Habimana, O.; Shah, N.P.; et al. The effect of lactic acid bacteria fermentation on physicochemical properties of starch from fermented proso millet flour. Food Chem. 2024, 437, 137764. [Google Scholar] [CrossRef]
- Silva, J.A.; Castañares, M.; Mouguelar, H.; Valenciano, J.A.; Pellegrino, M.S. Isolation of lactic acid bacteria from the reproductive tract of mares as potentially beneficial strains to prevent equine endometritis. Vet. Res. Commun. 2024; online ahead of print. [Google Scholar]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Catzeddu, K.; Zoumpopoulou, G.; Georgalaki, M.; Alexandraki, V.; Kazou, M.; Anastasiou, R.; Tsakalidou, E. Chapter 6—Sourdough Bread. In Innovations in Traditional Foods; Galanakis, C.M., Ed.; Woodhead Publishing: Sawston, UK, 2019; pp. 127–158. [Google Scholar]
- Katina, K.; Salmenkallio-Marttila, M.; Partanen, R.; Forssell, P.; Autio, K. Effects of sourdough and enzymes on staling of high-fibre wheat bread. LWT—Food Sci. Technol. 2006, 39, 479–491. [Google Scholar] [CrossRef]
- Garrity, G.M.; Bell, J.A.; Lilburn, T.G. Taxonomic outline of the prokaryotes. In Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA; Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Ercolini, D.; Pontonio, E.; De Filippis, F.; Minervini, F.; La Storia, A.; Gobbetti, M.; Di Cagno, R. Microbial Ecology Dynamics during Rye and Wheat Sourdough Preparation. Appl. Environ. Microbiol. 2013, 79, 7827–7836. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.D.J.; Lee, K.C.; Caruso, T.; Maki, T.; Lee, C.K.; Cary, S.C.; Cowan, D.A.; Maestre, F.T.; Pointing, S.B. Airborne microbial transport limitation to isolated Antarctic soil habitats. Nat. Microbiol. 2019, 4, 925–932. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for de-scribing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.C.; Lemos, Á.T.; Lopes, R.P.; Mota, M.J.; Inácio, R.S.; Gomes, A.M.P.; Sousa, S.; Delgadillo, I.; Saraiva, J.A. The Combined Effect of Pressure and Temperature on Kefir Production—A Case Study of Food Fermentation in Unconventional Conditions. Foods 2020, 9, 1133. [Google Scholar] [CrossRef]
- Irigoyen, A.; Arana, I.; Castiella, M.; Torre, P.; Ibáñez, F. Microbiological, physicochemical, and sensory characteristics of kefir during storage. Food Chem. 2005, 90, 613–620. [Google Scholar] [CrossRef]
- Kannangara, A.C.; Chandrajith, V.G.G.; Ranaweera, K.K.D.S. Comparative analysis of coconut water in four different maturity stages. J. Pharmacogn. Phytochem. 2018, 7, 1814–1817. [Google Scholar]
- Olieman, C.; Vries, E. Determination of D-and L-lactic acid in fermented dairy products with HPLC. Neth. Milk Dairy J. 1988, 42, 111. [Google Scholar]
- Liu, J.; Lin, C. Production of Kefir from Soymilk with or Without Added Glucose, Lactose, or Sucrose. J. Food Sci. 2000, 65, 716–719. [Google Scholar] [CrossRef]
- Farnworth, E.R.; Mainville, I. Kefir—A Fermented Milk Product Handbook of Fermented Functional Foods; CRC Press: Boca Raton, FL, USA, 2008; pp. 89–127. [Google Scholar]
- Uniacke-Lowe, T. Studies on Equine Milk and Comparative Studies on Equine and Bovine Milk Systems. 2011. Available online: https://cora.ucc.ie/items/08b0f256-4294-4ee5-aaa7-862080cf6f26 (accessed on 28 December 2019).
- Frías, J.; Martínez-Villaluenga, C.; Peñas, E. Fermented Foods in Health and Disease Prevention; Elsevier Science: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Suriasih, K. Microbiological and Chemical Properties of Kefir Made of Bali Cattle Milk. 2000. Available online: https://www.iiste.org/Journals/index.php/FSQM/article/view/2485 (accessed on 28 December 2019).
- Chen, T.-H.; Wang, S.-Y.; Chen, K.-N.; Liu, J.-R.; Chen, M.-J. Microbiological and chemical properties of kefir manufactured by entrapped microorganisms isolated from kefir grains. J. Dairy Sci. 2009, 92, 3002–3013. [Google Scholar] [CrossRef]
- Aponte, M.; Boscaino, F.; Sorrentino, A.; Coppola, R.; Masi, P.; Romano, A. Effects of fermentation and rye flour on microstructure and volatile compounds of chestnut flour based sourdoughs. LWT—Food Sci. Technol. 2014, 58, 387–395. [Google Scholar] [CrossRef]
- Corsetti, A.; Settanni, L. Lactobacilli in sourdough fermentation. Food Res. Int. 2007, 40, 539–558. [Google Scholar] [CrossRef]
- Minervini, F.; Di Cagno, R.; Lattanzi, A.; De Angelis, M.; Antonielli, L.; Cardinali, G.; Cappelle, S.; Gobbetti, M. Lactic Acid Bacterium and Yeast Microbiotas of 19 Sourdoughs Used for Traditional/Typical Italian Breads: Interactions between Ingredients and Microbial Species Diversity. Appl. Environ. Microbiol. 2012, 78, 1251–1264. [Google Scholar] [CrossRef]
- Robert, H.; Gabriel, V.; Fontagné-Faucher, C. Biodiversity of lactic acid bacteria in French wheat sourdough as determined by molecular characterization using species-specific PCR. Int. J. Food Microbiol. 2009, 135, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Vrancken, G.; De Vuyst, L.; Rimaux, T.; Allemeersch, J.; Weckx, S. Adaptation of Lactobacillus plantarum IMDO 130201, a Wheat Sourdough Isolate, to Growth in Wheat Sourdough Simulation Medium at Different pH Values through Differential Gene Expression. Appl. Environ. Microbiol. 2011, 77, 3406–3412. [Google Scholar] [CrossRef] [PubMed]
- Zotta, T.; Piraino, P.; Parente, E.; Salzano, G.; Ricciardi, A. Characterization of lactic acid bacteria isolated from sourdoughs for Cornetto, a traditional bread produced in Basilicata (Southern Italy). World J. Microbiol. Biotechnol. 2008, 24, 1785–1795. [Google Scholar] [CrossRef]
- Dos Santos, S.A.; Barbosa, G.M.; Bernardes, P.C.; Carneiro, J.C.S.; Tostes, M.d.G.V.; Coelho, J.M. Probiotification of whole grape juice by water kefir microorganisms. Braz. Arch. Biol. Technol. 2023, 66, 23210475. [Google Scholar] [CrossRef]
- Mohammed, S.; Orukotan, A.; Shittu, A.; Onuoha, E.; Moriki, A.; Mukhtar, A. Characterization and identification of lactic acid bacteria isolated from fermented milk sold at central market, Kaduna. KASU J. Biochem. Biotechnol. 2019, 3, 1–8. [Google Scholar]
- Diosma, G.; Romanin, D.E.; Rey-Burusco, M.F.; Londero, A.; Garrote, G.L. Yeasts from kefir grains: Isolation, identification, and probiotic characterization. World J. Microbiol. Biotechnol. 2014, 30, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Witthuhn, R.C.; Schoeman, T.; Britz, T. Characterisation of the microbial population at different stages of Kefir production and Kefir grain mass cultivation. Int. Dairy J. 2005, 15, 383–389. [Google Scholar] [CrossRef]
- Zanirati, D.F.; Abatemarco, M.; Sandes, S.H.d.C.; Nicoli, J.R.; Nunes, Á.C.; Neumann, E. Selection of lactic acid bacteria from Brazilian kefir grains for potential use as starter or probiotic cultures. Anaerobe 2015, 32, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Johnson, A.; Chen, X. Microbial Composition of Tibetan Kefir: Insights from Meta-genomic Analysis. J. Food Sci. 2023, 75, 123–135. [Google Scholar]
- Sun, Z.; Harris, H.M.B.; McCann, A.; Guo, C.; Argimón, S.; Zhang, W.; Yang, X.; Jeffery, I.B.; Cooney, J.C.; Kagawa, T.F.; et al. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat. Commun. 2015, 6, 8322. [Google Scholar] [CrossRef] [PubMed]
- Garrote, G.L.; Abraham, A.G.; DE Antoni, G.L. Chemical and microbiological characterisation of kefir grains. J. Dairy Res. 2002, 68, 639–652. [Google Scholar] [CrossRef]
- Gao, J.; Gu, F.; Abdella, N.H.; Ruan, H.; He, G. Investigation on Culturable Microflora in Tibetan Kefir Grains from Different Areas of China. J. Food Sci. 2012, 77, M425–M433. [Google Scholar] [CrossRef]
- Kurtzman, C.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Mainville, I.; Robert, N.; Lee, B.; Farnworth, E.R. Polyphasic characterization of the lactic acid bacteria in kefir. Syst. Appl. Microbiol. 2006, 29, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Wang, Y.; Ahmad, A.; Khan, S.T.; Nisa, M.; Ahmad, H.; Afreen, A. Kefir and health: A contemporary perspective. Crit. Rev. Food Sci. Nutr. 2013, 53, 422–434. [Google Scholar] [CrossRef]
- Kołakowski, P.; Ozimkiewicz, M. Restoration of kefir grains subjected to different treatments. Int. J. Dairy Technol. 2012, 65, 140–145. [Google Scholar] [CrossRef]
- Magalhães, K.T.; Pereira, M.A.; Nicolau, A.; Dragone, G.; Domingues, L.; Teixeira, J.A.; Silva, J.B.d.A.; Schwan, R.F. Production of fermented cheese whey-based beverage using kefir grains as starter culture: Evaluation of morphological and microbial variations. Bioresour. Technol. 2010, 101, 8843–8850. [Google Scholar] [CrossRef] [PubMed]
- Simova, E.; Beshkova, D.; Angelov, A.; Hristozova, T.; Frengova, G.; Spasov, Z. Lactic acid bacteria and yeasts in kefir grains and kefir made from them. J. Ind. Microbiol. Biotechnol. 2002, 28, 1–6. [Google Scholar] [CrossRef]
- Sakamoto, N.; Tanaka, S.; Sonomoto, K.; Nakayama, J. 16S rRNA pyrosequencing-based in-vestigation of the bacterial community in nukadoko, a pickling bed of fermented rice bran. Int. J. Food Microbiol. 2011, 144, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Yukphan, P.; Chaipitakchonlatarn, W.; Malimas, T.; Sugimoto, M.; Yoshino, M.; Kirtikara, K. 16S rRNA gene sequence analysis of lactic acid bacteria isolated from fermented foods in Thailand. Microbiol. Cult. Coll. 2012, 28, 1–9. [Google Scholar]
- De Pauw, P. Sourdough Compositions and Methods for Their Preparation. European Patent Application No. EP2741613B1, 18 April 2018. [Google Scholar]
- Rattray, F.P.; O’Connell, M.J. Fermented Milks|Kefir. In Encyclopedia of Dairy Sciences, 2nd ed.; Fuquay, J.W., Ed.; Academic Press: San Diego, CA, USA, 2011; pp. 518–524. [Google Scholar]
- Garofalo, C.; Osimani, A.; Milanović, V.; Aquilanti, L.; De Filippis, F.; Stellato, G.; Di Mauro, S.; Turchetti, B.; Buzzini, P.; Ercolini, D.; et al. Bacteria and yeast microbiota in milk kefir grains from different Italian regions. Food Microbiol. 2015, 49, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.R.; Blandón, L.M.; Vandenberghe, L.P.; Rodrigues, C.; Castro, G.R.; Thomaz-Soccol, V.; Soccol, C.R. Milk kefir: Composition, microbial cultures, biological activities, and related products. Front. Microbiol. 2015, 6, 1177. [Google Scholar] [CrossRef]
- Bourrie, B.C.T.; Willing, B.P.; Cotter, P.D. The microbiota and health promoting characteristics of the fermented beverage kefir. Front. Microbiol. 2016, 7, 647. [Google Scholar] [CrossRef]
- Magalhães, K.T.; Pereira, G.V.d.M.; Campos, C.R.; Dragone, G.; Schwan, R.F. Brazilian kefir: Structure, microbial communities and chemical composition. Braz. J. Microbiol. 2011, 42, 693–702. [Google Scholar] [CrossRef]
- De Vuyst, L.; Vrancken, G.; Ravyts, F.; Rimaux, T.; Weckx, S. Biodiversity, ecological determinants, and metabolic exploitation of sourdough microbiota. Food Microbiol. 2009, 26, 666–675. [Google Scholar] [CrossRef]
- Gänzle, M.G.; Vogel, R.F. Contribution of reutericyclin production to the stable persistence of Lactobacillus reuteri in an industrial sourdough fermentation. Int. J. Food Microbiol. 2003, 80, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Hammes, W.P.; Gänzle, M.G. Sourdough breads and related products. In Microbiology of Fermented Foods; Wood, B.B., Ed.; Springer: New York, NY, USA, 1998; pp. 199–216. [Google Scholar] [CrossRef]
- Bessmeltseva, M.; Viiard, E.; Simm, J.; Paalme, T.; Sarand, I. Evolution of bacterial consortia in spontaneously started rye sourdoughs during two months of daily propagation. PLoS ONE 2014, 9, e95449. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; Lattanzi, A.; De Angelis, M.; Celano, G.; Gobbetti, M. House microbiotas as sources of lactic acid bacteria and yeasts in traditional Italian sourdoughs. Food Microbiol. 2015, 52, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Rizzello, G.; Hodgins, M.; Naso, D.; York, A.; Seelecke, S. Modeling of the effects of the electrical dynamics on the electromechanical response of a DEAP circular actuator with a mass–spring load. Smart Mater. Struct. 2015, 24, 094003. [Google Scholar] [CrossRef]
- Van der Meulen, R.; Grosu-Tudor, S.; Mozzi, F.; Vaningelgem, F.; Zamfir, M.; de Valdez, G.F.; De Vuyst, L. Screening of lactic acid bacteria isolates from dairy and cereal products for exopolysaccharide production and genes involved. Int. J. Food Microbiol. 2007, 118, 250–258. [Google Scholar] [CrossRef]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- De Vuyst, L.; Vancanneyt, M. Biodiversity and identification of sourdough lactic acid bacteria. Food Microbiol. 2007, 24, 120–127. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Vogel, R.F. Molecular taxonomy and genetics of sourdough lactic acid bacteria. Trends Food Sci. Technol. 2005, 16, 31–42. [Google Scholar] [CrossRef]
- Gänzle, M.G. Lactic metabolism revisited: Metabolism of lactic acid bacteria in food fermentations and food spoilage. Curr. Opin. Food Sci. 2015, 2, 106–117. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Georgalaki, M.; Alexandraki, V.; Kazou, M.; Anastasiou, R.; Tsakalidou, E. Sourdough Bread. In Innovations in Traditional Foods; Elsevier: Amsterdam, The Netherlands, 2019; pp. 127–158. [Google Scholar]
- Valentino, V.; Magliulo, R.; Farsi, D.; Cotter, P.D.; O’Sullivan, O.; Ercolini, D.; De Filippis, F. Fermented foods, their microbiome and its potential in boosting human health. Microb. Biotechnol. 2024, 17, e14428. [Google Scholar] [CrossRef]
- Marco, M.L.; Heeney, D.; Binda, S.; Cifelli, C.J.; Cotter, P.D.; Foligné, B.; Gänzle, M.; Kort, R.; Pasin, G.; Pihlanto, A.; et al. Health benefits of fermented foods: Microbiota and beyond. Curr. Opin. Biotechnol. 2017, 44, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Minervini, F.; De Angelis, M.; Di Cagno, R.; Gobbetti, M. Ecological parameters influencing microbial diversity and stability of traditional sourdough. Int. J. Food Microbiol. 2014, 171, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-C.; Wang, S.-Y.; Chen, M.-J. Microbiological study of lactic acid bacteria in kefir grains by culture-dependent and culture-independent methods. Food Microbiol. 2008, 25, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-Y.; Chen, H.-C.; Liu, J.-R.; Lin, Y.-C.; Chen, M.-J. Identification of yeasts and evalu-ation of their distribution in Taiwanese kefir and viili starters. J. Dairy Sci. 2008, 91, 3798–3805. [Google Scholar] [CrossRef]
- Prakitchaiwattana, C.J.; Fleet, G.H.; Heard, G.M. Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res. 2004, 4, 865–877. [Google Scholar] [CrossRef]
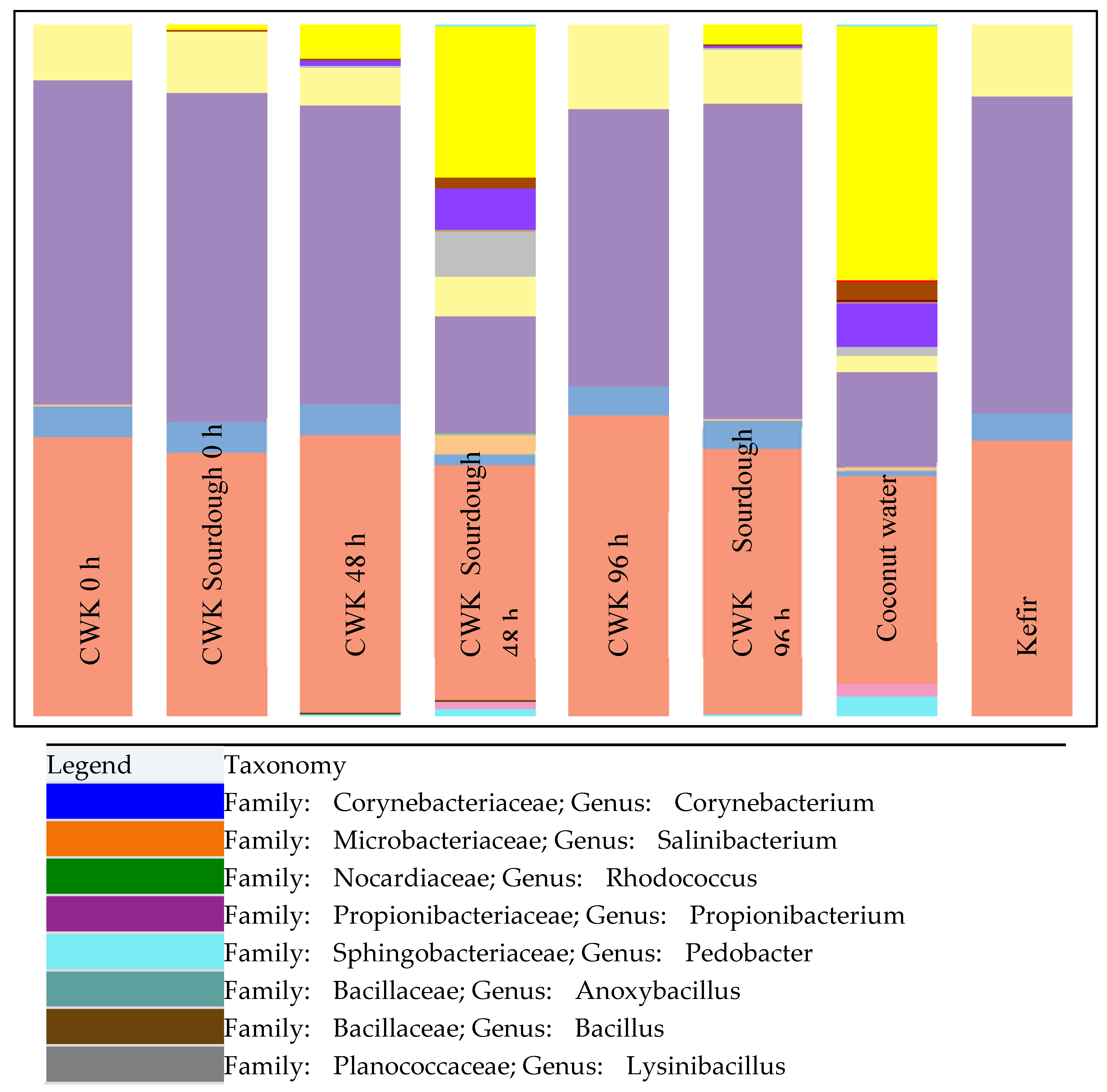
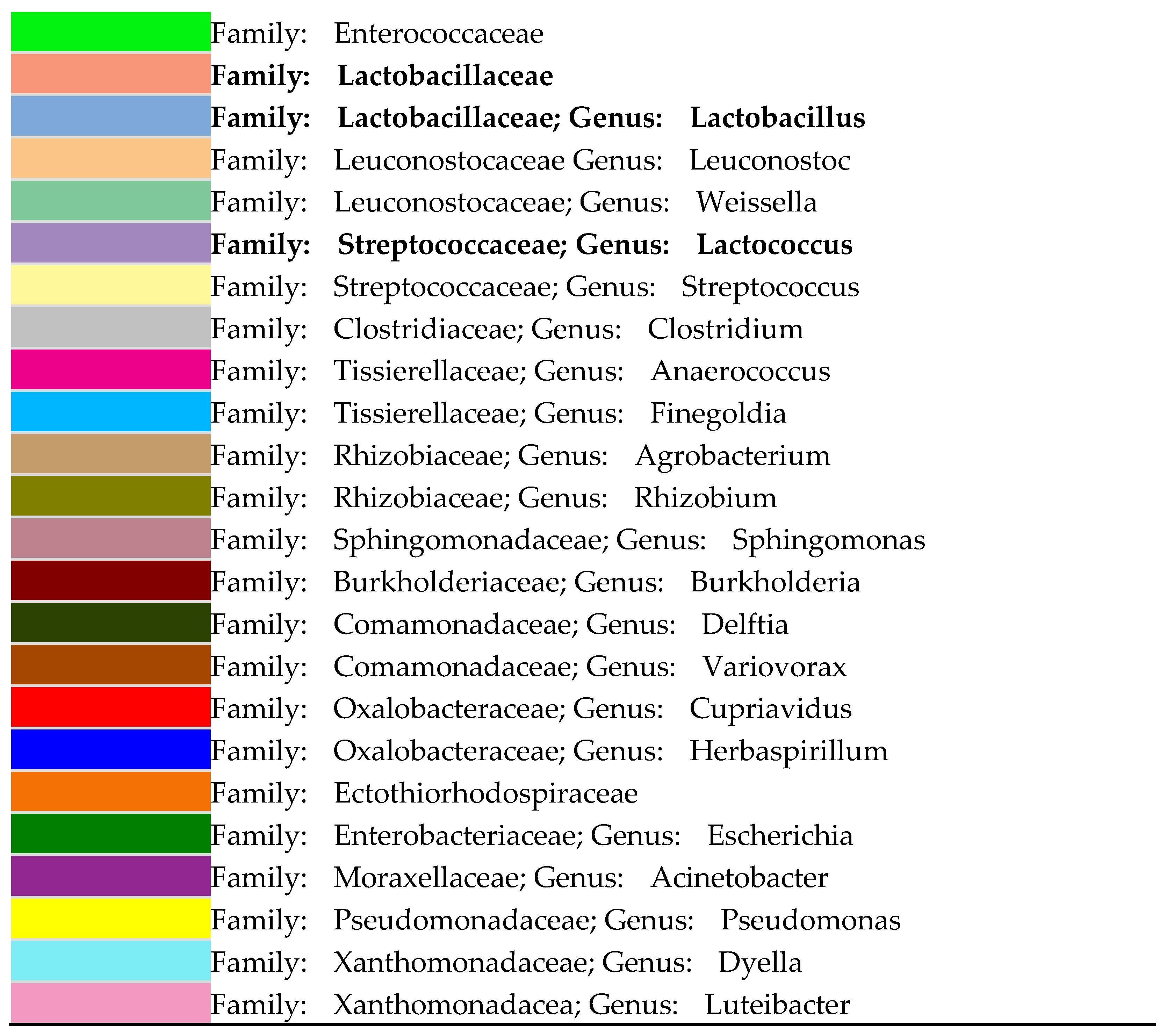
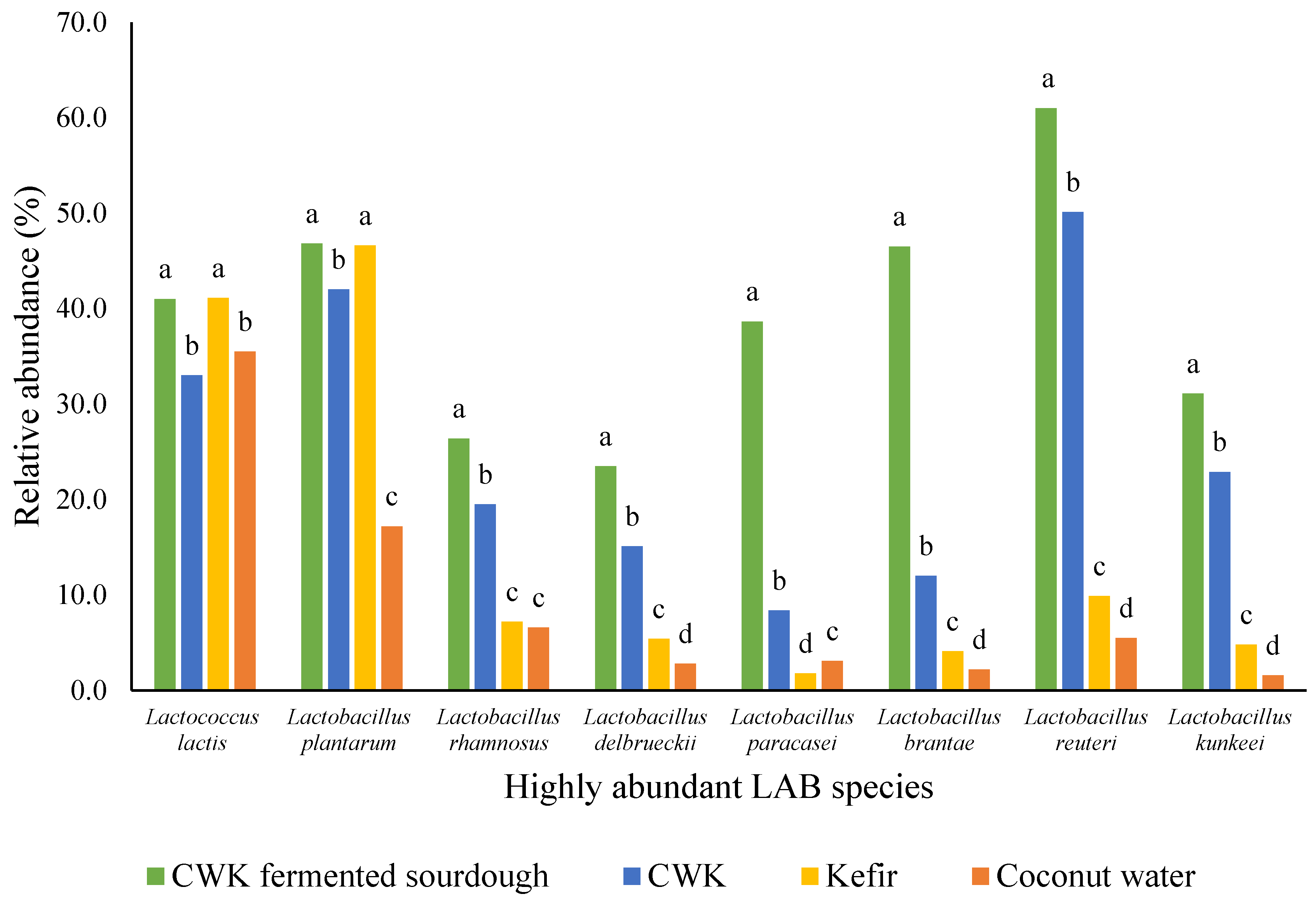
| Analysis | Kefir | Coconut Water | CWK | CWK-Fermented Sourdough | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 72 h | 96 h | 0 h | 24 h | 48 h | 72 h | 96 h | |||
| L-Lactic acid (g/L) | 0.55 ± 0.05 c | 2.55 ± 0.33 c | 2.29 ± 0.02 E,c | 2.59 ± 0.06 D,c | 3.03 ± 0.05 C,c | 4.85 ± 0.03 B,b | 5.56 ± 0.1 A,b | 2.1 ± 0.01 E,c | 3.92 ± 0.07 D,d | 4.34 ± 0.05 C,c | 6.25 ± 0.37 B,c | 8.08 ± 0.28 A,c |
| D-Lactic acid (g/L) | 0.16 ± 0.03 d | 1.68 ± 0.54 d | 1.52 ± 0.05 E,d | 2.07 ± 0.02 D,d | 2.69 ± 0.04 C,d | 2.94 ± 0.04 B,d | 3.17 ± 0.01 A,d | 2.14 ± 0.05 E,c | 4.18 ± 0.14 D,c | 6.6 ± 0.41 C,b | 7.79 ± 0.35 B,b | 10.26 ± 0.67 A,a |
| pH | 4.56 ± 0.66 a | 5.65 ± 0.21 a | 5.55 ± 0.01 A,a | 5.35 ± 0.01 B,b | 4.29 ± 0.01 C,b | 4.13 ± 0.01 D,c | 3.81 ± 0.05 E,c | 5.55 ± 0.09 A,a | 4.71 ± 0.33 B,b | 4.29 ± 0.22 C,c | 3.99 ± 0.14 D,d | 3.94 ± 0.14 E,d |
| TTA (0.1 M NaOH) | 6.14 ± 0.12 b | 5.34 ± 0.18 b | 5.21 ± 0.02 E,b | 6.50 ± 0.04 D,a | 8.85 ± 0.04 C,a | 10.26 ± 0.05 B,a | 11.71 ± 0.02 A,a | 5.18 ± 0.04 E,b | 6.94 ± 0.06 D,a | 7.3 ± 0.93 C,a | 9.44 ± 0.26 B,a | 9.92 ± 1.51 A,b |
| F-value | 100,457.100 | 151.866 | 13,672.771 | 1285.290 | 60,546.750 | 23,335.185 | 59,578.161 | 2347.616 | 148.945 | 671.822 | 1015.578 | 8019.417 |
| p-value | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
| Significant | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Limbad, M.; Gutierrez Maddox, N.; Hamid, N.; Kantono, K.; Higgins, C. Identification of the Microbiota in Coconut Water, Kefir, Coconut Water Kefir and Coconut Water Kefir-Fermented Sourdough Using Culture-Dependent Techniques and Illumina–MiSeq Sequencing. Microorganisms 2024, 12, 919. https://doi.org/10.3390/microorganisms12050919
Limbad M, Gutierrez Maddox N, Hamid N, Kantono K, Higgins C. Identification of the Microbiota in Coconut Water, Kefir, Coconut Water Kefir and Coconut Water Kefir-Fermented Sourdough Using Culture-Dependent Techniques and Illumina–MiSeq Sequencing. Microorganisms. 2024; 12(5):919. https://doi.org/10.3390/microorganisms12050919
Chicago/Turabian StyleLimbad, Mansi, Noemi Gutierrez Maddox, Nazimah Hamid, Kevin Kantono, and Colleen Higgins. 2024. "Identification of the Microbiota in Coconut Water, Kefir, Coconut Water Kefir and Coconut Water Kefir-Fermented Sourdough Using Culture-Dependent Techniques and Illumina–MiSeq Sequencing" Microorganisms 12, no. 5: 919. https://doi.org/10.3390/microorganisms12050919
APA StyleLimbad, M., Gutierrez Maddox, N., Hamid, N., Kantono, K., & Higgins, C. (2024). Identification of the Microbiota in Coconut Water, Kefir, Coconut Water Kefir and Coconut Water Kefir-Fermented Sourdough Using Culture-Dependent Techniques and Illumina–MiSeq Sequencing. Microorganisms, 12(5), 919. https://doi.org/10.3390/microorganisms12050919









