Nitrogen Application and Rhizosphere Effect Exert Opposite Effects on Key Straw-Decomposing Microorganisms in Straw-Amended Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Experimental Materials
2.2. Experimental Design
2.3. DNA Extraction and qPCR
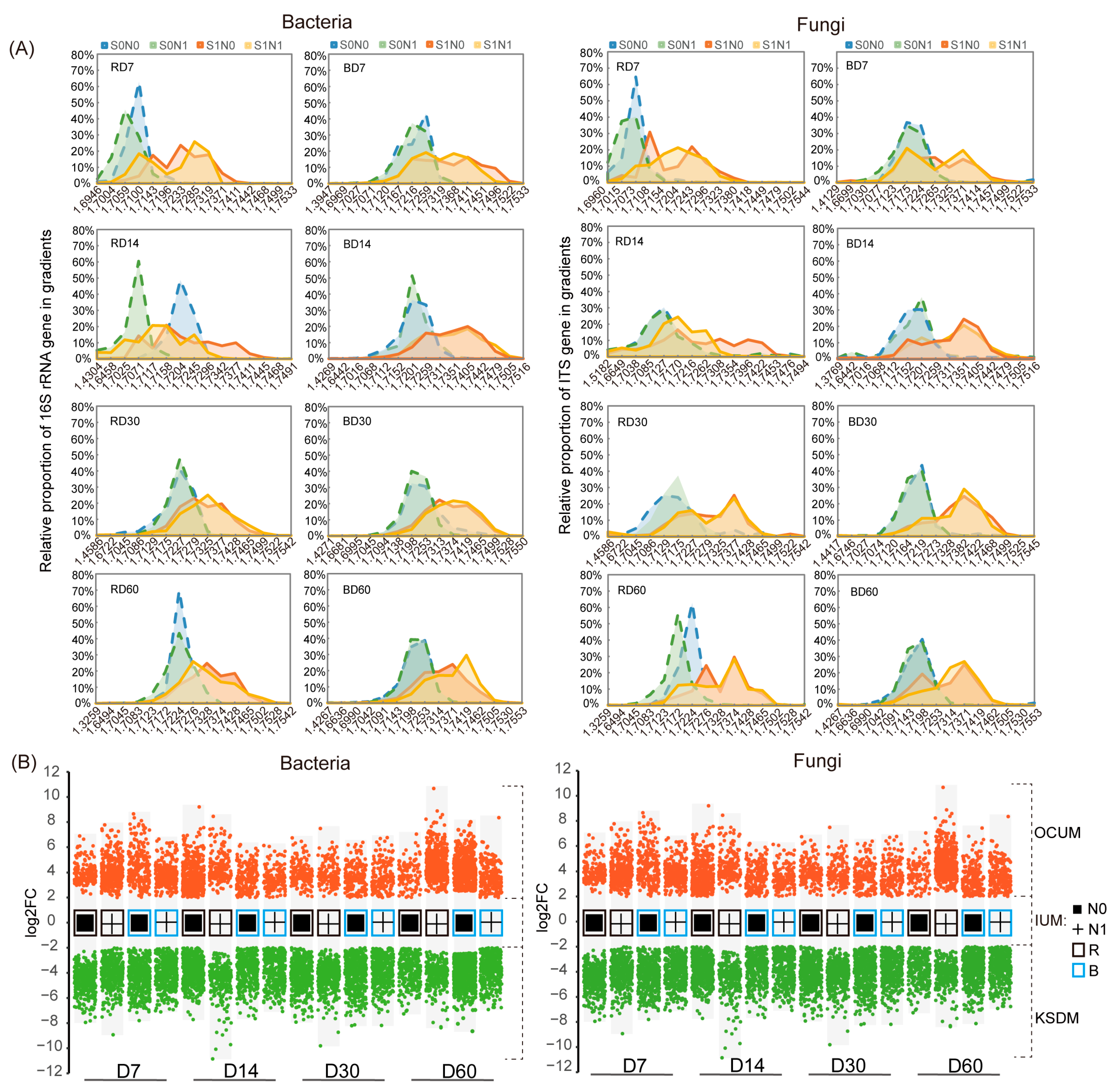
2.4. DNA-SIP
2.5. Amplicon Sequence and Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
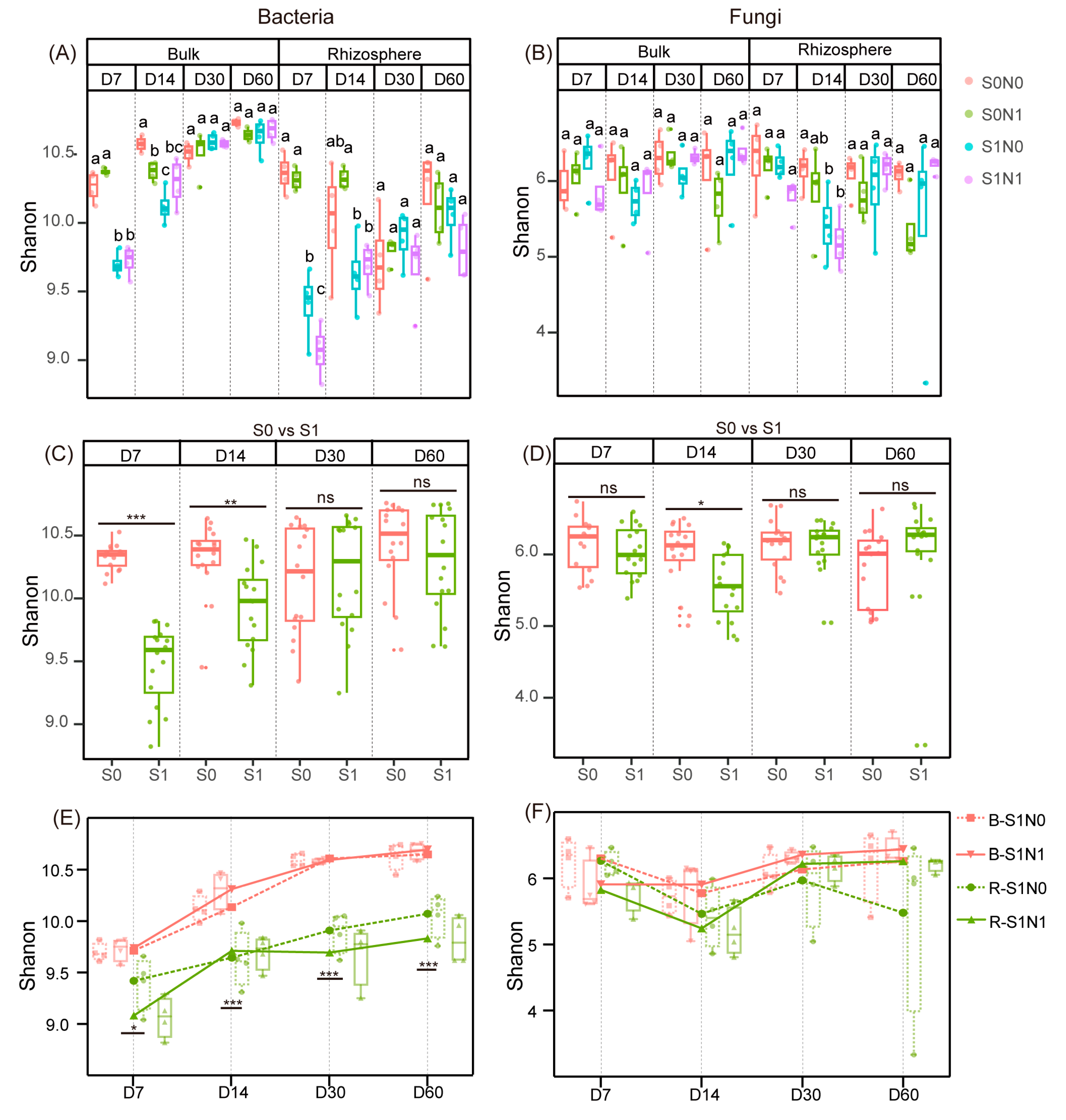
3.1. Changes of Microbial Alpha Diversity in Straw-Amended Soil
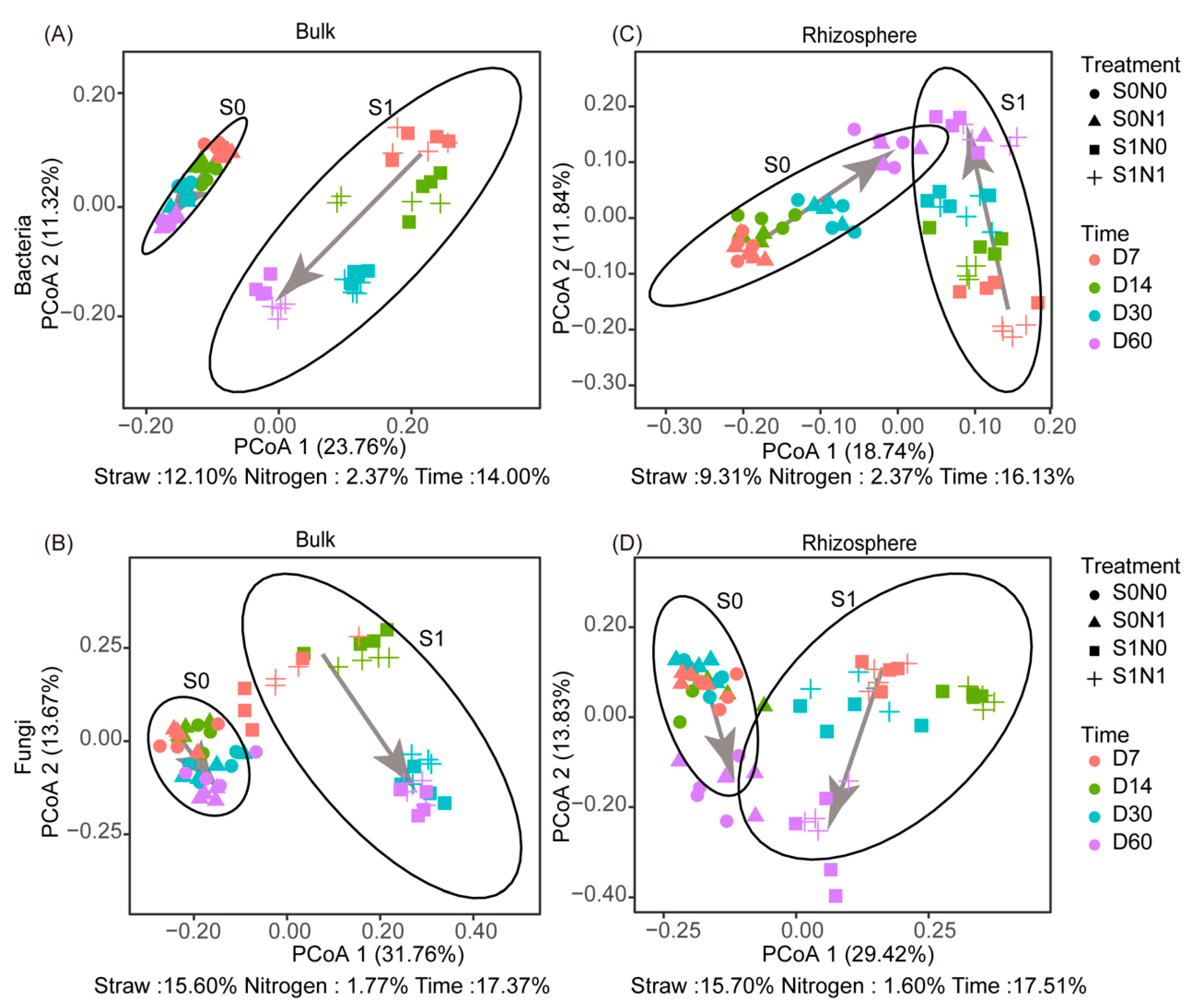
3.2. Shifts in Beta Diversity of Soil Microorganisms after Straw Addition
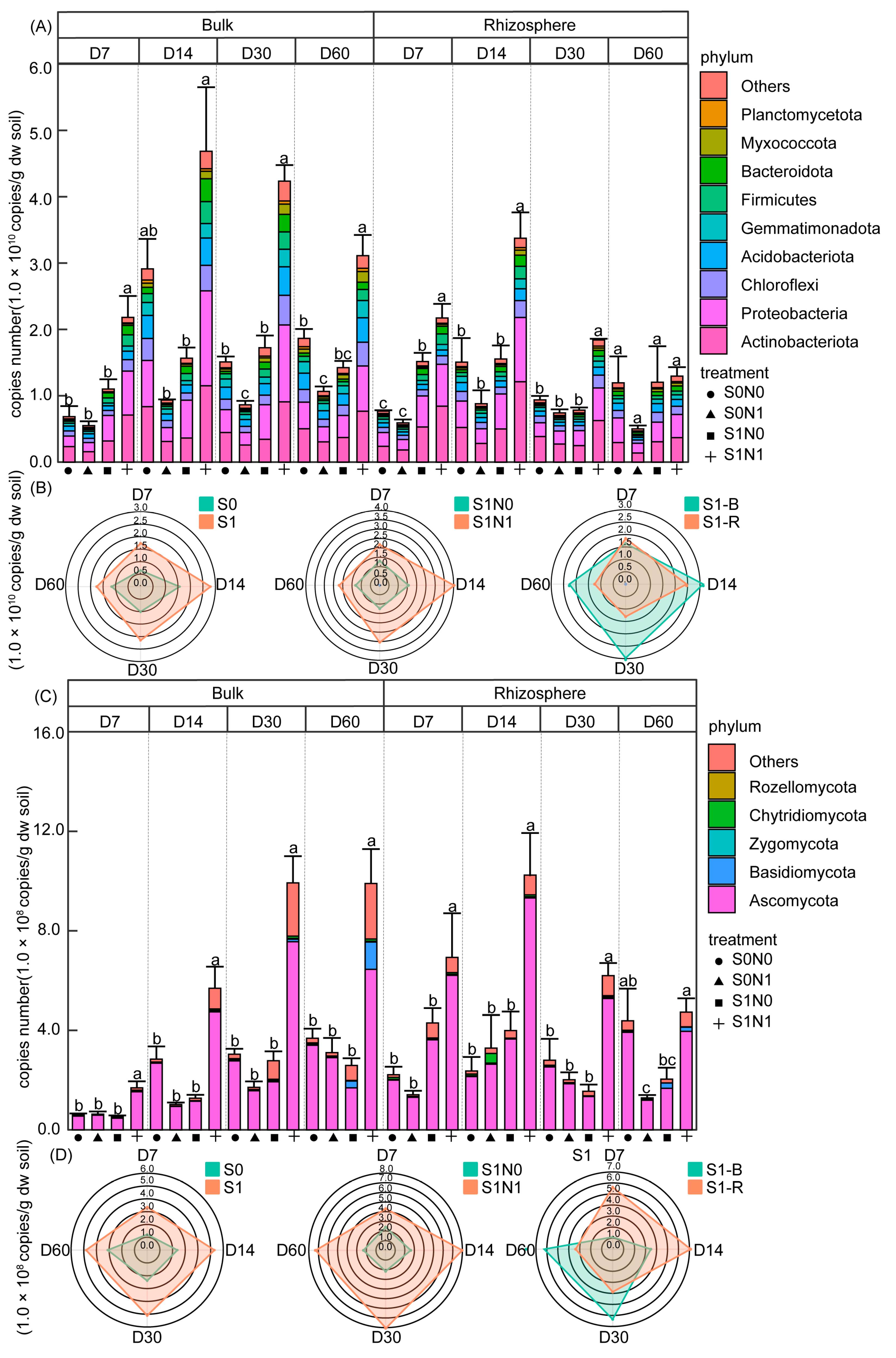
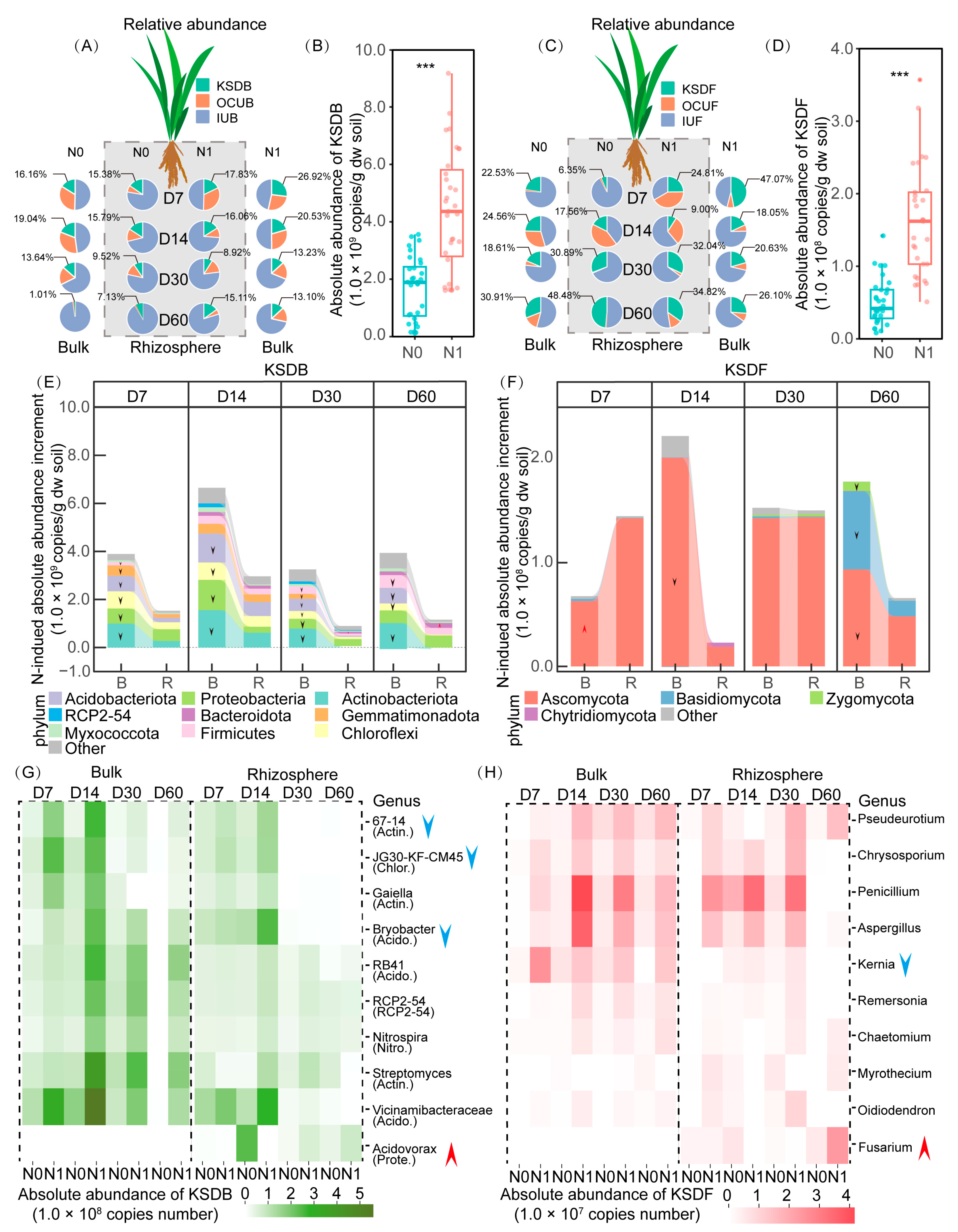
3.3. Succession Pattern of Taxa Composition and Absolute Abundance
3.4. Influence of N Application and Rhizosphere Effect on Key Straw-Decomposing Microorganisms Identified by DNA-SIP
4. Discussion
4.1. Effect of N Application and Rhizosphere Effect on Microbial Diversity and Taxa Composition in Straw-Amended Soil
4.2. KSDMs and Their Change in Absolute Abundance in Response to N Application and Rhizosphere Effects
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, J.; Qiu, T.; Peñuelas, J.; Sardans, J.; Tan, W.; Wei, X.; Cui, Y.; Cui, Q.; Wu, C.; Liu, L.; et al. Crop residue return sustains global soil ecological stoichiometry balance. Glob. Chang. Biol. 2023, 29, 2203–2226. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Xia, H.; Jiang, C.; Riaz, M.; Yang, L.; Chen, Y.; Fan, X.; Xia, X. 14 year applications of chemical fertilizers and crop straw effects on soil labile organic carbon fractions, enzyme activities and microbial community in rice-wheat rotation of middle China. Sci. Total Environ. 2022, 841, 156608. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, X.; Zhou, J.; Xu, Z.; Ma, Q.; Chu, J.; Zang, H.; Yang, Y.; Peixoto, L.; Zeng, Z.; et al. Transition of spatio-temporal distribution of soil enzyme activity after straw incorporation: From rhizosphere to detritusphere. Appl. Soil Ecol. 2023, 186, 104814. [Google Scholar] [CrossRef]
- Mo, F.; Yang, D.; Wang, X.; Crowther, T.W.; Vinay, N.; Luo, Z.; Yu, K.; Sun, S.; Zhang, F.; Xiong, Y.; et al. Nutrient limitation of soil organic carbon stocks under straw return. Soil Biol. Biochem. 2024, 192, 109360. [Google Scholar] [CrossRef]
- Li, H.; Dai, M.; Dai, S.; Dong, X. Current status and environment impact of direct straw return in China’s cropland—A review. Ecotoxicol. Environ. Saf. 2018, 159, 293–300. [Google Scholar] [CrossRef]
- Fang, Y.R.; Wu, Y.; Xie, G.H. Crop residue utilizations and potential for bioethanol production in China. Renew. Sustain. Energy Rev. 2019, 113, 109288. [Google Scholar] [CrossRef]
- Zhao, X.; Li, R.-C.; Liu, W.-X.; Liu, W.-S.; Xue, Y.-H.; Sun, R.-H.; Wei, Y.-X.; Chen, Z.; Lal, R.; Dang, Y.P.; et al. Estimation of crop residue production and its contribution to carbon neutrality in China. Resour. Conserv. Recycl. 2024, 203, 107450. [Google Scholar] [CrossRef]
- Jin, Z.; Shah, T.; Zhang, L.; Liu, H.; Peng, S.; Nie, L. Effect of straw returning on soil organic carbon in rice–wheat rotation system: A review. Food Energy Secur. 2020, 9, e200. [Google Scholar] [CrossRef]
- Zhou, G.; Zhang, J.; Zhang, C.; Feng, Y.; Chen, L.; Yu, Z.; Xin, X.; Zhao, B. Effects of changes in straw chemical properties and alkaline soils on bacterial communities engaged in straw decomposition at different temperatures. Sci. Rep. 2016, 6, 22186. [Google Scholar] [CrossRef]
- Bao, Y.; Dolfing, J.; Wang, B.; Chen, R.; Huang, M.; Li, Z.; Lin, X.; Feng, Y. Bacterial communities involved directly or indirectly in the anaerobic degradation of cellulose. Biol. Fertil. Soils 2019, 55, 201–211. [Google Scholar] [CrossRef]
- Duan, Y.; Chen, L.; Li, Y.; Li, J.; Zhang, C.; Ma, D.; Zhou, G.; Zhang, J. Nitrogen input level modulates straw-derived organic carbon physical fractions accumulation by stimulating specific fungal groups during decomposition. Soil Tillage Res. 2023, 225, 105560. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, Q.; Ai, C.; Liang, G.; He, P.; Zhou, W. Nitrogen enrichment regulates straw decomposition and its associated microbial community in a double-rice cropping system. Sci. Rep. 2018, 8, 1847. [Google Scholar] [CrossRef]
- Wang, J.; Chapman, S.J.; Yao, H. Incorporation of 13 C-labelled rice rhizodeposition into soil microbial communities under different fertilizer applications. Appl. Soil Ecol. 2016, 101, 11–19. [Google Scholar] [CrossRef]
- Zhong, Y.; Liu, J.; Jia, X.; Shangguan, Z.; Wang, R.; Yan, W. Microbial community assembly and metabolic function during wheat straw decomposition under different nitrogen fertilization treatments. Biol. Fertil. Soils 2020, 56, 697–710. [Google Scholar] [CrossRef]
- Gill, A.L.; Schilling, J.; Hobbie, S.E. Experimental nitrogen fertilisation globally accelerates, then slows decomposition of leaf litter. Ecol. Lett. 2021, 24, 802–811. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere bacteriome structure and functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-H.; Wang, N.; Yu, M.-K.; Yu, J.-G.; Xue, L.-H. Rhizosphere and Straw Return Interactively Shape Rhizosphere Bacterial Community Composition and Nitrogen Cycling in Paddy Soil. Front. Microbiol. 2022, 13, 945927. [Google Scholar] [CrossRef] [PubMed]
- Heredia-Acuña, C.; Semchenko, M.; De Vries, F.T. Root litter decomposition is suppressed in species mixtures and in the presence of living roots. J. Ecol. 2023, 111, 2519–2531. [Google Scholar] [CrossRef]
- Lee, C.G.; Watanabe, T.; Sato, Y.; Murase, J.; Asakawa, S.; Kimura, M. Bacterial populations assimilating carbon from 13C-labeled plant residue in soil: Analysis by a DNA-SIP approach. Soil Biol. Biochem. 2011, 43, 814–822. [Google Scholar] [CrossRef]
- Guo, T.; Zhang, Q.; Ai, C.; Liang, G.; He, P.; Lei, Q.; Zhou, W. Analysis of microbial utilization of rice straw in paddy soil using a DNA-SIP approach. Soil Sci. Soc. Am. J. 2020, 84, 99–114. [Google Scholar] [CrossRef]
- Kong, Y.; Kuzyakov, Y.; Ruan, Y.; Zhang, J.; Wang, T.; Wang, M.; Guo, S.; Shen, Q.; Ling, N. DNA Stable-Isotope Probing Delineates Carbon Flows from Rice Residues into Soil Microbial Communities Depending on Fertilization. Appl. Environ. Microbiol. 2020, 86, e02151-19. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Luo, Y.; Tang, C.; Li, Y.; Guggenberger, G.; Xu, J. Succession of the soil bacterial community as resource utilization shifts from plant residues to rhizodeposits. Soil Biol. Biochem. 2022, 173, 108785. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, L.; Huang, S.; Li, W.; Zhou, W.; Philippot, L.; Ai, C. Assessment of spike-AMP and qPCR-AMP in soil microbiota quantitative research. Soil Biol. Biochem. 2022, 166, 108570. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Alexander, I.J.; Eberhardt, U.; Erland, S.; Høiland, K.; Kjøller, R.; Larsson, E.; Pennanen, T.; et al. The UNITE database for molecular identification of fungi –recent updates and future perspectives. New Phytol. 2010, 186, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, M.; Huang, S.; Li, L.; Gao, Q.; Wang, Y.; Zhang, S.; Huang, S.; Yuan, L.; Wen, Y.; et al. A highly conserved core bacterial microbiota with nitrogen-fixation capacity inhabits the xylem sap in maize plants. Nat. Commun. 2022, 13, 3361. [Google Scholar] [CrossRef] [PubMed]
- Dang, P.; Li, C.; Lu, C.; Zhang, M.; Huang, T.; Wan, C.; Wang, H.; Chen, Y.; Qin, X.; Liao, Y.; et al. Effect of fertilizer management on the soil bacterial community in agroecosystems across the globe. Agric. Ecosyst. Environ. 2022, 326, 107795. [Google Scholar] [CrossRef]
- Wang, Q.; Cao, X.; Jiang, H.; Guo, Z. Straw Application and Soil Microbial Biomass Carbon Change: A Meta-Analysis. CLEAN Soil Air Water 2021, 49, 2000386. [Google Scholar] [CrossRef]
- Xie, Z.; Yu, Z.; Li, Y.; Wang, G.; Tang, C.; Mathesius, U.; Liu, X.; Liu, J.; Liu, J.; Chen, Y.; et al. Rhizosphere-induced shift in the composition of bacterial community favors mineralization of crop residue nitrogen. Plant Soil 2023. [Google Scholar] [CrossRef]
- Wang, J.-T.; Zheng, Y.-M.; Hu, H.-W.; Zhang, L.-M.; Li, J.; He, J.-Z. Soil pH determines the alpha diversity but not beta diversity of soil fungal community along altitude in a typical Tibetan forest ecosystem. J. Soils Sediments 2015, 15, 1224–1232. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, C.; Luo, Y. Meta-analysis of the impacts of global change factors on soil microbial diversity and functionality. Nat. Commun. 2020, 11, 3072. [Google Scholar] [CrossRef]
- Wu, A.-L.; Jiao, X.-Y.; Wang, J.-S.; Dong, E.-W.; Guo, J.; Wang, L.-G.; Sun, A.-Q.; Hu, H.-W. Sorghum rhizosphere effects reduced soil bacterial diversity by recruiting specific bacterial species under low nitrogen stress. Sci. Total Environ. 2021, 770, 144742. [Google Scholar] [CrossRef]
- Sun, R.; Zhang, X.-X.; Guo, X.; Wang, D.; Chu, H. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 2015, 88, 9–18. [Google Scholar] [CrossRef]
- Eo, J.; Park, K.-C. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Han, J.; Dong, Y.; Zhang, M. Chemical fertilizer reduction with organic fertilizer effectively improve soil fertility and microbial community from newly cultivated land in the Loess Plateau of China. Appl. Soil Ecol. 2021, 165, 103966. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Cheng, X.; Liu, G. Nitrogen addition stimulates litter decomposition rate: From the perspective of the combined effect of soil environment and litter quality. Soil Biol. Biochem. 2023, 179, 108992. [Google Scholar] [CrossRef]
- Xu, Z.; Sun, R.; He, T.; Sun, Y.; Wu, M.; Xue, Y.; Meng, F.; Wang, J. Disentangling the impact of straw incorporation on soil microbial communities: Enhanced network complexity and ecological stochasticity. Sci. Total Environ. 2023, 863, 160918. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Xu, H.; Shi, J.; Wang, Z.; Lv, J.; Li, L.; Wang, X. Soil microbial composition, diversity, and network stability in intercropping versus monoculture responded differently to drought. Agric. Ecosyst. Environ. 2024, 365, 108915. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, G.; Wang, D.; Liu, Q. Long-term straw return with N addition alters reactive nitrogen runoff loss and the bacterial community during rice growth stages. J. Environ. Manag. 2021, 292, 112772. [Google Scholar] [CrossRef]
- Ren, G.; Zhang, X.; Xin, X.; Yang, W.; Zhu, A.; Yang, J.; Li, M. Soil organic carbon and nitrogen fractions as affected by straw and nitrogen management on the North China Plain. Agric. Ecosyst. Environ. 2023, 342, 108248. [Google Scholar] [CrossRef]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Wang, J.; Liao, L.; Ye, Z.; Liu, H.; Zhang, C.; Zhang, L.; Liu, G.; Wang, G. Different bacterial co-occurrence patterns and community assembly between rhizosphere and bulk soils under N addition in the plant–soil system. Plant Soil 2022, 471, 697–713. [Google Scholar] [CrossRef]
- Zhang, L.; Yuan, L.; Wen, Y.; Zhang, M.; Huang, S.; Wang, S.; Zhao, Y.; Hao, X.; Li, L.; Gao, Q.; et al. Maize functional requirements drive the selection of rhizobacteria under long-term fertilization practices. New Phytol. 2024. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Ao, G.; Qin, W.; Han, M.; Shen, Y.; Liu, M.; Chen, Y.; Zhu, B. Globally nitrogen addition alters soil microbial community structure, but has minor effects on soil microbial diversity and richness. Soil Biol. Biochem. 2023, 179, 108982. [Google Scholar] [CrossRef]
- Averill, C.; Waring, B. Nitrogen limitation of decomposition and decay: How can it occur? Glob. Chang. Biol. 2018, 24, 1417–1427. [Google Scholar] [CrossRef] [PubMed]
- Kuzyakov, Y.; Xu, X. Competition between roots and microorganisms for nitrogen: Mechanisms and ecological relevance. New Phytol. 2013, 198, 656–669. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zou, H.; Qian, C.; Yu, Y.; Hao, Y.; Li, L.; Wang, Q.; Jiang, Y.; Ma, J. Construction of in situ degradation bacteria of corn straw and analysis of its degradation efficiency. Ann. Microbiol. 2020, 70, 62. [Google Scholar] [CrossRef]
- Dahal, B.; NandaKafle, G.; Perkins, L.; Brözel, V.S. Diversity of free-Living nitrogen fixing Streptomyces in soils of the badlands of South Dakota. Microbiol. Res. 2017, 195, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular Regulation of Antibiotic Biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef]
- Huber, K.J.; Overmann, J. Vicinamibacteraceae fam. nov., the first described family within the subdivision 6 Acidobacteria. Int. J. Syst. Evol. Microbiol. 2018, 68, 2331–2334. [Google Scholar] [CrossRef]
- Huang, B.; Chen, Y.; Pei, Z.; Jiang, L.; Zhang, Y.; Wang, J.; Wang, J. Application of microbial organic fertilizers promotes the utilization of nutrients and restoration of microbial community structure and function in rhizosphere soils after dazomet fumigation. Front. Microbiol. 2023, 13, 1122611. [Google Scholar] [CrossRef]
- Zhang, K.; Li, K.; Tong, M.; Xia, Y.; Cui, Y.; Liu, Z.; Chen, Q.; Li, Q.; Hu, F.; Yang, F. Distribution Pattern and Influencing Factors of Heavy Metal Resistance Genes in the Yellow River Sediments of Henan Section. Int. J. Environ. Res. Public Health 2022, 19, 10724. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guan, H.; Hu, J.; Feng, Y.; Li, X.; Yusef, K.K.; Gao, H.; Tian, D. Aspergillus niger Enhances Organic and Inorganic Phosphorus Release from Wheat Straw by Secretion of Degrading Enzymes and Oxalic Acid. J. Agric. Food Chem. 2022, 70, 10738–10746. [Google Scholar] [CrossRef] [PubMed]
- Strickland, M.S.; Rousk, J. Considering fungal:bacterial dominance in soils—Methods, controls, and ecosystem implications. Soil Biol. Biochem. 2010, 42, 1385–1395. [Google Scholar] [CrossRef]
- Raza, T.; Qadir, M.F.; Khan, K.S.; Eash, N.S.; Yousuf, M.; Chatterjee, S.; Manzoor, R.; Rehman, S.U.; Oetting, J.N. Unrevealing the potential of microbes in decomposition of organic matter and release of carbon in the ecosystem. J. Environ. Manag. 2023, 344, 118529. [Google Scholar] [CrossRef] [PubMed]
- Balmas, V.; Scherm, B.; Marcello, A.; Beyer, M.; Hoffmann, L.; Migheli, Q.; Pasquali, M. Fusarium species and chemotypes associated with fusarium head blight and fusarium root rot on wheat in Sardinia. Plant Pathol. 2015, 64, 972–979. [Google Scholar] [CrossRef]
- Chen, H.; Mommer, L.; Van Ruijven, J.; De Kroon, H.; Fischer, C.; Gessler, A.; Hildebrandt, A.; Scherer-Lorenzen, M.; Wirth, C.; Weigelt, A. Plant species richness negatively affects root decomposition in grasslands. J. Ecol. 2017, 105, 209–218. [Google Scholar] [CrossRef]
- Zheng, J.; Li, S.; Wang, H.; Dai, X.; Meng, S.; Jiang, L.; Ma, N.; Yan, H.; Fu, X.; Kou, L. Home-field advantage meets priming effect in root decomposition: Implications for belowground carbon dynamics. Funct. Ecol. 2023, 37, 676–689. [Google Scholar] [CrossRef]
- Yu, F.; Chen, Y.; Huang, X.; Shi, J.; Xu, J.; He, Y. Does straw returning affect the root rot disease of crops in soil? A systematic review and meta-analysis. J. Environ. Manag. 2023, 336, 117673. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Wang, S.; Zhang, M.; Zeng, L.; Zhang, L.; Huang, S.; Zhang, R.; Zhou, W.; Ai, C. Nitrogen Application and Rhizosphere Effect Exert Opposite Effects on Key Straw-Decomposing Microorganisms in Straw-Amended Soil. Microorganisms 2024, 12, 574. https://doi.org/10.3390/microorganisms12030574
Zhao Y, Wang S, Zhang M, Zeng L, Zhang L, Huang S, Zhang R, Zhou W, Ai C. Nitrogen Application and Rhizosphere Effect Exert Opposite Effects on Key Straw-Decomposing Microorganisms in Straw-Amended Soil. Microorganisms. 2024; 12(3):574. https://doi.org/10.3390/microorganisms12030574
Chicago/Turabian StyleZhao, Yuanzheng, Shiyu Wang, Meiling Zhang, Li Zeng, Liyu Zhang, Shuyu Huang, Rong Zhang, Wei Zhou, and Chao Ai. 2024. "Nitrogen Application and Rhizosphere Effect Exert Opposite Effects on Key Straw-Decomposing Microorganisms in Straw-Amended Soil" Microorganisms 12, no. 3: 574. https://doi.org/10.3390/microorganisms12030574
APA StyleZhao, Y., Wang, S., Zhang, M., Zeng, L., Zhang, L., Huang, S., Zhang, R., Zhou, W., & Ai, C. (2024). Nitrogen Application and Rhizosphere Effect Exert Opposite Effects on Key Straw-Decomposing Microorganisms in Straw-Amended Soil. Microorganisms, 12(3), 574. https://doi.org/10.3390/microorganisms12030574





