Abstract
The objective of this study was to characterize Cronobacter spp. and related organisms isolated from powder dairy products intended for consumption by adults and older adults using whole-genome sequencing (WGS), and to identify genes and traits that encode antibiotic resistance and virulence. Virulence (VGs) and antibiotic resistance genes (ARGs) were detected with the Comprehensive Antibiotic Resistance Database (CARD) platform, ResFinder, and MOB-suite tools. Susceptibility testing was performed using disk diffusion. Five presumptive strains of Cronobacter spp. were identified by MALDI–TOF MS and ribosomal MLST. Three C. sakazakii strains were of the clinical pathovar ST1, one was ST31, and the remaining isolate was C. malonaticus ST60. In addition, Franconibacter helveticus ST345 was identified. The C. sakazakii ST1 strains were further distinguished using core genome MLST based on 2831 loci. Moreover, 100% of the strains were resistant to cefalotin, 75% to ampicillin, and 50% to amikacin. The C. sakazakii ST1 strains were multiresistant (MDR) to four antibiotics. Additionally, all the strains adhered to the N1E-115 cell line, and two invaded it. Eighteen ARGs mainly involved in antibiotic target alteration and antibiotic efflux were detected. Thirty VGs were detected and clustered as flagellar proteins, outer membrane proteins, chemotaxis, hemolysins, and genes involved in metabolism and stress. The pESA3, pSP291-1, and pCMA1 plasmids were detected, and the prevalent mobile genetic elements (MGEs) were ISEsa1, ISEc52, and IS26. The isolates of C. sakazakii and C. malonaticus exhibited multiresistance to antibiotics, harbored genes encoding various antibiotic resistance proteins, and various virulence factors. Consequently, these contaminated powdered dairy products pose a risk to the health of hypersensitive adults.
1. Introduction
Cronobacter spp. is a genus of enteropathogenic bacteria comprising seven species: Cronobacter sakazakii, C. malonaticus, C. universalis, C. turicensis, C. muytjensii, C. dublinensis, and C. condimenti [1,2,3]. Among these species, C. sakazakii and C. malonaticus are especially important in clinical settings [4].
C. sakazakii primarily affects premature newborns and infants, while C. malonaticus is more common in older adults [5,6,7]. In infants, meningitis, septicemia, and necrotizing enterocolitis (NEC) have been reported, while urinary tract and respiratory infections are more common in adults. The mortality rates associated with C. sakazakii infection in neonatal meningitis and septicemia are 15% and 25%, respectively [6]. Neonatal meningitis cases are especially linked to C. sakazakii sequence type 4 (ST4/CC4) infections [8], while cases of infections in adults are often associated with C. malonaticus ST7 and ST60 [9,10]. In 2014, Holy et al. reported the presence of Cronobacter spp. in all samples from hospitalized patients between 2005 and 2011. The prevalence was higher in the under-one-year-old group at 7.7%, with an incidence of 8.7 per 1000 patients. For adults, the incidence of Cronobacter spp. was 0.5, 2.8, and 2.0 in patients aged 45–64, 65–74, and over 75 years, respectively [11]. Even though infections by C. sakazakii have been associated with particularly sensitive groups, in 2016, an outbreak of gastroenteritis affecting 156 Chinese students caused by Cronobacter sakazakii and C. malonaticus was reported [12]. Cronobacter spp. infections in infants are primarily attributed to the consumption of contaminated rehydrated powdered infant formula (PIF). Cronobacter spp. can be isolated from powdered infant formula (PIF), rehydrated powdered milk, infant cereals, water, and various surfaces within manufacturing facilities. Furthermore, international studies on Cronobacter spp. have reported prevalence values ranging from 3% to 30% [13,14]. However, the probable source of infection in adults has not been determined. Furthermore, the ability of these pathogens to persist in food environments and settings poses an additional risk to the presence of this pathogen in food [15].
The severity of the clinical condition has been associated with the presence of plasmid-encoded virulence factors [16,17], adherence and invasion in cell lines [18,19], and various other genes, such as aut, cpA, fliC, hly, ompA, sip, plas, and inv, that are important for the adherence of the microorganism to the epithelial surface, multiplication, colonization, evasion of the host’s natural or innate defenses, tissue invasion, and cellular damage [20,21,22]. Other factors include the utilization of sialic acid as a carbon source, capsule composition and presence, and endotoxin production [23]. Another important aspect is the resistance to β-lactam antibiotics such as cephalothin, cefotaxime, ceftazidime, and ampicillin, along with the presence of resistance genes such as marA, glpT, ampH, blaCSA, blaCMA, mrc, and the efflux pump system AcrAB-TolC [24,25,26].
The use of whole-genome sequencing (WGS) has allowed for more precise differentiation between species. For Cronobacter, WGS has shown high discrimination of conserved and variable genetic information, enabling differentiation between closely related species such as Franconibacter helveticus and Enterobacter hormaechei. Therefore, WGS is used as a tool for identification, genotyping (seven-loci multilocus sequence typing (MLST) and core genome multilocus sequence typing), CRISPR-Cas array profiling, serogrouping, SNP determination, detection of genes conferring resistance to antibiotics (ARGs) and/or virulence (VGs), and therefore considerably facilitates molecular epidemiological studies [27].
In this study, the objective was to use WGS to characterize Cronobacter spp. strains isolated from powdered dairy products intended for consumption by adults and older adults, and to identify genes and traits that encode antibiotic resistance and virulence.
2. Materials and Methods
2.1. Sampling
A total of 100 samples of dairy products intended for adults and older adults were analyzed between 2018 and 2020, from 2 commercially available brands. Each container or can served as a sample and was obtained monthly from supermarkets and pharmacies due to their monthly restocking practices. This allowed for greater variability in terms of the production batch origins of the samples. The samples were stored in their sealed containers following the manufacturer’s guidelines until analysis. Later, the containers were opened within a laminar flow hood. The tools used for sample collection were sterilized at 121 °C for 15 min, and the outer surfaces of the containers were treated with 70% alcohol.
2.2. Isolation and Identification Methods of Cronobacter spp.
Cronobacter spp. strains were isolated following the general methodology described by Iversen et al. [28]. Initially, food and environmental samples were pre-enriched in buffered peptone water (BPW) and then transferred to Enterobacteriaceae enrichment broth (BD Difco, Sparks, MD, USA). Subsequently, the samples were plated onto Brilliance CM 1035 chromogenic agar (Oxoid Thermo-Fisher, Basingstoke, UK) and purified on trypticase soy agar (BD Difco, Sparks, MD, USA). Before sequencing, the suspected strains underwent presumptive identification using Matrix-Assisted Laser Desorption/Ionization–Time of Flight Mass Spectrometry (MALDI–TOF MS) (Bruker, Billerica, MA, USA) and the MALDI Biotyper Compass IVD 4.1.60 software (Bruker, Billerica, MA, USA) as described by Lepuschitz et al. [29]. The confirmation of presumptive Cronobacter spp. strains was achieved using the ribosomal multilocus sequence typing (rMLST) software available at https://pubmlst.org/species-id (accessed on 28 September 2023) [30] and average nucleotide identity (ANI) analysis using JSpecies v3.9.7 [31]. The strains were annotated using software Proksee v1.3.0 [32] and Prokka v1.1.1 [33].
2.3. Whole-Genome Sequencing (WGS)
Cronobacter spp. isolates were cultured on Columbia blood agar plates (bioMérieux, Marcy-l’Étoile, France) at 37 °C for 24 h. Genomic DNA was extracted from bacterial cultures using the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The concentration of DNA was quantified using a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and the dsDNA BR assay kit (Thermo Fisher Scientific). For library preparation, Nextera XT chemistry (Illumina Inc., San Diego, CA, USA) was employed, and the libraries were subjected to a 2 × 300 bp paired-end sequencing run on an Illumina MiSeq sequencer. Sequencing was performed to achieve a minimum coverage of 80-fold using standard Illumina protocols. Raw reads were quality-controlled using FastQC v0.11.9, and adapter sequences were removed, while the last 10 bp of each sequence and sequences with quality scores below 20 were trimmed using Trimmomatic v0.36 [34]. The reads were then assembled using SPAdes v3.11.1 [35]. Finally, contigs were filtered based on a minimum coverage of 5× and a minimum length of 200 bp using SeqSphere+ software v9.0.8 (Ridom GmbH, Würzburg, Germany) [36]. Sequencing quality data as coverage, no. of the contigs, N50 were included Supplementary Table S1.
2.4. Sequence Type (ST) and Core Genome Multilocus Sequence Typing (cgMLST) of Cronobacter Isolates
Comprehensive core genome multilocus sequence typing (cgMLST) was carried out on Cronobacter isolates using a core genome comprising 2831 target genes using the Ridom SeqSphere+ software v.9.0.8 (Ridom, Münster, Germany) [36]. A minimum spanning tree (MST) was created from the allelic profiles of the isolates to determine genotypic relationships. Clusters were defined as isolates with a maximum difference of 10 alleles. Additionally, the sequences of the seven housekeeping genes commonly used in conventional MLST for C. sakazakii and C. malonaticus (atpD, fusA, glnS, gltB, gyrB, infB, and ppsA) were extracted and cross-checked against the Cronobacter MLST database available at https://pubmlst.org/organisms/cronobacter-spp/ (accessed on 1 September 2023).
2.5. Determination of Serotypes
The serotypes of Cronobacter spp. were determined by analyzing the profiles of the gnd and galF genes, which are specific to the serotype O region [37]. Whole-genome sequencing (WGS) data analysis was performed using the BIGSdb tool available in the PubMLST database (https://pubmlst.org/organisms/cronobacter-spp) (accessed on 10 September 2023) [38].
2.6. Antibiotic Susceptibility
Antibiotic susceptibility was assessed using the disk diffusion method, following the recommendations of the Clinical and Laboratory Standards Institute [39]. Commercial antibiotic disks include amikacin (30 µg), ampicillin (10 μg), amoxicillin-clavulanic acid (20/10 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), chloramphenicol (30 μg), cefotaxime (30 μg), cefepime (30 µg), gentamicin (10 μg), cephalothin (30 μg), and tetracycline (30 µg). The interpretation of resistance/susceptibility profiles was conducted according to the manufacturer’s instructions. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 strains were used as internal controls.
2.7. Adherence and Invasion Assays
The N1E-115 (ATCC®CRL-2263) cell line was cultured in Dulbecco’s Modified Eagle Medium (DMEM) high glucose 4.5 g/L (ATCC; Manassas, VA, USA), supplemented with 10% fetal bovine serum (FBS) from (ATCC; Manassas, VA, USA). Subsequently, the cells were induced to differentiate in DMEM medium supplemented with 2% FBS and 1.25% dimethyl sulfoxide over a period of five days. Cell cultures were established in 24-well plates (Corning Life Sciences, Tewksbury, USA) at a concentration of 1 × 105 cells/mL and subjected to infection with each C. sakazakii isolate at a multiplicity of infection ratio of 100:1 following prior cultivation in Luria broth. Infection was conducted for a duration of 4 h at 37 °C in an atmosphere containing 5% CO2. After the incubation period, the cells were rinsed with 1× phosphate-buffered saline (PBS), and the removal of C. sakazakii was achieved by introducing 1 mL of 0.1% Triton X-100 (Amresco, Solon, OH, USA). To determine the colony-forming units (CFU) of bacteria attached to the N1E-115 cells, various dilutions were carried out in Luria broth [14]. For the invasion assay, the preparation of the N1E-115 monolayers and the time for infection were as described for the adhesion assay. After 4 h of incubation, the infected monolayers were washed with 1× PBS and incubated with 1 mL of DMEM plus 300 µg/mL lysozyme (Sigma-Aldrich, Virginia Beach, USA) and 100 µg/mL gentamicin (Sigma-Aldrich, USA) for 2 h at 37 °C in 5% CO2. The cells were washed three times with 1× PBS, separated with 1 mL of 0.1% Triton X-100, and plated on Luria–Bertani agar. Invasion frequencies were calculated as the number of bacteria that survived incubation with gentamicin and lysozyme divided by the total number of bacteria present in the absence of this antibiotic (bacterial adherence) [18]. Both assays (adhesion, invasion) were repeated twice and performed in duplicate. The data were expressed as the means. The Cronobacter strain ATCC BAA-894 was used as the control.
2.8. Detection of Antibiotic Resistance and Virulence Genes
The presence of antibiotic resistance genes was determined using the ResFinder tool from the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org, accessed 28 September 23) [40]. Thresholds for target scanning were set with a required identity of ≥90% to the reference sequence and an aligned reference sequence ≥ 99%. For antimicrobial resistance genes, the Comprehensive Antibiotic Resistance Database (CARD) with the “perfect” and “strict” default settings [41], as well as the Task Template AMRFinderPlus 3.11.2 available in the Ridom SeqSphere+ v.9.0.8 software using the EXACT method at 100% and BLAST alignment for protein identification, were used [42].
2.9. Detection of Plasmids and Mobile Genetic Elements (MGEs)
The detection of plasmids was carried out using the MOB-suite tool v3.1.4 [43] integrated in Ridom SeqSphere v9.0.8, and the CGE MobileElementFinder v1.0.5 [44] (accessed on 10 September 2023).
3. Results
3.1. Sampling and Identification of Isolates
Out of the one-hundred samples subjected to analysis, five presumptive strains of Cronobacter spp. were isolated from separate samples. Subsequently, four tested positive for Cronobacter spp. using Matrix-Assisted Laser Desorption/Ionization–Time of Flight Mass Spectrometry (MALDI–TOF MS), and one was identified as Franconibacter helveticus. Further characterization through ribosomal multilocus sequence typing (rMLST), involving 53 genes, and whole-genome sequencing (WGS) data revealed three isolates as Cronobacter sakazakii, one as Cronobacter malonaticus, and the remaining isolate was confirmed as Franconibacter helveticus (Table 1).

Table 1.
Identification of Cronobacter spp. and F. helveticus strains isolated from powdered dairy products by MALDI–TOF MS and rMLST whole-genome sequencing.
Two strains of C. sakazakii ST1 (CC1) and one ST31, CC31 (serotypes Csak O:1 and O:2, respectively), and one strain of C. malonaticus ST60, CC60 (Cmal O:1) were identified by average nucleotide identity and cgMLST. Using the cgMLST scheme, it is observed that, between both ST1 strains, there were only four allele differences (Figure 1).
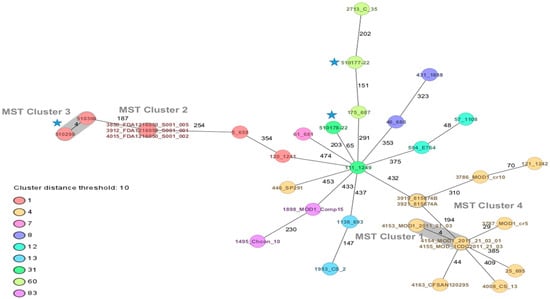
Figure 1.
Minimum spanning tree (MST) of three strains of Cronobacter sakazakii and one of Cronobacter malonaticus from dairy products, complemented with strains of C. sakazakii ST1, ST4, ST12, ST13, ST31, ST83 and C. malonaticus ST7, ST60 of clinical and food origin. The isolates are represented as colored circles according to their sequence type (ST) as defined using the 7-loci MLST scheme (STs). Black numbers on the connection lines indicate the number of allelic differences between isolates from the C. sakazakii/malonaticus cgMLST scheme comprising 2831 target genes. Isolates falling under the cluster threshold of 10 alleles are marked in grey as clusters. Strains of this study  .
.
 .
.
3.2. Antibiotic Resistance Profile
All strains of C. sakazakii and C. malonaticus were 100% resistant to cefalotin, 75% to ampicillin, 50% to amikacin and ceftazidime, and one strain was resistant to amoxicillin-clavulanic acid (Table 2). C. sakazakii ST1 strains exhibited multidrug resistance to four antibiotics (AK, AM, CAZ, and KF), while C. malonaticus ST60 strains displayed resistance to three antibiotics (AM, AMC, and KF). The strain of F. helveticus only showed resistance to ceftazidime.

Table 2.
Antibiotic resistance profiles of Cronobacter spp. strains and F. helveticus.
3.3. Adherence and Invasion Assays
In our study, 100% of the strains of C. sakazakii and C. malonaticus adhered to the N1E-115 cell line, with ranges from 1.4 to 8.7 × 106 CFU/mL. Strain 510178-22 (ST31) exhibited the highest level of adherence, while the strain 510177-22 (ST60) showed the lowest. In the invasion assay, only two strains, 510178-22 and 510177-22, invaded with frequencies ranging from 0.0002 to 0.0007%. The control strain ATCC BAA-894 achieved 22 × 106 CFU/mL and 0.15% for adherence and invasion, respectively.
3.4. Detection of Antibiotic Resistance and Virulence Genes
A total of 18 resistance genes were detected in C. sakazakii strains, and 17 in C. malonaticus strains. All C. sakazakii strains had blaCSA-1, while C. malonaticus exhibited blaCMA-1, both of which confer resistance to cephalosporins. Only C. sakazakii ST1 possessed mcr-9.1 genes, which confer resistance to colistin. All C. sakazakii and C. malonaticus strains shared the same efflux genes (adeF, H-NS, msbA, marA, kpnFE, emrRB, qacG, rsmA, and CRP), the antibiotic inactivation gene (fosA8), and five antibiotic target alteration genes (pBP3, glpT, eF-Tu, vanG, and AcrAB-TolC). The F. helveticus strain exhibited 13 resistance genes, with fosA5, qacJ, marA, and AcrAB-TolC being notable among them (Table 3).

Table 3.
Antibiotic resistance gene profiles in the strains under study.
C. sakazakii isolates encoded for 34 virulence genes that were detected by WGS and clustered as flagellar proteins, outer membrane proteins, chemotaxis, hemolysins, invasion, plasminogen activator (cpa), colonization, transcriptional regulator, survival in macrophages, utilization of sialic acid (nanA,K,T), desiccation tolerance (cheB, wzzB), and toxin–antitoxin genes (fic, relB). In the C. malonaticus strain, 30 virulence-related genes similar to C. sakazakii were detected, except for the cpa and nanAKT cassette genes (Table 4).

Table 4.
Putative virulence and distribution of other genes in the C. sakazakii and C. malonaticus strains by whole-genome sequencing.
3.5. Detection of Plasmids and Mobile Genetic Elements (MGEs)
The pESA3 plasmids and seven mobile genetic elements (MGEs) (IS5075, ISEsa2, ISEsa1, IS26, IS903, ISP-pu12, and IS102) were detected in C. sakazakii ST1. In the case of C. sakazakii ST31, the plasmid pSP291-1, the plasmid pCMA2, and one MGE (ISEsa1) were found. C. malonaticus strains harbored the pCMA1 plasmid and one MGE (ISSen4). F. helveticus harbored two plasmids, p24.751 and p14.9 with one MGE (ISPpu12) (Table 5 and Figure 2).

Table 5.
Plasmids and mobile genetic elements of Cronobacter spp. and Franconibacter helveticus strains.
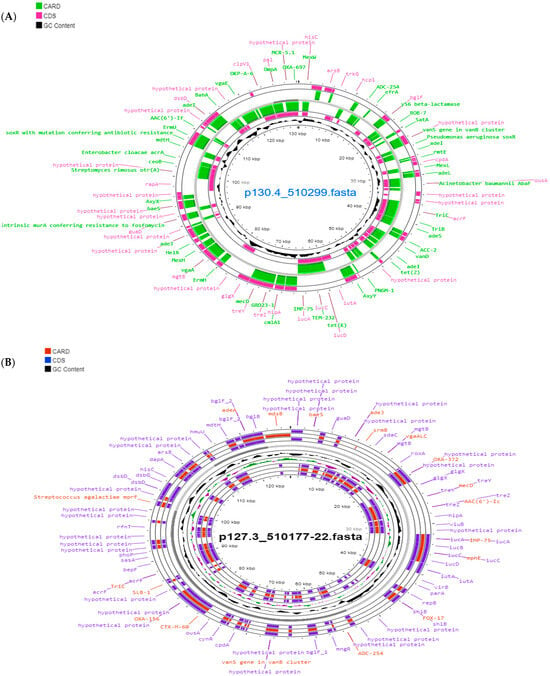
Figure 2.
Reconstruction of plasmid p130.4_510299 (pESA3) belonging to the strain C. sakazakii 510299 ST1 (A) and plasmid p127.3_510177 (pCMA1) of Cronobacter malonaticus 510177-22 ST60 (B) visualized using Proksee [32].
4. Discussion
The presence of C. sakazakii and C. malonaticus has been detected in a variety of foods, including infant formula and powdered dairy products intended for children under 2 years of age. However, there is a significant lack of information regarding the contamination of these pathogens in powdered dairy products intended for adult and older adult populations.
In our study, we found that the prevalence of Cronobacter spp. was 4%, and we isolated two strains of C. sakazakii ST1 (Csak O:1), one strain of C. sakazakii ST31 (Csak O:2), and one strain of C. malonaticus ST60 (Cmal O:1). These pathogenic strains have been associated with clinical cases in young and older adults by various authors [9,12,19,45].
CgMLST analysis allowed us to determine that the two isolates of C. sakazakii ST1 were closely related, with only a four-allele difference. In a previous study, the same cgMLST scheme enabled us to establish a genotypic relationship between three strains of C. sa-kazakii ST1 isolated in 2021 and four ST1 strains of C. sakazakii found in a food alert in Chile in 2017, with differences ranging from one to three alleles [46]. In another context, in 2019, as part of a multicenter European study that employed the cgMLST typing technique, eight C. sakazakii ST1 isolates were identified. Among these, two isolates from an outbreak in Austria in 2009, which affected two newborns with necrotizing enterocolitis, differed by only one allele. Furthermore, three ST1 isolates in Austria and one in Denmark differed by 203 alleles from the ATCC BAA-894 strain, isolated from an infant formula associated with a fatal case in the United States in 2001 [47]. The use of the standardized Cronobacter spp. cgMLST scheme allows for the establishment of genetic relationships even among strains that may not appear related initially. This tool has become essential in outbreak and case investigations, similar to how it is currently used for Listeria monocytogenes strains [48].
The overuse of antibiotics in the food chain has become a serious public health issue, leading to the implementation of global prevention campaigns [49]. Therefore, the presence of antibiotic-resistant microorganisms in dairy products consumed by older adults poses a significant risk that requires thorough study and documentation, especially due to the increasing immunological vulnerability associated with aging. In our study, we observed that all strains of C. sakazakii and the one of C. malonaticus were resistant to cephalothin, an intrinsic resistance that has been widely reported in various previous studies [50,51,52]. Additionally, the two strains of C. sakazakii ST1 exhibited multidrug resistance (MDR) to four antibiotics (AK, AM, CAZ, and KF), while the C. malonaticus strain was resistant to three antibiotics (AM, AMC, and KF). Bacteria are classified as MDR when they present resistance to three or more families of antibiotics to which they are usually sensitive, such as beta-lactams (penicillins and cephalosporins), carbapenems, aminoglycosides, and quinolones [53,54], as in this study. The presence of MDR strains has become increasingly concerning; for example, several cases of neonatal infection caused by MDR Cronobacter spp. strains have been documented, resulting in deaths or severe consequences [55,56]. In other similar studies with powdered dairy products intended for infants, it was found that 96% of C. sakazakii strains were MDR, primarily showing resistance to amoxicillin-clavulanic acid, amoxicillin, ampicillin, cefoxitin, cefepime, erythromycin, and ceftriaxone, while being susceptible to trimethoprim/sulfamethoxazole and levofloxacin [57]. In China, it was discovered that C. sakazakii strains isolated from powdered infant formulas and processing environments were also resistant to amoxicillin-clavulanic acid, ampicillin, and cefazolin [58], and, in a more recent study, they exhibited resistance to clarithromycin, ampicillin, and trimethoprim/sulfamethoxazole [59].
One of the crucial initial steps in the bacterial pathogenesis process is cellular adhesion [60]. In our study, all strains of C. sakazakii and C. malonaticus demonstrated the ability to adhere to the N1E-115 ATCC CRL-2263 cell line. Similar results have been observed in previous studies, where it was found that strains of C. sakazakii isolated from clinical cases exhibited higher adherence rates (0.915%) compared to strains from other sources of the pathogen (0.0002%) [19]. These values are consistent with what we obtained in our study. However, in terms of the cellular invasion assay, only the strains of C. sakazakii ST31 and C. malonaticus isolated in our study showed the capacity for cellular invasion, with extremely low invasion frequencies ranging from 0.0002% to 0.0007%. These results contrast significantly with previous reports [61,62,63,64], where higher invasion rates were recorded.
Genomic analysis revealed the presence of several antibiotic resistance genes, primarily grouped into the categories of antibiotic target alteration and antibiotic efflux. All strains of Cronobacter spp. examined showed resistance genes for beta-lactamases CSA-1 and CMA-1, which was consistent with the observed in vitro resistance to the cephalosporin KF. Since the initial report on the blaCSA and blaCMA gene family responsible for cephalosporin resistance in Cronobacter spp., resistance to KF has been documented in various clinical and environmental strains in different countries worldwide [65,66,67,68,69]. Additionally, we detected the presence of the mrc 9.1 gene, which confers resistance to colistin (polymyxin), considered a gene of significant concern in public health. This gene has been reported in clinical cases associated with Cronobacter sakazakii ST1, as well as isolates from foods like powdered infant formula (PIF) [70,71].
The isolates of C. sakazakii and C. malonaticus shared 30 similar virulence genes identified through whole-genome sequencing (WGS), which were grouped into various categories, such as flagellar proteins, outer membrane proteins, chemotaxis, hemolysins, invasion, colonization, transcriptional regulation, survival in macrophages, desiccation tolerance, and toxin–antitoxin genes. In the C. malonaticus strain, the cpa gene, which is specifically associated with the species C. sakazakii and is related to serum resistance and invasion, was not found [72]. Additionally, the nanAKT gene cassette, which encodes exogenous sialic acid utilization, was absent [73]. Sialic acid, a nine-carbon monosaccharide, is an essential component of glycoconjugates and glycoproteins found in various mammalian tissues and plays a crucial role in numerous physiological processes, including brain development and host–pathogen interactions [4]. It also regulates the expression of key enzymes, such as sialidase and adhesins, and its ability to inhibit transcription factors involved in the fimB gene, which mediates adhesion and invasion of epithelial cells [74,75].
Flagellum synthesis is crucial for motility, adhesion, and invasion in bacteria, and the detection of flagellar proteins such as FlgE, FlgL, and FliC in the studied strains of C. sakazakii and C. malonaticus plays a crucial role in their pathogenesis [22,76], as well as the presence of outer membrane proteins like OmpA and OmpX [77,78]. Additionally, we identified the presence of the fic gene, which encodes a toxin, and the relB gene, responsible for the production of the antitoxin relE, both forming part of a bicistronic toxin–antitoxin (TA) operon. These TA systems are small genetic elements found in plasmids, phage genomes, and the chromosomes of various bacterial species. TA genes play a fundamental role in the physiology of bacterial stress response, contributing to the stabilization of horizontally acquired mobile genetic elements and participating in persistence phenotypes in some species, including E. coli and Salmonella [79]. It has been reported that fic and hipA follow specific evolutionary patterns in C. sakazakii, highlighting that C. sakazakii ST1 strains were the only ones containing TA homologs [80].
The two strains of C. sakazakii ST1 that we evaluated carried plasmids homologous to pESA3, which have been previously reported due to their association with clinical cases caused by C. sakazakii ST1 [81]. Franco et al. [16] reported that plasmid pESA3 encodes a replication origin gene similar to RepFIB (incompatibility class) both unique and shared (repA), as well as virulence genes for iron acquisition, a siderophore aerobactin, a type VI secretion system, and the cpa-producing gene, a protease capable of degrading host serum, found only in C. sakazakii strains. Additionally, we detected the presence of mobile genetic elements (MGEs) such as IS26, which acts as an adapter mediating recombination and transposition events, playing a key role in the formation of large multidrug-resistant (MDR) regions, as found in these ST1 strains [63]. On the other hand, the C. sakazakii ST31 strain harbored the pSP291-1 plasmid, which had been previously isolated from the C. sakazakii SP291 (ST4) strain. The latter strain is known for its persistent thermotolerance, harboring various heavy metal resistance genes, disinfectants, stress, cpa virulence gene, and it was isolated from a powdered infant formula factory [82]. Regarding the C. malonaticus ST60 strain, we found the pCMA1 plasmid, which was first documented in 2016. This plasmid is homologous to another one found in the C. malonaticus ST7 strain (LMG23826T), which was isolated from a clinical case in 1977 [83].
Prior to the era of taxonomy based on whole-genome sequencing, Franconibacter helveticus was named as Enterobacter helveticus, before being placed temporarily in the Cronobacter genus as Cronobacter helveticus. It was reclassified in 2014 as F. helveticus [3]. The misidentification as Cronobacter can not only affect epidemiological statistics but also generate false positives when analyzing powdered infant formula or other dairy products [84].
C. sakazakii exhibits a notable capacity to adapt to extremely dry environments, such as powdered dairy food production facilities. Despite the lack of water, these microorganisms have developed strategies to cope with such unfavorable conditions [85]. Therefore, gathering information on the presence of microorganisms like C. sakazakii and C. malonaticus in dairy products provides robust evidence to understand the severity of the problem and establish strategies aimed at improving food safety. The implementation of Hazard Analysis and Critical Control Points (HACCP) systems, along with preventive measures in the industry, such as regular monitoring of dry products, rigorous cleaning and disinfection protocols, and the use of packaging and storage techniques that minimize moisture, are essential to mitigate the risk of contamination by C. sakazakii and C. malonaticus [86]. Further research directions could focus on understanding the molecular mechanisms underlying the adaptability of C. sakazakii and C. malonaticus to dry conditions and developing more effective control strategies to ensure the safety of dairy products.
5. Conclusions
The isolates of C. sakazakii and C. malonaticus analyzed in this study exhibited high resistance to multiple antibiotics, carrying genes encoding various proteins related to antibiotic resistance, as well as a variety of virulence factors. As a result, powdered dairy products intended for consumption by immunocompromised adults pose a significant health risk. Therefore, strict adherence to good manufacturing practices, good hygiene practices, and the implementation of HACCP in the production of powdered dairy products, along with epidemiological surveillance by health authorities, form the basis of measures to mitigate the risks of future infections.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11122841/s1, Table S1: Process data of genomic sequencing of the strains studied.
Author Contributions
Conceptualization, J.P.-F., S.F. and W.R.; methodology, J.P.-F., B.D.-P., A.C.-R., F.F.-S., C.F.-B. and W.R.; software, J.P.-F., B.D.-P. and F.F.-S.; validation, J.P.-F., B.D.-P. and W.R.; formal analysis, W.R. and A.C.-R.; investigation, F.F.-S., C.F.-B., M.P.A.-L., B.D.-P., B.S. and J.P.-F.; resources, J.P.-F. and J.L.-C.; data curation, J.P.-F., S.F. and W.R.; writing—original draft preparation, J.P.-F., A.C.-C., A.C.-R., S.F. and W.R.; writing—review and editing, J.P.-F., W.R., S.F. and J.L.-C.; visualization, J.P.-F. and B.D.-P.; supervision, J.P.-F. and A.C.-C.; project administration, J.P.-F.; funding acquisition, J.P.-F., M.P.A.-L., B.S. and W.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Research Directorate of the Universidad del Bío-Bío, Project RE2340221 and code 2240216-FP.
Data Availability Statement
The Cronobacter sakazakii, Cronobacter malonaticus, and Franconibacter helveticus isolates were submitted to https://pubmlst.org/organisms/cronobacter-spp (accessed on 23 September 2023) and have the ID numbers 4183, 4184, 4187, 4188, and 4189.
Acknowledgments
We thank the Institute of Medical Microbiology and Hygiene AGES-Austrian Agency for Health and Food Safety for their support.
Conflicts of Interest
Author Prof. SJ Forsythe was employed by the company Foodmicrobe.com Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Iversen, C.; Mullane, N.; Mc Cardell, B.; Tall, B.; Lehner, A.; Fanning, S.; Stephan, R.; Joosten, H. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov.comb. nov., C. malonaticus sp. nov., C. turicensis sp. nov., C. muytjensii sp. nov., C. dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, C. dublinensis sp. nov.subsp. dublinensis subsp. nov., C. dublinensis sp. nov.subsp. lausannensis subsp. nov., and C. dublinensis sp. nov.subsp. lactaridi subsp. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 1442–1447. [Google Scholar] [PubMed]
- Joseph, S.; Cetinkaya, E.; Drahovska, H.; Levican, A.; Figueras, M.; Forsythe, S. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int. J. Syst. Evol. Microbiol. 2012, 62, 1277–1283. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Grim, C.; Gopinath, G.; Mammel, M.; Sathyamoorthy, V.; Trach, L.; Chase, H.; Fanning, S.; Tall, B. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2014, 64, 3402–3410. [Google Scholar] [PubMed]
- Forsythe, S.J. Updates on the Cronobacter genus. Annu. Rev. Food Sci. Technol. 2018, 25, 23–44. [Google Scholar] [CrossRef]
- Hariri, S.; Joseph, S.; Forsythe, S.J. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg. Infect. Dis. 2013, 19, 175–177. [Google Scholar] [CrossRef]
- Holý, O.; Forsythe, S. Cronobacter spp. as emerging causes of healthcare-associated infection. J. Hosp. Infect. 2014, 86, 169–177. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Al Mousa, W.; Elbetieha, A.; Al Nabulsi, A.; Tall, B.D. Cronobacter spp. opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits. J. Med. Microbiol. 2014, 63, 1023–1037. [Google Scholar] [CrossRef]
- Joseph, S.; Forsythe, S.J. Predominance of Cronobacter sakazakii sequence type 4 in neonatal infections. Emerg. Infect. Dis. 2011, 17, 1713–1715. [Google Scholar] [CrossRef]
- Alsonosi, A.; Hariri, S.; Kajsík, M.; Oriešková, M.; Hanulík, V.; Röderová, M.; Petrželová, J.; Kollárová, H.; Drahovská, H.; Forsythe, S.; et al. The speciation and genotyping of Cronobacter isolates from hospitalised patients. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1979–1988. [Google Scholar] [CrossRef]
- Ling, N.; Li, C.; Zhang, J.; Wu, Q.; Zeng, H.; He, W.; Ye, Y.; Wang, J.; Ding, Y.; Chen, M.; et al. Prevalence and molecular and antimicrobial characteristics of Cronobacter spp. isolated from raw vegetables in China. Front. Microbiol. 2018, 9, 1149. [Google Scholar] [CrossRef]
- Holý, O.; Petrželová, J.; Hanulík, V.; Chromá, M.; Matoušková, I.; Forsythe, S. Epidemiology of Cronobacter spp. isolates from patients admitted to the Olomouc University Hospital (Czech Republic). Epidemiol. Mikrobiol. Imunol. 2014, 63, 69–72. [Google Scholar] [PubMed]
- Yong, W.; Guo, B.; Shi, X.; Cheng, T.; Chen, M.; Jiang, X.; Ye, Y.; Wang, J.; Xie, G.; Ding, J. An investigation of an acute gastroenteritis outbreak: Cronobacter sakazakii, a potential cause of food-borne illness. Front. Microbiol. 2018, 9, 2549. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Maury-Sintjago, E.; Rodriguez-Fernández, A.; Acuña, S.; Cerda, F.; Aguirre, J.; Holy, O. Microbiological quality of powdered infant formula in Latin America. J. Food Prot. 2020, 83, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Brandão, M.L.; Umeda, N.S.; Jackson, E.; Forsythe, S.J.; de Filippis, I. Isolation, molecular and phenotypic characterization, and antibiotic susceptibility of Cronobacter spp. from Brazilian retail foods. Food Microbiol. 2017, 63, 129–138. [Google Scholar] [CrossRef]
- Vojkovska, H.; Karpiskova, R.; Orieskova, M.; Drahovska, H. Characterization of Cronobacter spp. isolated from food of plant origin and environmental samples collected from farms and from supermarkets in the Czech Republic. Int. J. Food Microbiol. 2016, 217, 130–136. [Google Scholar] [CrossRef]
- Franco, A.A.; Hu, L.; Grim, C.J.; Gopinath, G.; Sathyamoorthy, V.; Jarvis, K.G.; Lee, C.; Sadowski, J.; Kim, J.; Kothary, M.H.; et al. Characterization of putative virulence genes on the related RepFIB plasmids harbored by Cronobacter spp. Appl. Environ. Microbiol. 2011, 77, 3255–3267. [Google Scholar] [CrossRef]
- Aly, M.A.; Domig, K.; Kneifel, W.; Reimhult, E. Whole genome sequencing-based comparison of food isolates of Cronobacter sakazakii. Front. Microbiol. 2019, 10, 1464. [Google Scholar] [CrossRef]
- Cruz, A.; Xicohtencatl, J.; Gonzalez, B.; Bobadilla, M.; Eslava, C.; Rosas, I. Virulence traits in Cronobacter species isolated from different sources. Can. J. Microbiol. 2011, 57, 735–744. [Google Scholar] [CrossRef]
- Holý, O.; Cruz-Cordova, A.; Xicohtencatl-Cortés, J.; Hochel, I.; Parra-Flores, J.; Petrzelova, J.; Facevicova, K.; Forsythe, S.; Alsonosi, A. Occurrence of virulence factors in Cronobacter sakazakii and Cronobacter malonaticus originated from clinical samples. Microb. Pathog. 2019, 127, 250–256. [Google Scholar] [CrossRef]
- Singh, N.; Goel, G.; Raghav, M. Insights into virulence factors determining the pathogenicity of Cronobacter sakazakii. Virulence 2015, 6, 433–440. [Google Scholar] [CrossRef]
- Cruz-Córdova, A.; Rocha-Ramírez, L.; Ochoa, S.; Gónzalez-Pedrajo, B.; Espinosa, N.; Eslava, C.; Hernández-Chiñas, U.; Mendoza-Hernández, G.; Rodríguez-Leviz, A.; Valencia-Mayoral, P.; et al. Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS ONE 2012, 7, e52091. [Google Scholar] [CrossRef] [PubMed]
- Aldubyan, M.; Almami, I.; Benslimane, F.; Alsonosi, A.; Forsythe, S. Comparative outer membrane protein analysis of high and low-invasive strains of Cronobacter malonaticus. Front. Microbiol. 2017, 8, 2268. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, H.; Winter, J.; Boocock, D.; De Girolamo, L.; Forsythe, S.J. Characterization of outer membrane vesicles from a neonatal meningitic strain of Cronobacter sakazakii. FEMS Microbiol. Lett. 2015, 362, fnv085. [Google Scholar] [CrossRef] [PubMed]
- Flores, J.P.; Arvizu, S.; Silva, J.; Fernández, E. Two cases of hemorrhagic diarrhea caused by Cronobacter sakazakii in hospitalized nursing infants associated with the consumption of powdered infant formula. J. Food Prot. 2011, 74, 2177–2181. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.D.; Park, J.; Chang, H. Detection, antibiotic susceptibility and biofilm formation of Cronobacter spp. from various foods in Korea. Food Control 2012, 24, 225–230. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Ling, N.; Shen, Y.; Zhang, D.; Liu, D.; Ou, D.; Wu, Q.; Ye, Y. Effects of tolC on tolerance to bile salts and biofilm formation in Cronobacter malonaticus. J. Dairy Sci. 2021, 104, 9521–9531. [Google Scholar] [CrossRef]
- Leopold, S.; Goering, R.; Witten, A.; Harmsen, D.; Mellmann, A. Bacterial whole-genome sequencing revisited: Portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J. Clin. Microbiol. 2014, 52, 2365–2370. [Google Scholar] [CrossRef]
- Iversen, C.; Forsythe, S.J. Isolation of Enterobacter sakazakii and other Enterobacteriaceae from powdered infant formula milk and related products. Food Microbiol. 2004, 21, 771–776. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Sorschag, S.; Springer, B.; Allerberger, F.; Ruppitsch, W. Draft genome sequence of carbapenemase-producing Serratia marcescens isolated from a patient with chronic obstructive pulmonary disease. Genome Announc. 2017, 5, e01288-17. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bliss, C.M.; Bennett, J.S.; Bratcher, H.B.; Brehony, C.; Colles, F.M.; Wimalarathna, H.; Harrison, O.B.; Sheppard, S.K.; Cody, A.J.; et al. Ribosomal multilocus sequence typing: Universal characterization of bacteria from domain to strain. Microbiology 2012, 158, 1005–1015. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Jünemann, S.; Sedlazeck, F.J.; Prior, K.; Albersmeier, A.; John, U.; Kalinowski, J.; Mellmann, A.; Goesmann, A.; von Haesler, A.; Stoye, J.; et al. Updating benchtop sequencing performance comparison. Nat. Biotechnol. 2013, 31, 294–296. [Google Scholar] [CrossRef]
- Ogrodzki, P.; Forsythe, S. Capsular profiling of the Cronobacter genus and the association of specific Cronobacter sakazakii and C. malonaticus capsule types with neonatal meningitis and necrotizing enterocolitis. BMC Genom. 2015, 16, 758. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 30th ed.; CLSI supplement M100; Clinical and Laboratory Standars Institute: Wayne, PA, USA, 2020; pp. 1–263. [Google Scholar]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.; Lago, B.; Dave, B.; Pereira, S.; Sharma, A.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.H.; McDermott, P.F.; et al. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef] [PubMed]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018, 4, e000206. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Kadlicekova, V.; Kajsik, M.; Soltys, K.; Szemes, T.; Slobodnikova, L.; Janosikova, L.; Hubenakova, Z.; Ogrodzki, P.; Forsythe, S.; Turna, J.; et al. Characterisation of Cronobacter strains isolated from hospitalised adult patients. Antonie van Leeuwenhoek 2018, 111, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Cerda-Leal, F.; Contreras, A.; Valenzuela-Riffo, N.; Rodriguez, A.; Aguirre, J. Cronobacter sakazakii and microbiological parameters in dairy formulas associated with a food alert in Chile. Front. Microbiol. 2018, 9, 1708. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Ruppitsch, W.; Pekard-Amenitsch, S.; Forsythe, S.J.; Cormican, M.; Mach, R.L.; Piérard, D.; Allerberger, F.; The EUCRONI Study Group. Multicenter study of Cronobacter sakazakii infections in humans, Europe, 2017. Emerg. Infect. Dis. 2019, 25, 515–522. [Google Scholar] [CrossRef]
- Ruppitsch, W.; Pietzka, A.; Prior, K.; Bletz, S.; Fernandez, H.L.; Allerberger, F.; Harmsen, D.; Mellmann, A. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J. Clin. Microbiol. 2015, 53, 2869–2876. [Google Scholar] [CrossRef]
- Serwecińska, L. Antimicrobials and Antibiotic-Resistant Bacteria: A risk to the environment and to Public Health. Water 2020, 12, 3313. [Google Scholar] [CrossRef]
- Molloy, C.; Cagney, C.; O’Brien, S.; Iversen, C.; Fanning, S.; Duffy, G. Surveillance and characterization by Pulsed-Field Gel Electrophoresis of Cronobacter spp in farming and domestic environments, food production animals and retails foods. Int. J. Food Microbiol. 2009, 136, 198–238. [Google Scholar] [CrossRef]
- Arslan, S.; Ertürk, H.G. Occurrence, virulence and antimicrobial susceptibility profiles of Cronobacter spp. from ready-to-eat foods. Curr. Microbiol. 2021, 78, 3403–3416. [Google Scholar] [CrossRef]
- Li, Q.; Li, C.; Ye, Q.; Gu, Q.; Wu, S.; Zhang, Y.; Wei, X.; Xue, L.; Chen, M.; Zeng, H.; et al. Occurrence, molecular characterization and antibiotic resistance of Cronobacter spp. isolated from wet rice and flour products in Guangdong, China. Curr. Res. Food Sci. 2023, 7, 100554. [Google Scholar] [CrossRef] [PubMed]
- López-Pueyo, M.J.; Barcenilla-Gaite, F.; Amaya-Villar, R.; Garnacho-Montero, J.J. Multirresistencia antibiótica en unidades de críticos. Med. Intensiva 2011, 35, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Wolfensberger, A.; Kuster, S.P.; Marchesi, M.; Zbinden, R.; Hombach, M. The effect of varying multidrug-resistence (MDR) definitions on rates of MDR gram-negative rods. Antimicrob. Resist. Infect. Control 2019, 8, 193. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Lei, T.; He, W.; Zhang, J.; Liang, B.; Li, C.; Ling, N.; Ding, Y.; Wu, S.; Wang, J.; et al. Novel multidrug-resistant Cronobacter sakazakii causing meningitis in neonate, China, 2015. Emerg. Infect. Dis. 2018, 24, 2121–2124. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.H.; Yu, B.; Xiang, Y.; Zhang, Z.; Zhang, T.; Zeng, Y.C.; Cui, Z.G.; Huo, X.X. Two cases of multi-antibiotic resistant Cronobacter spp. infections of infants in China. Biomed. Environ. Sci. 2017, 30, 601–605. [Google Scholar]
- Pakbin, B.; Brück, W.M.; Allahyari, S.; Rossen, J.W.A.; Mahmoudi, R. Antibiotic resistance and molecular characterization of Cronobacter sakazakii strains isolated from powdered infant formula milk. Foods 2022, 11, 1093. [Google Scholar] [CrossRef]
- Fei, P.; Jiang, Y.; Feng, J.; Forsythe, S.J.; Li, R.; Zhou, Y.; Man, C. Antibiotic and desiccation resistance of Cronobacter sakazakii and C. malonaticus isolates from powdered infant formula and processing environments. Front. Microbiol. 2017, 8, 316. [Google Scholar] [CrossRef]
- Gan, X.; Li, M.; Xu, J.; Yan, S.; Wang, W.; Li, F. Emerging of multidrug-resistant Cronobacter sakazakii isolated from infant supplementary food in China. Microbiol. Spectr. 2022, 10, e0119722. [Google Scholar] [CrossRef]
- Quintero-Villegas, M.; Wittke, A.; Hutkins, R. Adherence inhibition of Cronobacter sakazakii to intestinal epithelial cells by lactoferrin. Curr. Microbiol. 2014, 69, 574–579. [Google Scholar] [CrossRef]
- Mange, J.P.; Stephan, R.; Borel, L.; Wild, P.; Kim, K.S.; Pospischil, A.; Lenher, A. Adhesive properties of Enterobacter sakazakii to human epithelial and brain microvascular endothelial cells. BMC Microbiol. 2006, 6, 58–68. [Google Scholar] [CrossRef]
- Townsend, S.; Hurrell, E.; Gonzalez-Gomez, I.; Lowe, J.; Frye, J.; Forsythe, S.; Badger, J. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology 2007, 153, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Parra-Flores, J.; Aguirre, J.; Juneja, V.; Jackson, E.; Cruz, A.; Silva, J.; Forsythe, S. Virulence and antibiotic resistance profiles of Cronobacter sakazakii and Enterobacter spp. involved in the diarrheic hemorrhagic outbreak in Mexico. Front. Microbiol. 2018, 9, 2206. [Google Scholar] [CrossRef] [PubMed]
- Holý, O.; Parra-Flores, J.; Lepuschitz, S.; Alarcón-Lavín, M.P.; Cruz-Córdova, A.; Xicohtencatl-Cortes, J.; Mancilla-Rojano, J.; Ruppitsch, W.; Forsythe, S. Molecular characterization of Cronobacter sakazakii strains isolated from powdered milk. Foods 2021, 10, 20. [Google Scholar] [CrossRef]
- Müller, A.; Hächler, H.; Stephan, R.; Lehner, A. Presence of AmpC beta-lactamases, CSA-1, CSA-2, CMA-1, and CMA-2 conferring an unusual resistance phenotype in Cronobacter sakazakii and Cronobacter malonaticus. Microb. Drug Resist. 2014, 20, 275–280. [Google Scholar] [CrossRef]
- Jang, H.; Chase, H.R.; Gangiredla, J.; Grim, C.J.; Patel, I.R.; Kothary, M.H.; Jackson, S.A.; Mammel, M.K.; Carter, L.; Negrete, F.; et al. Analysis of the molecular diversity among Cronobacter species isolated from filth flies using targeted PCR, pan genomic DNA microarray, and whole genome sequencing analyses. Front. Microbiol. 2020, 11, 561204. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Eshwar, A.; Lehner, A.; Gangiredla, J.; Patel, I.R.; Beaubrun, J.J.-G.; Chase, H.R.; Negrete, F.; Finkelstein, S.; Weinstein, L.M.; et al. Characterization of Cronobacter sakazakii strains originating from plant-origin foods using comparative genomic analyses and zebrafish infectivity studies. Microorganisms 2022, 10, 1396. [Google Scholar] [CrossRef]
- Fei, P.; Jing, H.; Ma, Y.; Dong, G.; Chang, Y.; Meng, Z.; Jiang, S.; Xie, Q.; Li, S.; Chen, X.; et al. Cronobacter spp. in commercial powdered infant formula collected from nine provinces in China: Prevalence, genotype, biofilm formation, and antibiotic susceptibility. Front. Microbiol. 2022, 13, 900690. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Al-Mousa, W.A.; Elbetieha, A.M.; Ababneh, Q.O.; Al-Nabulsi, A.A.; Jang, H.; Gangiredla, J.; Patel, I.R.; Gopinath, G.R.; Tall, B.D. Virulence, antimicrobial susceptibility and phylogenetic analysis of Cronobacter sakazakii isolates of food origins from Jordan. J. Appl. Microbiol. 2022, 133, 2528–2546. [Google Scholar] [CrossRef]
- Shi, L.; Liang, Q.; Zhan, Z.; Feng, J.; Zhao, Y.; Chen, Y.; Huang, M.; Tong, Y.; Wu, W.; Chen, W.; et al. Co-occurrence of 3 different resistance plasmids in a multi-drug resistant Cronobacter sakazakii isolate causing neonatal infections. Virulence 2018, 9, 110–112. [Google Scholar] [CrossRef]
- Parra-Flores, J.; Holý, O.; Riffo, F.; Lepuschitz, S.; Ruppitsch, W.; Forsythe, S. Draft Genome Sequences of seven Cronobacter sakazakii strains carrying the mcr 9.1 gene isolated in Chile. Microbiol. Resour. Announc. 2021, 10, e0050621. [Google Scholar] [CrossRef]
- Franco, A.A.; Kothary, M.; Gopinath, G.; Jarvis, K.; Grim, C.J.; Hu, L.; Datta, R.; McCardell, B.; Tall, B. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect. Immun. 2011, 79, 1578–1587. [Google Scholar] [CrossRef] [PubMed]
- Joseph, S.; Hariri, S.; Masood, N.; Forsythe, S. Sialic acid utilization by Cronobacter sakazakii. Microb. Inform. Exp. 2013, 3, e3. [Google Scholar] [CrossRef] [PubMed]
- Severi, E.; Hood, D.W.; Thomas, G.H. Sialic acid utilization by bacterial pathogens. Microbiology 2007, 153, 2817–2822. [Google Scholar] [CrossRef]
- Sohanpal, B.K.; Friar, S.; Roobol, J.; Plumbridge, J.A.; Blomfield, I.C. Multiple co-regulatory elements and IHF are necessary for the control of fimB expression in response to sialic acid and N-acetylglucosamine in Escherichia coli K-12. Mol. Microbiol. 2007, 63, 1223–1236. [Google Scholar] [CrossRef]
- Esteban-Kenel, V.; Ochoa, S.; Curiel-Quesada, E.; Quezada, H.; Medina-Contreras, O.; Fernández-Rendón, E.; Rosas-Pérez, I.; Arellano-Galindo, J.; Cisneros, B.; Hernandez-Castro, R.; et al. Proteomics profiles of Cronobacter sakazakii and a fliF mutant: Adherence and invasion in mouse neuroblastoma cells. Microb. Pathog. 2020, 149, 104595. [Google Scholar]
- Kim, K.; Kim, K.P.; Choi, J.; Lim, J.A.; Lee, J.; Hwang, S.; Ryu, S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 2010, 76, 5188–5198. [Google Scholar] [CrossRef] [PubMed]
- Mohan Nair, M.K.; Venkitanarayanan, K. Role of bacterial OmpA and host cytoskeleton in the invasion of human intestinal epithelial cells by Enterobacter sakazakii. Pediatr. Res. 2007, 62, 664–669. [Google Scholar] [CrossRef]
- Walling, L.R.; Butler, J.S. Toxins targeting transfer RNAs: Translation inhibition by bacterial toxin–antitoxin systems. Wiley Interdiscip. Rev. RNA 2019, 10, e1506. [Google Scholar] [CrossRef]
- Finkelstein, S.; Negrete, F.; Jang, H.; Gangiredla, J.; Mammel, M.; Patel, I.R.; Chase, H.R.; Woo, J.; Lee, Y.; Wang, C.Z.; et al. Prevalence, distribution, and phylogeny of type two toxin-antitoxin genes possessed by Cronobacter species where C. sakazakii homologs follow sequence type lineages. Microorganisms 2019, 7, 554. [Google Scholar] [CrossRef]
- Lachowska, M.; Izdebski, R.; Urbanowicz, P.; Żabicka, D.; Królak-Olejnik, B. Infection of Cronobacter sakazakii ST1 producing SHV-12 in a premature infant born from triplet pregnancy. Microorganisms 2021, 9, 1878. [Google Scholar] [CrossRef]
- Yan, Q.; Power, K.A.; Cooney, S.; Fox, E.; Gopinath, G.R.; Grim, C.J.; Tall, B.D.; McCusker, M.P.; Fanning, S. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: A persistent isolate cultured from a powdered infant formula production facility. Front. Microbiol. 2013, 4, 256. [Google Scholar] [CrossRef] [PubMed]
- Moine, D.; Kassam, M.; Baert, L.; Tang, Y.; Barretto, C.; Bru, C.N.; Klijn, C.; Descombes, P. Fully closed genome sequences of five type strains of the genus Cronobacter and one Cronobacter sakazakii strain. Genome Announc. 2016, 4, e00142-16. [Google Scholar] [CrossRef] [PubMed]
- Svobodová, B.; Vlach, J.; Junková, P.; Karamonová, L.; Blažková, M.; Fukal, L. Novel method for reliable identification of Siccibacter and Franconibacter strains: From "Pseudo-Cronobacter" to New Enterobacteriaceae genera. Appl. Environ. Microbiol. 2017, 83, e00234-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Forsythe, S.J.; Yang, X.; Fu, S.; Man, C.; Jiang, Y. Invited review: Stress resistance of Cronobacter spp. affecting control of its growth during food production. J. Dairy Sci. 2021, 104, 11348–11367. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, Z.E.; Hunt, K.; Koolman, L.; Butler, F.; Fanning, S. Cronobacter species in the built food production environment: A review on persistence, pathogenicity, regulation and detection methods. Microorganisms 2023, 11, 1379. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).