Antibiotic Resistance Profile of Salmonella sp. Isolates from Commercial Laying Hen Farms in Central-Western Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Location
2.2. Sampling
2.3. Antibiotic Susceptibility Profiling
2.4. Gene Detection
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, A.; Zhi, W.; Qiu, Y.; Wei, L.; Tian, J.; Pan, Z.; Duan, L. Surveillance study of the prevalence and antimicrobial resistance of Salmonella in pork from open markets in Xuzhou, China. Food Control 2019, 98, 474–480. [Google Scholar] [CrossRef]
- Jajere, S.M. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Vet. World 2019, 12, 504–521. [Google Scholar] [CrossRef] [PubMed]
- Manafi, L.; Aliakbarlu, J.; DastmalchiSaei, H. Antibiotic resistance and biofilm formation ability of Salmonella serotypes isolated from beef, mutton, and meat contact surfaces at retail. J. Food Sci. 2020, 85, 2516–2522. [Google Scholar] [CrossRef] [PubMed]
- Marchello, C.S.; Carr, S.D.; Crump, J.A. A Systematic Review on Antimicrobial Resistance among Salmonella Typhi Worldwide. Am. J. Trop. Med. Hyg. 2020, 103, 2518–2527. [Google Scholar] [CrossRef]
- Khan, M.; Shamim, S. Understanding the Mechanism of Antimicrobial Resistance and Pathogenesis of Salmonella enterica Serovar Typhi. Microorganisms 2022, 10, 2006. [Google Scholar] [CrossRef] [PubMed]
- Ayoola, M.B.; Das, A.R.; Krishnan, B.S.; Smith, D.R.; Nanduri, B.; Ramkumar, M. Predicting Salmonella MIC and Deciphering Genomic Determinants of Antibiotic Resistance and Susceptibility. Microorganisms 2024, 12, 134. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gu, D. Determination of Antimicrobial Resistance of Salmonella in Pork. Methods Mol. Biol. 2021, 21, 179–186. [Google Scholar]
- Magwedere, K.; Rauff, D.; De Klerk, G.; Keddy, K.H.; Dziva, F. Incidence of Nontyphoidal Salmonella in Food-Producing Animals, Animal Feed, and the Associated Environment in South Africa, 2012–2014. Clin. Infect Dis. 2015, 61, S283–S289. [Google Scholar] [CrossRef]
- Campos, J.; Mourão, J.; Silveira, L.; Saraiva, M.; Correia, C.B.; Maçãs, A.P.; Peixe, L.; Antunes, P. Imported poultry meat as a source of extended-spectrum cephalosporin-resistant CMY-2-producing Salmonella Heidelberg and Salmonella Minnesota in the European Union, 2014–2015. Int. J. Antimicrob. Agents 2018, 51, 151–154. [Google Scholar] [CrossRef] [PubMed]
- Viana, C.; Sereno, M.J.; Pegoraro, K.; Yamatogi, R.S.; Call, D.R.; Dos Santos Bersot, L.; Nero, L.A. Distribution, diversity, virulence genotypes and antibiotic resistance for Salmonella isolated from a Brazilian pork production chain. Int. J. Food Microbiol. 2019, 310, 108310. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Zeng, X.; Zhang, P.; Zhang, D.; Wang, C.; Lin, J. Characterization of the emerging multidrug-resistant Salmonella enterica serovar Indiana strains in China. Emerg. Microbes Infect. 2019, 8, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Castro-Vargas, R.E.; Herrera-Sánchez, M.P.; Rodríguez-Hernández, R.; Rondón-Barragán, I.S. Antibiotic resistance in Salmonella sp. isolated from poultry: A global overview. Vet. World 2020, 13, 2070–2084. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Salazar, E.; Gudiño, M.E.; Sevillano, G.; Zurita, J.; Guerrero-López, R.; Jaramillo, K.; Calero-Cáceres, W. Antibiotic resistance of Salmonella strains from layer poultry farms in central Ecuador. J. Appl. Microbiol. 2020, 128, 1347–1354. [Google Scholar] [CrossRef] [PubMed]
- Sodagari, H.R.; Habib, I.; Whiddon, S.; Wang, P.; Mohammed, A.B.; Robertson, I.; Goodchild, S. Occurrence and Characterization of Salmonella Isolated from Table Egg Layer Farming Environments in Western Australia and Insights into Biosecurity and Egg Handling Practices. Pathogens 2020, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, W.; Li, W.; Ding, Y.; Zhang, Y.; Yang, Q.; Wang, J.; Wang, X. A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 2020, 132, 109–111. [Google Scholar] [CrossRef] [PubMed]
- Wright, G.D. Q&A: Antibiotic resistance: Where does it come from and what can we do about it? BMC Biol. 2010, 8, 123. [Google Scholar]
- Torres, C.; Alonso, C.A.; Ruiz-Ripa, L.; León-Sampedro, R.; Del Campo, R.; Coque, T.M. Antimicrobial Resistance in Enterococcus spp. of animal origin. Microbiol. Spectr. 2018, 6, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Paveenkittiporn, W.; Kamjumphol, W.; Kerdsin, A. Draft Genome Sequence of Invasive Salmonella enterica Serovar Cannstatt Harboring mcr-1.1, Isolated from a Fatal Sepsis Case. Microbiol. Resour. Announc. 2021, 10, e01270-20. [Google Scholar] [CrossRef] [PubMed]
- Gudipati, S.; Zervos, M.; Herc, E. Can the One Health Approach Save Us from the Emergence and Reemergence of Infectious Pathogens in the Era of Climate Change: Implications for Antimicrobial Resistance? Antibiotics 2020, 9, 599. [Google Scholar] [CrossRef] [PubMed]
- Ojo, O.E.; Amosun, E.A.; Opebiyi, O.O.; Oyekunle, M.A.; Dipeolu, M.A.; Otesile, E.B. Multidrug resistant enterohaemorrhagic Escherichia coli serogroups in the faeces of hunted Wildlife, Abeokuta, Nigeria. Vet. Ital. 2022, 58, 173–179. [Google Scholar]
- Song, H.; Zou, S.; Huang, Y.; Jian, C.; Liu, W.; Tian, L.; Gong, L.; Chen, Z.; Sun, Z.; Wang, Y. Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid. Microorganisms 2024, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Bonciu, E. Aspects of the involvement of biotechnology in functional food and nutraceuticals. University of Craiova, Faculty of Agronomy. Sci. Pap.-Ser. A Agron. 2020, 2, 261–266. [Google Scholar]
- Iannetti, S.; Scarpone, R.; Dall’Acqua, F.; Rosato, R.; Chiumiento, F.; Cioci, D.; Migliorati, G.; Morelli, D.; Calistri, P. Estimation of the risk of fipronil ingestionthrough the consumption of contaminated table eggs for the Italian consumer. Vet. Ital. 2022, 58, 181–188. [Google Scholar]
- Barlow, R.S.; Debess, E.E.; Winthrop, K.L.; Lapidus, J.A.; Vega, R.; Cieslak, P.R. Travel-associated antimicrobial drug-resistant nontyphoidal Salmonellae, 2004–2009. Emerg. Infect Dis. 2014, 20, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Linke, L.; Doster, E.; Hyatt, D.; Burgess, B.A.; Magnuson, R.; Pabilonia, K.L.; Morley, P.S. Genomic diversity of class I integrons from antimicrobial resistant strains of Salmonella Typhimurium isolated from livestock, poultry and humans. PLoS ONE 2020, 15, e0243477. [Google Scholar] [CrossRef] [PubMed]
- Katzouraki, G.; Vasiliadis, E.S.; Marougklianis, V.; Evangelopoulos, D.S.; Pneumaticos, S.G. A Systematic Review of the Diagnosis and Treatment of Non-Typhoid Salmonella Spondylodiscitis in Immunocompetent Children. Children 2022, 9, 1852. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.J.I.; Islam, M.; Sikder, T.; Rubaya, R.; Halder, J.; Alam, J. Multidrug-resistant Escherichia coli and Salmonella spp. isolated from pigeons. Vet. World 2020, 13, 2156–2165. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). The European Union Summary Report on Antimicrobial Resistance in zoonotic and indicator bacteria from humans, animals and food in 2020/2021. EFSA J. 2023, 21, e07867. [Google Scholar]
- Velilla, A.V.; Terzolo, H.R. Biología molecular aplicada a La detección y caracterización de Salmonella enterica. In Proceedings of the XXI Congresso Latinoamericano de Avicultura, La Habana, Cuba, 6–9 October 2009; Volume 1, pp. 328–334. [Google Scholar]
- Heymans, R.; Vila, A.; Heerwaarden, C.A.M.; Jansen, C.C.C.; Castelijn, G.A.A.; Voort, M.; Biesta-Peters, E.G. Rapid detection and differentiation of Salmonella species, Salmonella Typhimurium and Salmonella Enteritidis by multiplex quantitative PCR. PLoS ONE 2018, 13, e0206316. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, C.; Wu, G.; Zhao, H.; He, T.; Cao, X.; Dai, L.; Xia, L.; Qin, S.; Shen, J. Prevalence of antimicrobial resistance among Salmonella isolates from chicken in China. Foodborne Pathog. Dis. 2011, 8, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Hernandez, A.; Carrascal-Camacho, A.K.; Varón-García, A.; Brashears, M.M.; Sanchez-Plata, M.X. Genotypic Characterization of Antimicrobial Resistant Salmonella spp. Strains from Three Poultry Processing Plants in Colombia. Foods 2021, 25, 491. [Google Scholar] [CrossRef] [PubMed]
- Doublet, B.; Douard, G.; Targant, H.; Meunier, D.; Madec, J.; Cloeckaert, A. Antibiotic marker modifications of λ Red and FLP helper plasmids, pKD46 and pCP20, for inactivation of chromosomal genes using PCR products in multidrug-resistant strains. J. Microbiol. Methods 2008, 75, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Parker, A.M.; Mohler, V.L.; Gunn, A.A.; House, J.K. Development of a qPCR for the detection and quantification of Salmonella spp. in sheep feces and tissues. J. Vet. Diagn. Investig. 2020, 32, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Chen, K.; Chan, E.W.; Yao, W.; Chen, S. Transmission of Chromosomal MDR DNA Fragment Encoding Ciprofloxacin Resistance by a Conjugative Helper Plasmid in Salmonella. Front Microbiol. 2020, 11, 556227. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Su, L.H.; Janapatla, R.P.; Lin, C.Y.; Chiu, C.H. Genetic analysis of virulence and antimicrobial-resistant plasmid pOU7519 in Salmonella enterica serovar Choleraesuis. J. Microbiol. Immunol. Infect. 2020, 53, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI supplement M100; Clinical Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Santos, L.R.; Nascimento, V.P.; de Oliveira, S.D.; Flores, M.L.; Pontes, A.P.; Ribeiro, A.R.; Salle, C.T.; Lopes, R.F. Polymerase chain reaction (PCR) for the detection of Salmonella in artificially inoculated chicken meat. Rev. Inst. Med. Trop. São Paulo 2001, 43, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Bugarel, M.; Granier, S.A.; Weill, F.X.; Fach, P.; Brisabois, A. A multiplex real-time PCR assay targeting virulence and resistance genes in Salmonella enterica serotype Typhimurium. BMC Microbiol. 2011, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Arkali, A.; Çetinkaya, B. Molecular identification and antibiotic resistance profiling of Salmonella species isolated from chickens in eastern Turkey. BMC Vet. Res. 2020, 19, 205. [Google Scholar] [CrossRef] [PubMed]
- Tawyabur, M.; Islam, M.S.; Sobur, M.A.; Hossain, M.J.; Mahmud, M.M.; Paul, S.; Hossain, M.T.; Ashour, H.M.; Rahman, M.T. Isolation and Characterization of Multidrug-Resistant Escherichia coli and Salmonella spp. from Healthy and Diseased Turkeys. Antibiotics 2020, 9, 770. [Google Scholar] [CrossRef]
- Ince, S.S.; Akan, M. Phenotypic and genotypic characterization of antimicrobial resistance in commonly isolated Salmonella serovars from chickens. Turk. J. Vet. Anim. Sci. 2023, 47, 19–25. [Google Scholar] [CrossRef]
- Garcia, J.S.; Gast, R.K.; Guard, J.Y.; Karcher, D.M.; Jones, D. Tissue Colonization and Egg and Environmental Contamination Associated with the Experimental Infection of Cage-Free Laying Hens with Salmonella Braenderup. Avian Dis. 2022, 66, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, M.; Hendriksen, R.S.; Nochi, Z.; Zali, M.R.; Aarestrup, F.M.; Garcia-Migura, L. Antimicrobial resistance in Salmonella spp. recovered from patients admitted to six different hospitals in Tehran, Iran from 2007 to 2008. Folia Microbiol. 2012, 57, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Villegas, K.J.; Herrera-Sánchez, M.P.; Beltrán-Martínez, M.A.; Cárdenas-Moscoso, S.; Rondón-Barragán, I.S. Molecular Detection of Virulence Factors in Salmonella serovars Isolated from Poultry and Human Samples. Vet. Med. Int. 2023, 2, 1875253. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Cao, G.; Brown, E.W.; Allard, M.W.; Ma, L.M.; Khan, A.A.; Zhang, G. Antimicrobial resistance and related gene analysis of Salmonella from egg and chicken sources by whole-genome sequencing. Poult. Sci. 2020, 99, 7076–7083. [Google Scholar] [CrossRef] [PubMed]
- Galdino, V.M.C.; Melo, R.T.; Oliveira, R.P.; Mendonça, E.P.; Nalevaiko, P.C.; Rossi, D.A. Virulência de Salmonella spp. de origem avícola e resistência a antimicrobianos. Biosci. J. 2013, 29, 932–939. [Google Scholar]
- Khoo, E.; Roslee, R.; Zakaria, Z.; Ahmad, N.I. Virulence gene profiles and antimicrobial susceptibility of Salmonella Brancaster from chicken. J. Vet. Sci. 2023, 24, e82. [Google Scholar] [CrossRef]
- Penha-Filho, R.A.C.; Ferreira, J.C.; Galetti, R.; Kanashiro, A.M.I.; Berchieri, A.J.; Costa Darini, A.L. The rise of multidrug resistant Salmonella isolates in healthy chickens in Brazil by successful establishment of plasmid IncHI2A carrying several antibiotic resistance genes. Braz. J. Microbiol. 2023, 54, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; Norrung, B. Scientific Opinion of the Panel on Biological Hazards on a request from the European Food Safety Authority on foodborne antimicrobial resistance as a biological hazard in Question No EFSA-Q-2007-089. Eur. Food Saf. Auth. 2008, 765, 1–84. [Google Scholar]
- WHO—World Healthy Organization. Critically Important Antibacterial Agents for Human Medicine for Risk Management Strategies of Non-Human Use: Report of a WHO Working Group Consultation. 6th Revision 2018; World Health Organization: Genebra, Switzerland, 2019; pp. 1–52. [Google Scholar]
- Ribeiro, A.R.; Kellermann, A.; Santos, L.R.; Nascimento, V.P. Resistência antimicrobiana em Salmonella Enteritidis isoladas de amostras clínicas e ambientais de frangos de corte e matrizes pesadas. Arq. Bras. Med. Veterinária Zootec. 2008, 60, 1259–1262. [Google Scholar] [CrossRef]
- Alcântara, J.B.; Martins, P.C.; Nascente, E.P.; Café, M.B.; Pascoal, L.M.; Teles, A.V.; Jayme, V.S.; Andrade, M.A. Salmonella enterica diversity and antimicrobial resistance profile in broiler slaughterhouse by-products. Vet. Ital. 2022, 58, 209–213. [Google Scholar]
- Mohammed, Y.; Dubie, T. Isolation, identification and antimicrobial susceptibility profile of Salmonella isolated from poultry farms in Addis Ababa, Ethiopia. Vet. Med. Sci. 2022, 8, 1166–1173. [Google Scholar] [CrossRef] [PubMed]
- Urban-Chmiel, R.; Marek, A.; Stępień-Pyśniak, D.; Wieczorek, K.; Dec, M.; Nowaczek, A.; Osek, J. Antibiotic Resistance in Bacteria-A Review. Antibiotics 2022, 11, 1079. [Google Scholar] [CrossRef] [PubMed]
- Guillén, S.; Marcén, M.; Álvarez, I.; Mañas, P.; Cebrián, G. Stress resistance of emerging poultry-associated Salmonella serovars. Int. J. Food Microbiol. 2020, 16, 108884. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Li, J.; Sun, Z.; Hu, C.; Jin, S.; Li, F.; Guo, Y.; Ran, L.; Ma, Y. Characterization of Salmonella enterica isolates from infants and toddlers in Wuhan, China. J. Antimicrob. Chemother. 2009, 63, 87–94. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hawkey, J.; Le Hello, S.; Doublet, B.; Granier, S.A.; Hendriksen, R.S.; Fricke, W.F.; Ceyssens, P.J.; Gomart, C.; Billman-Jacobe, H.; Holt, K.E.; et al. Global phylogenomics of multidrug-resistant Salmonella enterica serotype Kentucky ST198. Microb. Genom. 2019, 5, e000269. [Google Scholar] [CrossRef] [PubMed]
- Hsu, Y.; Tang, C.; Lin, H.; Chen, Y.; Chen, Y.; Su, Y.; Chen, D.S.; Lin, J.; Chang, C. Comparative study of class 1 integron, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline (ACSSuT) and fluoroquinolone resistance in various Salmonella serovars from humans and animals. Comp. Immunol. Microbiol. Infect. Dis. 2013, 36, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Chauhan, S.L.; Kumar, S.; Jindal, N.; Mahajan, N.K.; Joshi, V.G. Carriage of Class 1 integrons and molecular characterization of intI1 gene in multidrug-resistant Salmonella spp. isolates from broilers. Vet. World 2019, 12, 609–613. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Su, J.; Xu, Z. Antibiotic Susceptibility, Biofilm-Forming Ability, and Incidence of Class 1 Integron of Salmonella spp., Escherichia coli, and Staphylococcus aureus isolated from various foods in a school canteen in China. Foodborne Pathog. Dis. 2020, 17, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Monte, D.F.M.; Sellera, F.P.; Lopes, R.; Keelara, S.; Landgraf, M.; Greene, S.; Fedorka-Cray, P.J.; Thakur, S. Class 1 integron-borne cassettes harboring blaCARB-2 gene in multidrug-resistant and virulent Salmonella Typhimurium ST19 strains recovered from clinical human stool samples, United States. PLoS ONE 2020, 30, e0240978. [Google Scholar] [CrossRef] [PubMed]
- Eshaghi-Zadeh, S.H.; Fahimi, H.; Fardsanei, F.; Soltan-Dallal, M.M. Antimicrobial Resistance and Presence of Class 1 Integrons Among Different Serotypes of Salmonella spp. Recovered from Children with Diarrhea in Tehran, Iran. Infect. Disord. Drug Targets 2020, 20, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Curraize, C.; Siebor, E.; Neuwirth, C.; Hall, R.M. SGI0, a relative of Salmonella genomic islands SGI1 and SGI2, lacking a class 1 integron, found in Proteus mirabilis. Plasmid 2020, 107, 102453. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, S.; Duong, V.T.; Tuyen, H.T.; Campbell, J.I.; Thomson, N.R.; Parkhill, J.; Le Phuc, H.; Chau, T.T.H.; Maskell, D.J.; Perron, G.G.; et al. Mobility of antimicrobial resistance across serovars and disease presentations in non-typhoidal Salmonella from animals and humans in Vietnam. Microb. Genom. 2022, 8, 000798. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.B.; Mahmud, M.; Akter, R.; Hasan, M.; Sobur, A.; Nazir, K.N.H.; Noreddin, A.; Rahman, T.; El Zowalaty, M.E.; Rahman, M. Molecular Detection of Multidrug Resistant Salmonella Species Isolated from Broiler Farm in Bangladesh. Pathogens 2020, 9, 201. [Google Scholar] [CrossRef]
- Yang, C.M.; Lin, M.F.; Lin, C.H.; Huang, Y.T.; Hsu, C.T.; Liou, M.L. Characterization of antimicrobial resistance patterns and integrons in human fecal Escherichia coli in Taiwan. Jpn. J. Infect. Dis. 2009, 62, 177–181. [Google Scholar] [CrossRef]
- Firoozeh, F.; Zahraei-Salehi, T.; Shahcheraghi, F.; Karimi, V.; Aslani, M.M. Characterization of class I integrons among Salmonella enterica serovar Enteritidis isolated from humans and poultry. FEMS Immunol. Med. Microbiol. 2012, 64, 237–243. [Google Scholar] [CrossRef]
- Ramatla, T.; Mileng, K.; Ndou, R.; Mphuti, N.; Syakalima, M.; Lekota, K.E.; Thekisoe, O.M.M. Molecular Detection of Integrons, Colistin and β-lactamase Resistant Genes in Salmonella enterica Serovars Enteritidis and Typhimurium Isolated from Chickens and Rats Inhabiting Poultry Farms. Microorganisms 2022, 10, 313. [Google Scholar] [CrossRef] [PubMed]
- Wannaprasat, W.; Padungtod, P.; Chuanchuen, R. Class 1 integrons and virulence genes in Salmonella enterica isolates from pork and humans. Int. J. Antimicrob. Agents 2011, 37, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Bonilla-Caballero, M.A.; Lozano-Puentes, M.P.; Ospina, M.A.; Varón-López, M. First report of multidrug-resistant Salmonella Infantis in broiler litter in Tolima, Colombia. Vet. World 2022, 15, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.A.; Hossain, M.T.; Islam, M.S.; Islam, M.Z.; Islam, P.; Shaha, S.N.; Sikder, M.H.; Rafiq, K. Isolation of multidrug-resistant Escherichia coli and Salmonella spp. from sulfonamide-treated diarrheic calves. Vet. World 2022, 15, 2870–2876. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Sidjabat, H.E.; Davis, S.; Vong da Silva, P.G.; Alves, A.; Dos Santos, C.; Jong, J.B.D.C.; da Conceição, F.; Felipe, N.D.J.; Ximenes, A.; et al. Prevalence of Antimicrobial Resistance in Escherichia coli and Salmonella Species Isolates from Chickens in Live Bird Markets and Boot Swabs from Layer Farms in Timor-Leste. Antibiotics 2024, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Ministério da Agricultura Pecuária e Abastecimento. Resultados do Plano Nacional de Controle de Resíduos e Contaminantes—PNCRC 2022; MAPA: Brasília, Brasil, 2023; pp. 1–13. [Google Scholar]
- ABPA. Associação Brasileira de Proteína Animal; Relatório Anual ABPA 2023; ABPA: São Paulo, Brazil, 2023; pp. 1–75. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions, and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions, or products referred to in the content. |

| Antibiotic Class | Antibiotic Tested | Concentration |
|---|---|---|
| Cephalosporins | Ceftiofur | 30 μg |
| Fluoroquinolones | Enrofloxacin | 5 μg |
| Ciprofloxacin | 10 μg | |
| Penicillins | Amoxicillin + Clavulanic acid | 3 μg |
| Ampicillin | 20 μg | |
| Tetracyclines | Doxycycline | 30 μg |
| Tetracycline | 30 μg | |
| Oxytetracycline | 30 μg | |
| Aminoglycosides | Gentamicin | 10 μg |
| Neomycin | 30 μg | |
| Apramycin | 15 μg | |
| Sulfonamides | Trimethoprim + sulfamethoxazole | 25 μg |
| Sulfamethoxazole | 25 μg | |
| Sulfonamides | 300 μg |
| Antibiotic | Susceptibility Profile | ||
|---|---|---|---|
| Resistance (%) | Intermediate Resistance (%) | Sensitivity (%) | |
| Sulfamethoxazole | 91 | 6.7 | 2.3 |
| Sulfonamides | 51 | 9 | 40 |
| Ceftiofur | 28.9 | 42.2 | 28.9 |
| Apramycin | 6.7 | 48.9 | 44.4 |
| Amoxicillin/clavulanic acid | 4.4 | 20 | 75.5 |
| Enrofloxacin | 4.4 | 6.7 | 88.9 |
| Gentamicin | 4.4 | - | 95.6 |
| Neomycin | 4.4 | 75.6 | 20 |
| Doxycycline | 4.4 | 60 | 35.5 |
| Tetracycline | 2.2 | 15.5 | 82.3 |
| Ampicillin | 2.2 | 2.2 | 95.6 |
| Trimethoprim-sulfamethoxazole | 2.2 | - | 97.8 |
| Oxytetracycline | 2.2 | 8.9 | 88.9 |
| Ciprofloxacin | - | 11.1 | 88.9 |
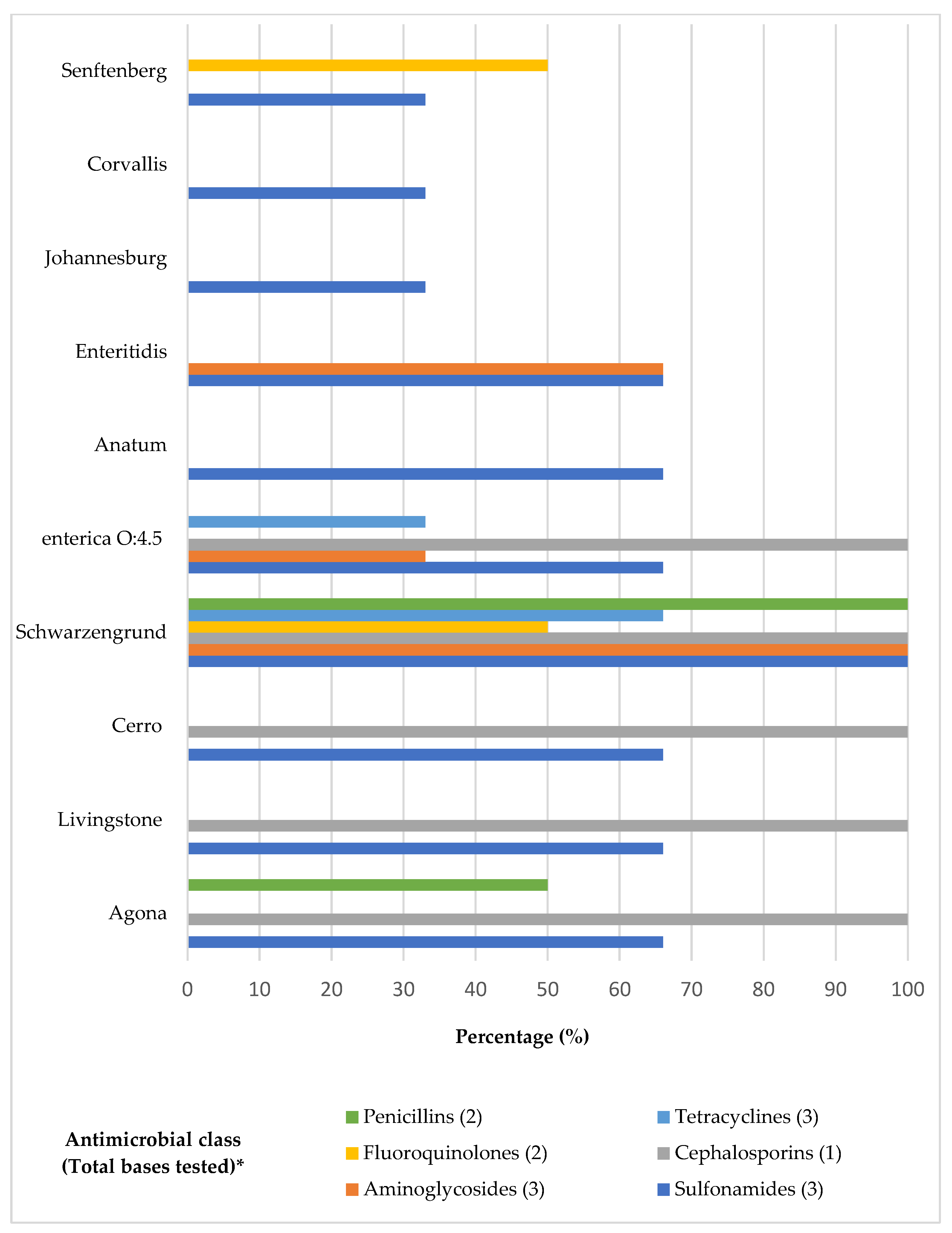
| Serovar (n) | Agona (14) | Livingstone (8) | Cerro (6) | Schwarzengrund (5) | Enterica O:4.5 (4) | Anatum (3) | Enteritidis (2) | Johannesburg (1) | Corvallis (1) | Senftenberg (1) | Total (45) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SMT | 11/14 DFPA | 8/8 DFPA | 5/6 DFPA | 5/5 DFPA | 4/4 DFPA | 3/3 DFPA | 2/2 DFPA | 1/1 DFPA | 1/1 DFPA | 1/1 DFPA | 41 |
| STX | - | - | - | 1/5 DFPA | - | - | - | - | - | - | 1 |
| ENR | - | - | - | 1/5 DFPA | - | - | - | - | - | 1/1 DFPA | 2 |
| TET | - | - | - | 1/5 DFPA | - | - | - | - | - | - | 1 |
| SUL | 6/14 DFPA | 3/8 DFPA | 2/6 DFPA | 4/5 DFPA | 4/4 DFPA | 2/3 DFPA | 2/2 DFPA | - | - | - | 23 |
| CIP | - | - | - | - | - | - | - | - | - | - | - |
| AMC | 1/14 DFPA | - | - | 1/5 DFPA | - | - | - | - | - | - | 2 |
| AMP | - | - | - | 1/5 DFPA | - | - | - | - | - | - | 1 |
| CEF | 4/14 DFPA | 2/8 DFPA | 1/6 DFPA | 3/5 DFPA | 3/4 DFPA | - | - | - | - | - | 13 |
| GEN | - | - | - | 1/5 DFPA | - | - | 1/2 DFPA | - | - | - | 2 |
| OXI | - | - | - | 1/5 DFPA | - | - | - | - | - | - | 1 |
| NEO | - | - | - | 1/5 DFPA | - | - | 1/2 DFPA | - | - | - | 2 |
| DOX | - | - | - | 1/5 DFPA | 1/4 DFPA | - | - | - | - | - | 2 |
| APR | - | - | - | 1/5 DFPA | 1/4 DFPA | - | 1/2 DFPA | - | - | - | 3 |
| Salmonella Serovar | n | AMP | AMC | SUL | Resistance Gene | ||
|---|---|---|---|---|---|---|---|
| intl1 | sul1 | blaTEM | |||||
| Agona | 14 | - | 1/14 DFPA | 6/14 DFPA | 1/6 DFPA | 1/6 DFPA | - |
| Livingstone | 8 | - | - | 3/8 DFPA | 3/8 DFPA | 3/8 DFPA | - |
| Cerro | 6 | - | - | 2/6 DFPA | 1/6 DFPA | - | - |
| Schwarzengrund | 5 | 1/5 DFPA | 1/5 DFPA | 4/5 DFPA | 2/5 DFPA | 2/5 DFPA | - |
| enterica O:4.5 | 4 | - | - | 4/4 DFPA | 1/4 DFPA | - | - |
| Anatum | 3 | - | - | 2/3 DFPA | - | - | - |
| Enteritidis | 2 | - | - | 2/2 DFPA | - | - | - |
| Johannesburg | 1 | - | - | - | 1/1 DFPA | - | - |
| Corvallis | 1 | - | - | - | - | - | - |
| Senftenberg | 1 | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moraes, D.M.C.; Almeida, A.M.D.S.; Andrade, M.A.; Nascente, E.d.P.; Duarte, S.C.; Nunes, I.A.; Jayme, V.D.S.; Minafra, C. Antibiotic Resistance Profile of Salmonella sp. Isolates from Commercial Laying Hen Farms in Central-Western Brazil. Microorganisms 2024, 12, 669. https://doi.org/10.3390/microorganisms12040669
Moraes DMC, Almeida AMDS, Andrade MA, Nascente EdP, Duarte SC, Nunes IA, Jayme VDS, Minafra C. Antibiotic Resistance Profile of Salmonella sp. Isolates from Commercial Laying Hen Farms in Central-Western Brazil. Microorganisms. 2024; 12(4):669. https://doi.org/10.3390/microorganisms12040669
Chicago/Turabian StyleMoraes, Dunya Mara Cardoso, Ana Maria De Souza Almeida, Maria Auxiliadora Andrade, Eduardo de Paula Nascente, Sabrina Castilho Duarte, Iolanda Aparecida Nunes, Valéria De Sá Jayme, and Cíntia Minafra. 2024. "Antibiotic Resistance Profile of Salmonella sp. Isolates from Commercial Laying Hen Farms in Central-Western Brazil" Microorganisms 12, no. 4: 669. https://doi.org/10.3390/microorganisms12040669
APA StyleMoraes, D. M. C., Almeida, A. M. D. S., Andrade, M. A., Nascente, E. d. P., Duarte, S. C., Nunes, I. A., Jayme, V. D. S., & Minafra, C. (2024). Antibiotic Resistance Profile of Salmonella sp. Isolates from Commercial Laying Hen Farms in Central-Western Brazil. Microorganisms, 12(4), 669. https://doi.org/10.3390/microorganisms12040669






