Exploring Cereal Metagenomics: Unravelling Microbial Communities for Improved Food Security
Abstract
1. Introduction
2. Microbial Communities Interaction with Cereal Plants
2.1. Beneficial Interactions
2.2. Non-Beneficial Interactions
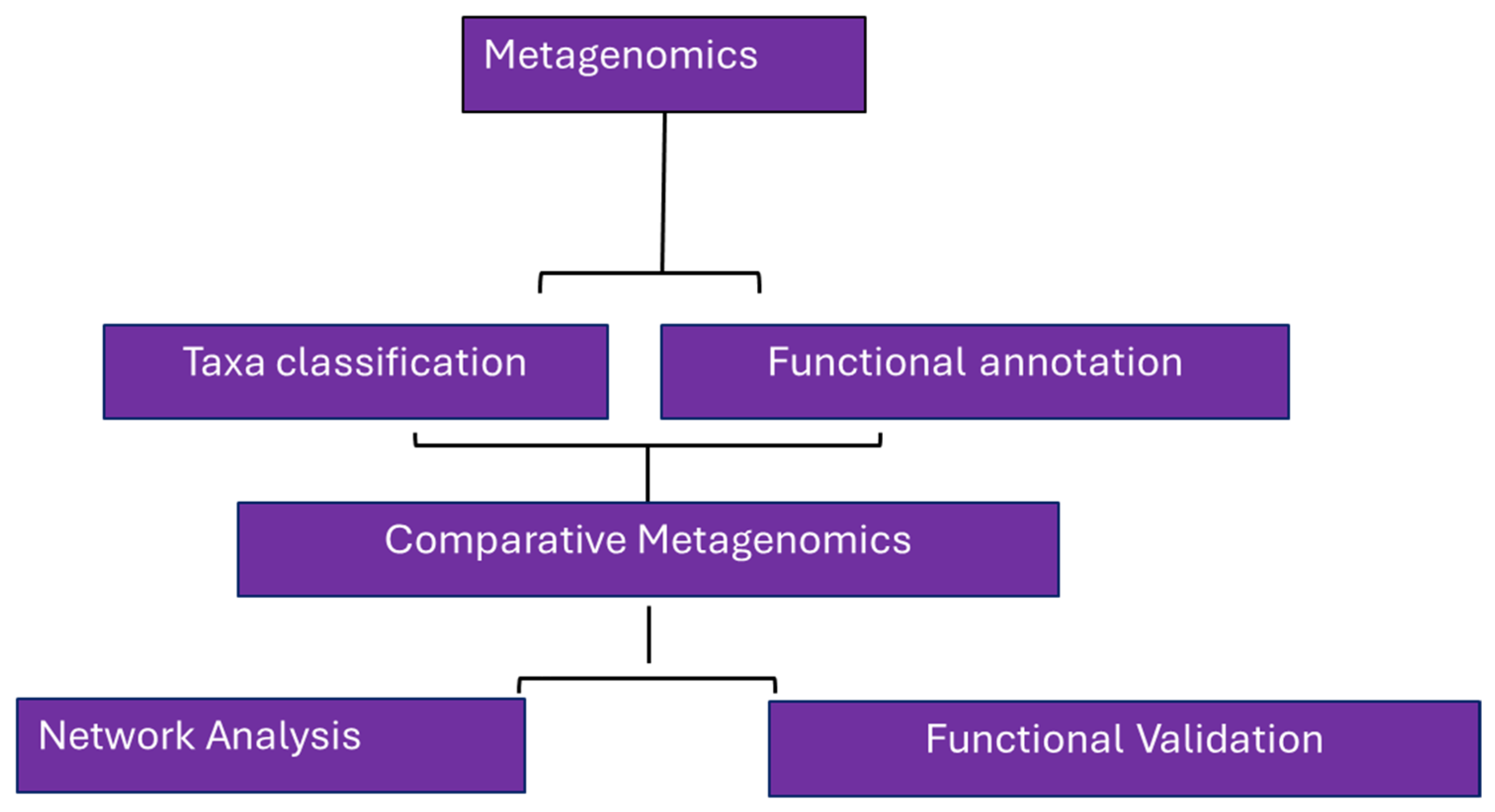
3. Metagenomics: An Overview
3.1. Metagenomics Approaches for Studying Agricultural Microbiomes
3.2. Utilization of Metagenome Studies to Identify Candidate Microbial Taxa and Genes
| Taxa Classification | Gene Identification | Host | Reference |
|---|---|---|---|
| Ascomycota, Basidiomycota, Mortierellomycota, Actinobacteria, Alphaproteobacteria, Bacteriodota, Gammaproteobacteria | Plant pathogen interactions. 3-Indol Acetic Acid (IAA) pathways, tryptophan metabolism, aminobenzoyl-glutamate. ACC deaminase pathway. | Wheat rhizosphere | [96] |
| Actinobacteria, Chloroflexi, Cyanobacteria, Firmicutes, Bacteroidetes, Proteobacteria, Acidobacteria, Gemmatimonadetes, Nitrospirae, Planctomycetes, Tenericutes, TM7 | Iron metabolism. Ferritin1, Oxoglutarate/iron-dependent oxygenase Stabilizer of iron transporter SufD/Polynucleotidyl transferase. | Maize rhizosphere | [97] |
| Plant growth promoting taxa. Planctomycetes, Bacteroidetes, Verrucomicrobia, Cyanobacteria, Gemmatimonadetes, Chloroflexi, and Firmicute | Genes mitigating salt stress. Sulfur and glutathione metabolism bacterial chemotaxis, Sulfate reduction (cysNC, cysQ, sat, and sir), sulfur reduction (fsr), SOX systems (soxB), sulfur oxidation (sqr), organic sulfur transformation (tpa, mdh, gdh, and betC). | Grapevine rhizosphere | [98] |
| Streptomyces renae, Streptomyces flavovariabilis, Streptomyces variegatus, Streptomyces chartreusis and Streptomyces cellvibrio | Genes for metabolism of plant polysaccharides, iron, sulfur, trehalose, and vitamins, β-glucosidase Cellulose-hydrolyzing enzyme. | Tomato rhizosphere | [99] |
| Actinomycetia, Anaerolineae, Chloroflexia, and Nitrospira | Catalyzation of the transfer of oligosaccharides, dentification, nitrification, nitrate reduction genes, ureB, ureA, glnA, nxrB, amoA_A, amoC_A, amoB_B, norC, nirS, nirK, nirD, narJ, narH, napC nirA, narC nitrate reductase (Anr) and the gene pmoA. | Forest deep soil | [100] |
| Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria | Carbohydrate metabolic processing, cell adhesion, pathogenesis, response to abiotic stimulus, and responses to chemicals. | Barley Rhizosphere | [101] |
| Pseudomonas, Agrobacterium, Cupriavidus, Bradyrhizobium, Rhizobium, Mesorhizobium, Burkholderia, Cellvibrio, Sphingomonas, Variovorax and Paraburkholderia | Plant-microbe and microbe-microbe interactions, nutrition acquisition, and plant growth promotion genes, pqqB, appA, phnCEF, nrtABC, phoRPA, senX3, regX3, pmoA/amoA, ics, irp9, nagG, nagH, udC, nirK. | Citrus rhizosphere | [102] |
| Rhizophagus, Burkholderia, Trichoderma, Fusarium, Ochrobactrum phage POA1180, Blastococcus, Microvirga, Nocardioides, Geodermatophilus, Belnapia, Solirubrobacter, Arthrobacter, Mycobacterium phage Edugator, and Mycobacterium phage Kratio | Not identified. | Cleome pallida (Desert plant) rhizosphere | [103] |
| Kaistobacter and Rubrobacter Bacillus Nocardioides, Cellulomonas, Skermanella, Methylobacterium, Modestobacter and Aeromicrobium, Rhizobiales, Kaistobacter, Rubrobacter or Bacillus | Metabolism of carbohydrate (especially C degradation) and membrane transporters. Carbohydrate degradation metabolism, carbohydrate synthesis, and its related energy metabolism. | Chickpea, wheat | [104] |
3.3. Applications of Metagenomics in Enhancing Food Security
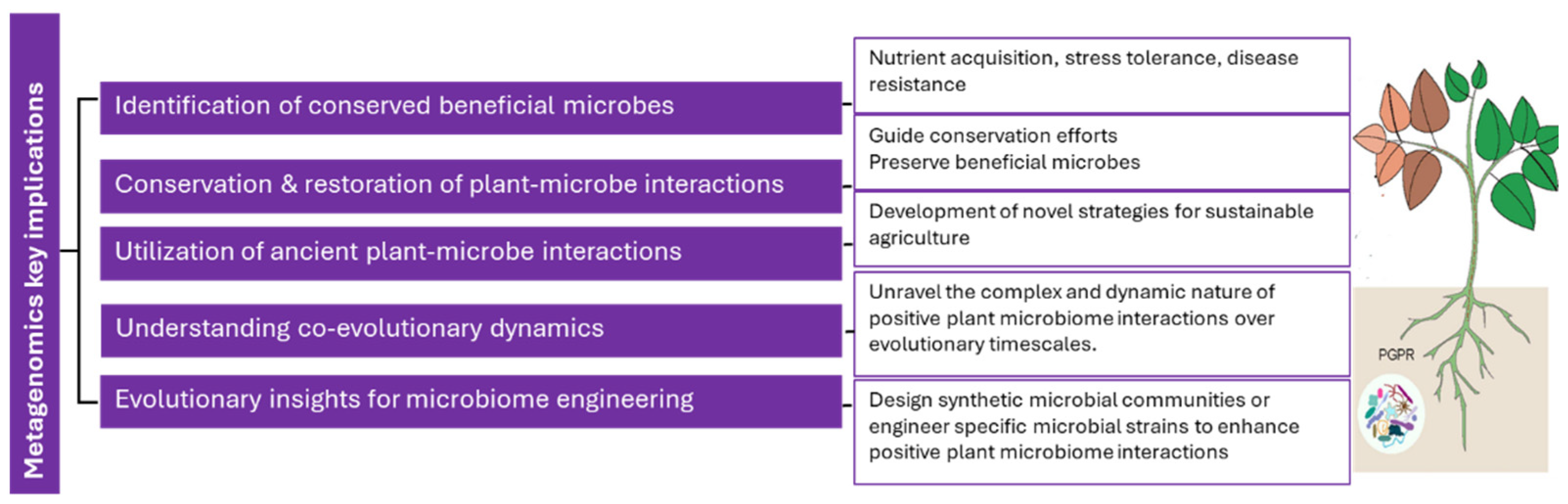
3.4. Implications of Metagenomic Studies on Positive Plant Microbiome Interactions
4. Metagenomics and Integrated Epigenetics and Machine Learning Analysis
4.1. Practical Applications and Benefits of Employing Machine Learning in Epigenomic and Metagenomic Analysis
4.2. Machine Learning Coupled with Epigenomics in Identifying Differentially Methylated Regions
5. Metagenomics Workflow for Studying Agricultural Microbiomes
5.1. Sample Collection and DNA Analysis
5.2. Library Preparation and Sequencing
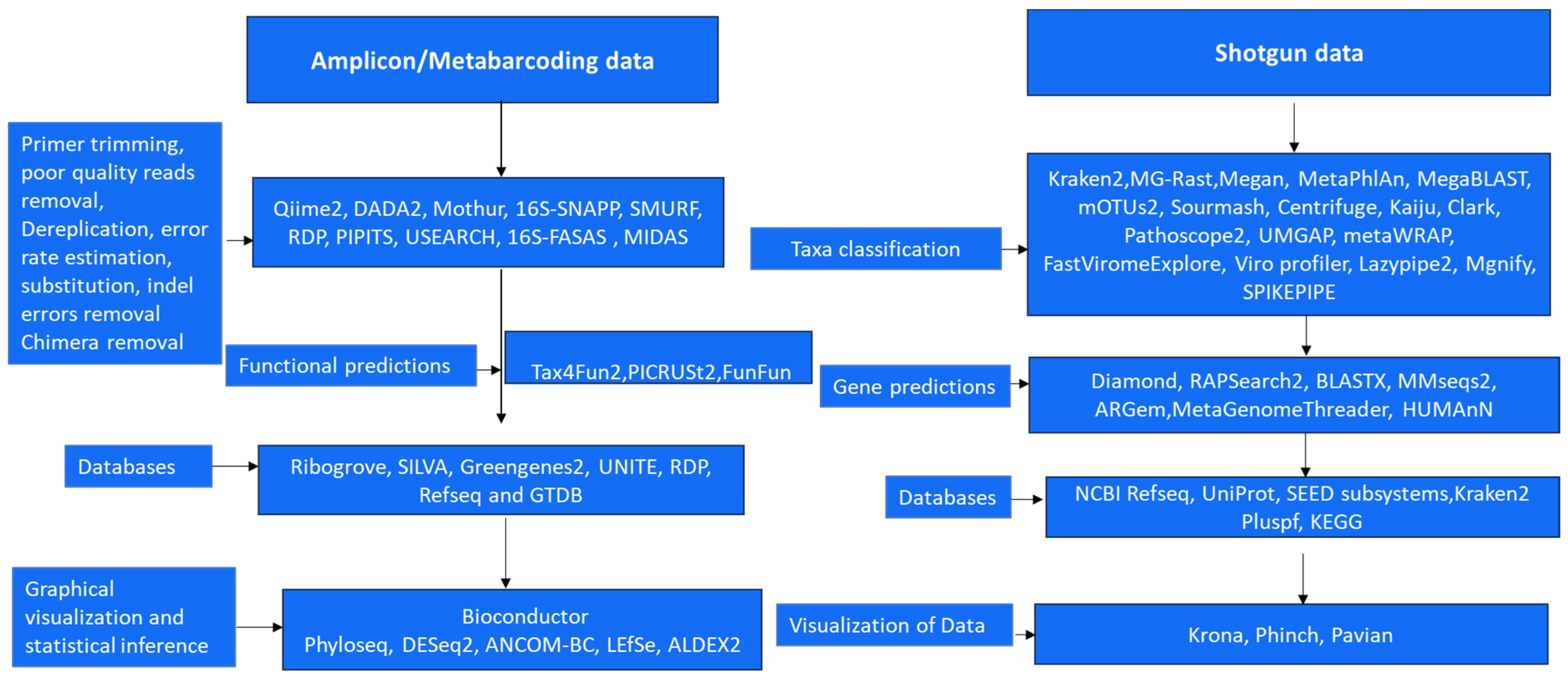
5.3. Bioinformatics Analysis
6. Challenges and Limitations in Metagenomics Studies
6.1. Sample Preparation Biases
6.2. Bias in DNA Extraction
6.3. PCR Biases
6.4. Reference Database Limitations
6.5. Detection Limits
6.6. Taxonomic Resolution
6.7. Fragmented Genomes
6.8. Difficulty in Functional Annotation
6.9. Computational and Storage Requirements
6.10. Challenges Associated with Identifying Primary Cereals Loci
7. Reliability and Reproducibility
8. Contribution of Large-Scale Cereal Microbe Genetic Datasets to the Advancement of Knowledge
9. Advances Facilitated by HTS Technologies in Understanding Cereals-Associated Microorganisms
9.1. Long Read Sequencing
9.2. Hi-C
9.3. CRISPR
9.4. Machine Learning
10. Future Directions and Emerging Technologies
11. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grote, U.; Fasse, A.; Nguyen, T.T.; Erenstein, O. Food Security and the Dynamics of Wheat and Maize Value Chains in Africa and Asia. Front. Sustain. Food Syst. 2021, 4, 617009. [Google Scholar] [CrossRef]
- Tedersoo, L.; Sánchez-Ramírez, S.; Kõljalg, U.; Bahram, M.; Döring, M.; Schigel, D.; May, T.; Ryberg, M.; Abarenkov, K. High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 2018, 90, 135–159. [Google Scholar] [CrossRef]
- Kuittinen, S.; Hietaharju, J.; Bhattarai, I.; Hassan, M.K.; Kupiainen, L.; Kangas, J.; Tanskanen, J.; Pappinen, A. Technoeconomic analysis and environmental sustainability estimation of bioalcohol production from barley straw. Biocatal. Agric. Biotechnol. 2022, 43, 102427. [Google Scholar] [CrossRef]
- Knief, C. Analysis of plant microbe interactions in the era of next generation sequencing technologies. Front. Plant Sci. 2014, 5, 216. [Google Scholar] [CrossRef] [PubMed]
- Rosselli, R.; Squartini, A. Metagenomics of Plant–Microbe Interactions. In Advances in the Understanding of Biological Sciences Using Next Generation Sequencing (NGS) Approaches; Springer: Berlin/Heidelberg, Germany, 2015; pp. 135–153. [Google Scholar]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems Biology of Plant-Microbiome Interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef] [PubMed]
- Ngah, N.; Thomas, R.; Shaw, M.; Fellowes, M. Asymptomatic host plant infection by the widespread pathogen Botrytis cinerea alters the life histories, behaviors, and interactions of an aphid and its natural enemies. Insects 2018, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Fatima, U.; Senthil-Kumar, M. Plant and pathogen nutrient acquisition strategies. Front. Plant Sci. 2015, 6, 750. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host–multi-pathogen warfare: Pathogen interactions in co-infected plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef] [PubMed]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Jamir, A.; Jamir, S.; Uikey, P.; Singh, B.V.; Sulochna Saikanth, D.R.K. Harnessing Microorganisms for Sustainable Agriculture: Promoting Environmental Protection and Soil Health. Bionature 2023, 43, 26–29. [Google Scholar] [CrossRef]
- Agrahari, R.K.; Singh, P.; Koyama, H.; Panda, S.K. Plant-microbe Interactions for Sustainable Agriculture in the Postgenomic Era. Curr. Genom. 2020, 21, 168–178. [Google Scholar] [CrossRef]
- Taş, N.; de Jong, A.E.; Li, Y.; Trubl, G.; Xue, Y.; Dove, N.C. Metagenomic tools in microbial ecology research. Curr. Opin. Biotechnol. 2021, 67, 184–191. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gomez-Valero, L.; Buchrieser, C. Metagenomic approaches in microbial ecology: An update on whole-genome and marker gene sequencing analyses. Microb. Genom. 2020, 6, e000409. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2005, 69, 195. [Google Scholar] [CrossRef]
- Trivedi, P.; Duan, Y.; Wang, N. Huanglongbing, a systemic disease, restructures the bacterial community associated with citrus roots. Appl. Environ. Microbiol. 2010, 76, 3427–3436. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Cruz, D.; Troyee, A.N.; Becker, C. Epigenetics in plant organismic interactions. Curr. Opin. Plant Biol. 2021, 61, 102060. [Google Scholar] [CrossRef] [PubMed]
- D’Elia, D.; Truu, J.; Lahti, L.; Berland, M.; Papoutsoglou, G.; Ceci, M.; Zomer, A.; Lopes, M.B.; Ibrahimi, E.; Gruca, A.; et al. Advancing microbiome research with machine learning: Key findings from the ML4Microbiome COST action. Front. Microbiol. 2023, 14, 1257002. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Dai, R.; Xu, H.; Liu, Y.; Bai, B.; Meng, Y.; Li, H.; Cao, X.; Bai, Y.; Song, X.; et al. The rice histone methylation regulates hub species of the root microbiota. J. Genet. Genom. 2021, 48, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Gamalero, E. Recent developments in the study of plant microbiomes. Microorganisms 2021, 9, 1533. [Google Scholar] [CrossRef] [PubMed]
- Afridi, M.S.; Javed, M.A.; Ali, S.; De Medeiros, F.H.V.; Ali, B.; Salam, A.; Sumaira Marc, R.A.; Alkhalifah, D.H.M.; Selim, S.; Santoyo, G. New opportunities in plant microbiome engineering for increasing agricultural sustainability under stressful conditions. Front. Plant Sci. 2022, 13, 899464. [Google Scholar] [CrossRef]
- Michl, K.; Berg, G.; Cernava, T. The microbiome of cereal plants: The current state of knowledge and the potential for future applications. Environ. Microbiome 2023, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Kavamura, V.N.; Mendes, R.; Bargaz, A.; Mauchline, T.H. Defining the wheat microbiome: Towards microbiome-facilitated crop production. Comput. Struct. Biotechnol. J. 2021, 19, 1200–1213. [Google Scholar] [CrossRef] [PubMed]
- Mehta, S.; Singh, B.; Patra, A.; Tripathi, A.; Easwaran, M.; Choudhary, J.R.; Choudhary, M.; Aggarwal, S.K. Maize microbiome: Current insights for the sustainable agriculture. In Microbiomes and Plant Health: Panoply and Their Applications; Elsevier: Amsterdam, The Netherlands, 2020; pp. 267–297. [Google Scholar]
- Masenya, K.; Thompson, G.D.; Tekere, M.; Makhalanyane, T.P.; Pierneef, R.E.; Rees, D.J.G. Pathogen infection influences a distinct microbial community composition in sorghum RILs. Plant Soil 2021, 463, 555–572. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Woźniak, M.; Furtak, K.; Gałązka, A.; Dziadczyk, E.; Skórzyńska-Polit, E.; Wolińska, A. New insight into the composition of wheat seed microbiota. Int. J. Mol. Sci. 2020, 21, 4634. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C.; Fitt, B.D.L.; Atkins, S.D.; Walters, D.R.; Daniell, T.J. Pathogenesis, parasitism and mutualism in the trophic space of microbe–plant interactions. Trends Microbiol. 2010, 18, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; van Themaat, E.V.L.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2013, 501, S25. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Schröder, P.; Vestergaard, G.; Schloter, M. Response of barley plants to drought might be associated with the recruiting of soil-borne endophytes. Microorganisms 2020, 8, 1414. [Google Scholar] [CrossRef] [PubMed]
- Milazzo, C.; Zulak, K.G.; Muria-Gonzalez, M.J.; Jones, D.; Power, M.; Bransgrove, K.; Bunce, M.; Lopez-Ruiz, F.J. High-Throughput Metabarcoding Characterizes Fungal Endophyte Diversity in the Phyllosphere of a Barley Crop. Phytobiomes J. 2021, 5, 316–325. [Google Scholar] [CrossRef]
- Links, M.G.; Demeke, T.; Gräfenhan, T.; Hill, J.E.; Hemmingsen, S.M.; Dumonceaux, T.J. Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 2014, 202, 542–553. [Google Scholar] [CrossRef]
- Xing, H.Q.; Ma, J.C.; Xu, B.L.; Zhang, S.W.; Wang, J.; Cao, L.; Yang, X.M. Mycobiota of maize seeds revealed by rDNA-ITS sequence analysis of samples with varying storage times. Microbiologyopen 2018, 7, e00609. [Google Scholar] [CrossRef] [PubMed]
- Solanki, M.K.; Abdelfattah, A.; Britzi, M.; Zakin, V.; Wisniewski, M.; Droby, S.; Sionov, E. Shifts in the composition of the microbiota of stored wheat grains in response to fumigation. Front. Microbiol. 2019, 10, 1098. [Google Scholar] [CrossRef] [PubMed]
- Pilo, P.; Lawless, C.; Tiley, A.M.M.; Karki, S.J.; Burke, J.I.; Feechan, A. Comparison of microscopic and metagenomic approaches to identify cereal pathogens and track fungal spore release in the field. Front. Plant Sci. 2022, 13, 1039090. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Różewicz, M.; Wyzińska, M.; Grabiński, J. The most important fungal diseases of cereals—Problems and possible solutions. Agronomy 2021, 11, 714. [Google Scholar] [CrossRef]
- Redila, C.D.; Prakash, V.; Nouri, S. Metagenomics analysis of the wheat virome identifies novel plant and fungal-associated viral sequences. Viruses 2021, 13, 2457. [Google Scholar] [CrossRef]
- Wamaitha, M.J.; Nigam, D.; Maina, S.; Stomeo, F.; Wangai, A.; Njuguna, J.N.; Holton, T.A.; Wanjala, B.W.; Wamalwa, M.; Lucas, T.; et al. Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virol. J. 2018, 15, 90. [Google Scholar] [CrossRef]
- Lappe, R.R.; Elmore, M.G.; Lozier, Z.R.; Jander, G.; Miller, W.A.; Whitham, S.A. Metagenomic identification of novel viruses of maize and teosinte in North America. BMC Genom. 2022, 23, 767. [Google Scholar] [CrossRef]
- Boukari, W.; Mollov, D.; Wei, C.; Tang, L.; Nouman Tahir, M.; Mulandesa, E.; Hincapie, M.; Beiriger, R.; Rott, P. Screening for sugarcane yellow leaf virus in sorghum in Florida revealed its occurrence in mixed infections with sugarcane mosaic virus and a new marafivirus. Crop. Prot. 2021, 139, 105373. [Google Scholar] [CrossRef]
- Ravisankar, D.; Nithya, C. Significance and Applications of Plant Growth Promoting Rhizobacteria (PGPR) In Agriculture: A Review. Res. Rev. J. Agric. Sci. Technol. 2018, 1, 9–22. [Google Scholar]
- Gao, Y.; Xu, G. Development of an effective nonchemical method against Plasmodiophora brassicae on Chinese cabbage. Int. J. Agron. 2014, 2014, 307367. [Google Scholar] [CrossRef]
- Gerhardson, B. Biological substitutes for pesticides. Trends Biotechnol. 2002, 20, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Postma, J.; Montanari, M.; van den Boogert, P.H.J.F. Microbial enrichment to enhance the disease suppressive activity of compost. Eur. J. Soil Biol. 2003, 39, 157–163. [Google Scholar] [CrossRef]
- Welbaum, G.E.; Sturz, A.V.; Dong, Z.; Nowak, J. Managing soil microorganisms to improve productivity of agro-ecosystems. CRC Crit. Rev. Plant Sci. 2004, 23, 175–193. [Google Scholar] [CrossRef]
- Berg, G. Plant–microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, B.O.; Schafer, J.T.; Moura, A.B. Spectrum of biocontrol bacteria to control leaf, root and vascular diseases of dry bean. Biol. Control. 2014, 72, 71–75. [Google Scholar] [CrossRef]
- Guetskyl, R.; Shtienberg, D.; Dinoor, A.; Elad, Y. Establishment, survival and activity of the biocontrol agents Pichia guilermondii and Bacillus mycoides applied as a mixture on strawberry plants. Biocontrol. Sci. Technol. 2002, 12, 705–714. [Google Scholar] [CrossRef]
- Dash, N.; Dangar, T.K. Perspectives of Phosphate Solubilizing Microbes for Plant Growth Promotion, Especially Rice-A Review. Int. J. Biochem. Res. Rev. 2017, 18, 1–16. [Google Scholar] [CrossRef]
- Hayat, W.; Aman, H.; Irshad, U.; Azeem, M.; Iqbal, A.; Nazir, R. Analysis of ecological attributes of bacterial phosphorus solubilizers, native to pine forests of Lower Himalaya. Appl. Soil Ecol. 2017, 112, 51–59. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, L.; Tan, X.; Kong, X.; Yang, J.; Wang, D.; Zhang, D.; Jin, D.; Liu, Y. Pumpkin powdery mildew disease severity influences the fungal diversity of the phyllosphere. PeerJ 2018, 6, e4559. [Google Scholar] [CrossRef]
- Morrissey, J.; Guerinot, M.L. Iron uptake and transport in plants: The good, the bad, and the ionome. Chem. Rev. 2009, 109, 4553–4567. [Google Scholar] [CrossRef]
- Sharma, R.; Thakur, R.P.; Senthilvel, S.; Nayak, S.; Reddy, S.V.; Rao, V.P.; Varshney, R.K. Identification and characterization of toxigenic Fusaria associated with sorghum grain mold complex in India. Mycopathologia 2011, 171, 223–230. [Google Scholar] [CrossRef][Green Version]
- Besset-Manzoni, Y.; Rieusset, L.; Joly, P.; Comte, G.; Prigent-Combaret, C. Exploiting rhizosphere microbial cooperation for developing sustainable agriculture strategies. Environ. Sci. Pollut. Res. 2018, 25, 29953–29970. [Google Scholar] [CrossRef]
- Luján, A.M.; Gómez, P.; Buckling, A. Siderophore cooperation of the bacterium Pseudomonas fluorescens in soil. Biol. Lett. 2015, 11, 20140934. [Google Scholar] [CrossRef] [PubMed]
- Hanudin, H.; Budiarto, K.; Marwoto, B. Application of PGPR and antagonist fungi-based biofungicide for white rust disease control and its economyc analysis in chrysanthemum production. AGRIVITA J. Agric. Sci. 2017, 39, 266–278. [Google Scholar] [CrossRef][Green Version]
- Kurepin, L.V.; Park, J.M.; Lazarovits, G.; Bernards, M.A. Burkholderia phytofirmans-induced shoot and root growth promotion is associated with endogenous changes in plant growth hormone levels. Plant Growth Regul. 2015, 75, 199–207. [Google Scholar] [CrossRef]
- Ludwig-Müller, J. Bacteria and fungi controlling plant growth by manipulating auxin: Balance between development and defense. J. Plant Physiol. 2015, 172, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Nihorimbere, V.; Ongena, M.; Smargiassi, M.; Thonart, P. Beneficial effect of the rhizosphere microbial community for plant growth and health. Biotechnol. Agron. Société Environ. 2011, 15, 327–337. [Google Scholar]
- Liu, J.L.; Xie, B.M.; Shi, X.H.; Ma, J.M.; Guo, C.H. Effects of two plant growth-promoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase on oat growth in petroleum-contaminated soil. Int. J. Environ. Sci. Technol. 2015, 12, 3887–3894. [Google Scholar] [CrossRef][Green Version]
- Ravanbakhsh, M.; Sasidharan, R.; Voesenek, L.A.C.J.; Kowalchuk, G.A.; Jousset, A. Microbial modulation of plant ethylene signaling: Ecological and evolutionary consequences. Microbiome 2018, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Raghuwanshi, R.; Prasad, J.K. Perspectives of Rhizobacteria with ACC Deaminase Activity in Plant Growth Under Abiotic Stress. In Root Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 303–321. [Google Scholar]
- Saleem, M.; Ali, M.S.; Hussain, S.; Jabbar, A.; Ashraf, M.; Lee, Y.S. Marine natural products of fungal origin. Nat. Prod. Rep. 2007, 24, 1142–1152. [Google Scholar] [CrossRef]
- Klein, R.R.; Rodriguez-Herrera, R.; Schlueter, J.A.; Klein, P.E.; Yu, Z.H. Identification of genomic regions that affect grain-mould incidence and other traits of agronomic importance in sorghum. Theor. Appl. Genet. 2001, 102, 307–319. [Google Scholar] [CrossRef]
- Ghimire, B.; Sapkota, S.; Bahri, B.A.; Martinez-Espinoza, A.D.; Buck, J.W.; Mergoum, M. Fusarium Head Blight and Rust Diseases in Soft Red Winter Wheat in the Southeast United States: State of the Art, Challenges and Future Perspective for Breeding. Front. Plant Sci. 2020, 11, 1080. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, J.; Zhang, H.; Ji, G.; Zeng, L.; Li, Y.; Yu, C.; Fernando, W.G.D.; Chen, W. Bacterial Blight Induced Shifts in Endophytic Microbiome of Rice Leaves and the Enrichment of Specific Bacterial Strains With Pathogen Antagonism. Front. Plant Sci. 2020, 11, 963. [Google Scholar] [CrossRef]
- Su, J.; Zhao, J.; Zhao, S.; Li, M.; Pang, S.; Kang, Z.; Zhen, W.; Chen, S.; Chen, F.; Wang, X. Genetics of Resistance to Common Root Rot (Spot Blotch), Fusarium Crown Rot, and Sharp Eyespot in Wheat. Front. Genet. 2021, 12, 699342. [Google Scholar] [CrossRef] [PubMed]
- Kokhmetova, A.; Sehgal, D.; Ali, S.; Atishova, M.; Kumarbayeva, M.; Leonova, I.; Dreisigacker, S. Genome-Wide Association Study of Tan Spot Resistance in a Hexaploid Wheat Collection From Kazakhstan. Front. Genet. 2021, 11, 581214. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Lakshman, D.K.; Roberts, D.P.; Ismaiel, A.; Abhishek, A.; Kumar, S.; Hooda, K.S. Fungal species causing maize leaf blight in different agro-ecologies in India. Pathogens 2021, 10, 1621. [Google Scholar] [CrossRef] [PubMed]
- Broccanello, C.; Ravi, S.; Deb, S.; Bolton, M.; Secor, G.; Richards, C.; Maretto, L.; Lucia, M.C.D.; Bertoldo, G.; Orsini, E.; et al. Bacterial endophytes as indicators of susceptibility to Cercospora Leaf Spot (CLS) disease in Beta vulgaris L. Sci. Rep. 2022, 12, 10719. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, S.M.; Sidhu, J.S.; Ali, S.; Kaur, N.; Wu, J.; Sehgal, S.K. Molecular characterization of bacterial leaf streak resistance in hard winter wheat. PeerJ 2019, 7, e7276. [Google Scholar] [CrossRef] [PubMed]
- Crouch, J.A.; Beirn, L.A. Anthracnose of Cereals and Grasses. 2009. Available online: http://www.broadinsti (accessed on 27 December 2023).
- Ababa, G.; Kesho, A.; Tadesse, Y.; Amare, D. Reviews of taxonomy, epidemiology, and management practices of the barley scald (Rhynchosporium graminicola) disease. Heliyon 2023, 9, e14315. [Google Scholar] [CrossRef]
- Ackerman, A.; Wenndt, A.; Boyles, R. The Sorghum Grain Mold Disease Complex: Pathogens, Host Responses, and the Bioactive Metabolites at Play. Front. Plant Sci. 2021, 12, 660171. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.W.; Liu, F.H.; Tan, X.L.; Zhang, Z.F.; Zhu, J.Y.; Tian, H.G.; Liu, T.X. Infection of powdery mildew reduces the fitness of grain aphids (Sitobion avenae) through restricted nutrition and induced defense response in wheat. Front. Plant Sci. 2018, 9, 778. [Google Scholar] [CrossRef]
- Dracatos, P.M.; Haghdoust, R.; Singh, D.; Park, R.F. Exploring and exploiting the boundaries of host specificity using the cereal rust and mildew models. New Phytol. 2018, 218, 453–462. [Google Scholar] [CrossRef]
- Riesenfeld, C.S.; Schloss, P.D.; Handelsman, J. Metagenomics: Genomic analysis of microbial communities. Annu. Rev. Genet. 2004, 38, 525–552. [Google Scholar] [CrossRef]
- Singh, J.; Behal, A.; Singla, N.; Joshi, A.; Birbian, N.; Singh, S.; Bali, V.; Batra, N. Metagenomics: Concept, methodology, ecological inference and recent advances. Biotechnol. J. Healthc. Nutr. Technol. 2009, 4, 480–494. [Google Scholar] [CrossRef]
- Oulas, A.; Pavloudi, C.; Polymenakou, P.; Pavlopoulos, G.A.; Papanikolaou, N.; Kotoulas, G.; Arvanitidis, C.; Iliopoulos, I. Metagenomics: Tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform. Biol. Insights 2015, 9, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Lin, R.; Chang, Y.; Zhou, Q.; Zhang, Z. Advances in Metagenomics and Its Application in Environmental Microorganisms. Front. Microbiol. 2021, 12, 766364. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to sequencing and analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Ranjan, R.; Rani, A.; Metwally, A.; McGee, H.S.; Perkins, D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem. Biophys. Res. Commun. 2016, 469, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Fajarningsih, N.D. Internal Transcribed Spacer (ITS) as DNA barcoding to identify fungal species: A review. Squalen Bull. Mar. Fish. Postharvest Biotechnol. 2016, 11, 37–44. [Google Scholar] [CrossRef]
- Peterson, D.; Bonham, K.S.; Rowland, S.; Pattanayak, C.W.; Klepac-Ceraj, V.; Deoni, S.C.L.; D’Sa, V.; Bruchhage, M.; Volpe, A.; Beauchemin, J.; et al. Comparative Analysis of 16S rRNA Gene and Metagenome Sequencing in Pediatric Gut Microbiomes. Front. Microbiol. 2021, 12, 670336. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Wemheuer, F.; Taylor, J.A.; Daniel, R.; Johnston, E.; Meinicke, P.; Thomas, T.; Wemheuer, B. Tax4Fun2, Prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiomes 2020, 15, 11. [Google Scholar] [CrossRef]
- Buytaers, F.E.; Fraiture, M.A.; Berbers, B.; Vandermassen, E.; Hoffman, S.; Papazova, N.; Vanneste, K.; Marchal, K.; Roosens, N.H.C.; De Keersmaecker, S.C.J. A shotgun metagenomics approach to detect and characterize unauthorized genetically modified microorganisms in microbial fermentation products. Food Chem. Mol. Sci. 2021, 2, 100023. [Google Scholar] [CrossRef]
- Walsh, L.H.; Coakley, M.; Walsh, A.M.; O’Toole, P.W.; Cotter, P.D. Bioinformatic approaches for studying the microbiome of fermented food. Crit. Rev. Microbiol. 2023, 49, 693–725. [Google Scholar] [CrossRef]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- Zaheer, R.; Noyes, N.; Ortega Polo, R.; Cook, S.R.; Marinier, E.; Van Domselaar, G.; Belk, K.E.; Morley, P.S.; McAllister, T.A. Impact of sequencing depth on the characterization of the microbiome and resistome. Sci. Rep. 2018, 8, 5890. [Google Scholar] [CrossRef]
- Navgire, G.S.; Goel, N.; Sawhney, G.; Sharma, M.; Kaushik, P.; Mohanta, Y.K.; Mohanta, T.K.; Al-Harrasi, A. Analysis and Interpretation of metagenomics data: An approach. Biol. Proced. Online 2022, 24, 18. [Google Scholar] [CrossRef]
- Chouhan, U.; Gamad, U.; Choudhari, J.K. Metagenomic analysis of soybean endosphere microbiome to reveal signatures of microbes for health and disease. J. Genet. Eng. Biotechnol. 2023, 21, 84. [Google Scholar] [CrossRef] [PubMed]
- Khoiri, A.N.; Cheevadhanarak, S.; Jirakkakul, J.; Dulsawat, S.; Prommeenate, P.; Tachaleat, A.; Kusonmano, K.; Wattanachaisaereekul, S.; Sutheeworapong, S. Comparative Metagenomics Reveals Microbial Signatures of Sugarcane Phyllosphere in Organic Management. Front. Microbiol. 2021, 12, 623799. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.H.; Lu, Z.M.; Zhang, X.J.; Wang, Z.M.; Yu, Y.J.; Shi, J.S.; Xu, Z.H. Metagenomics reveals flavour metabolic network of cereal vinegar microbiota. Food Microbiol. 2017, 62, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Franzosa, E.A.; McIver, L.J.; Rahnavard, G.; Thompson, L.R.; Schirmer, M.; Weingart, G.; Lipson, K.S.; Knight, R.; Caporaso, J.G.; Segata, N.; et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 2018, 15, 962–968. [Google Scholar] [CrossRef]
- Quiza, L.; Tremblay, J.; Pagé, A.P.; Greer, C.W.; Pozniak, C.J.; Li, R.; Haug, B.; Hemmingsen, S.M.; St-Arnaud, M.; Yergeau, E. The effect of wheat genotype on the microbiome is more evident in roots and varies through time. ISME Commun. 2023, 3, 32. [Google Scholar] [CrossRef]
- Xu, W.; Whitman, W.B.; Gundale, M.J.; Chien, C.C.; Chiu, C.Y. Functional response of the soil microbial community to biochar applications. GCB Bioenergy 2021, 13, 269–281. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.; Wang, Z.; Zhu, K.; Wu, W. Comparative metagenomic analysis reveals rhizosphere microbial community composition and functions help protect grapevines against salt stress. Front. Microbiol. 2023, 14, 1102547. [Google Scholar] [CrossRef] [PubMed]
- Oyserman, B.O.; Flores, S.S.; Griffioen, T.; Pan, X.; van der Wijk, E.; Pronk, L.; Lokhorst, W.; Nurfikari, A.; Paulson, J.N.; Movassagh, M. Disentangling the genetic basis of rhizosphere microbiome assembly in tomato. Nat. Commun. 2022, 13, 3228. [Google Scholar] [CrossRef] [PubMed]
- Frey, B.; Varliero, G.; Qi, W.; Stierli, B.; Walthert, L.; Brunner, I. Shotgun Metagenomics of Deep Forest Soil Layers Show Evidence of Altered Microbial Genetic Potential for Biogeochemical Cycling. Front. Microbiol. 2022, 13, 828977. [Google Scholar] [CrossRef] [PubMed]
- Alegria Terrazas, R.; Robertson-Albertyn, S.; Corral, A.M.; Escudero-Martinez, C.; Kapadia, R.; Balbirnie-Cumming, K.; Morris, J.; Hedley, P.E.; Barret, M.; Torres-Cortes, G. Defining Composition and Function of the Rhizosphere Microbiota of Barley Genotypes Exposed to Growth-Limiting Nitrogen Supplies. mSystems 2022, 7, e0093422. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D.; et al. The structure and function of the global citrus rhizosphere microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef] [PubMed]
- Shami, A.; Jalal, R.S.; Ashy, R.A.; Abuauf, H.W.; Baz, L.; Refai, M.Y.; Barqawi, A.A.; Baeissa, H.M.; Tashkandi, M.A.; Alshareef, S.; et al. Use of Metagenomic Whole Genome Shotgun Sequencing Data in Taxonomic Assignment of Dipterygium glaucum Rhizosphere and Surrounding Bulk Soil Microbiomes, and Their Response to Watering. Sustainability 2022, 14, 8764. [Google Scholar] [CrossRef]
- Zhou, Y.; Coventry, D.R.; Gupta, V.V.S.R.; Fuentes, D.; Merchant, A.; Kaiser, B.N.; Li, J.; Wei, Y.; Liu, H.; Wang, Y.; et al. The preceding root system drives the composition and function of the rhizosphere microbiome. Genome Biol. 2020, 21, 89. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Al Rwahnih, M.; Daubert, S.; Golino, D.; Rowhani, A. Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology 2009, 387, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Sun, H.; Zhang, X.; Cai, B. Next-generation sequencing of root fungal communities in continuous cropping soybean. Chil. J. Agric. Res. 2018, 78, 528–538. [Google Scholar] [CrossRef]
- Gu, L.; Bai, Z.; Jin, B.; Hu, Q.; Wang, H.; Zhuang, G.; Zhang, H. Assessing the impact of fungicide enostroburin application on bacterial community in wheat phyllosphere. J. Environ. Sci. 2010, 22, 134–141. [Google Scholar] [CrossRef]
- Hong, C.; Si, Y.; Xing, Y.; Li, Y. Illumina MiSeq sequencing investigation on the contrasting soil bacterial community structures in different iron mining areas. Environ. Sci. Pollut. Res. 2015, 22, 10788–10799. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-K.; Lee, S.-H.; Han, S.; Kim, J.-C.; Kim, Y.C.; Gardener, B.M. Marker-Assisted Selection of Novel Bacteria Contributing to Soil-Borne Plant Disease Suppression. Mol. Microb. Ecol. Rhizosphere 2013, 1, 637–642. [Google Scholar]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Bziuk, N.; Maccario, L.; Sørensen, S.J.; Schikora, A.; Smalla, K. Barley Rhizosphere Microbiome Transplantation—A Strategy to Decrease Susceptibility of Barley Grown in Soils With Low Microbial Diversity to Powdery Mildew. Front. Microbiol. 2022, 13, 830905. [Google Scholar] [CrossRef]
- De Vries, F.T.; Griffiths, R.I.; Knight, C.G.; Nicolitch, O.; Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [CrossRef]
- Hou, S.; Thiergart, T.; Vannier, N.; Mesny, F.; Ziegler, J.; Pickel, B.; Hacquard, S. A microbiota–root–shoot circuit favours Arabidopsis growth over defence under suboptimal light. Nat. Plants 2021, 7, 1078–1092. [Google Scholar] [CrossRef]
- Verhagen, B.W.M.; Glazebrook, J.; Zhu, T.; Chang, H.-S.; Van Loon, L.C.; Pieterse, C.M.J. The Transcriptome of Rhizobacteria-Induced Systemic Resistance in Arabidopsis. Mol. Plant-Microbe Interact. 2004, 17, 895–908. [Google Scholar] [CrossRef]
- du Jardin, P. Plant biostimulants: Definition, concept, main categories and regulation. Sci. Hortic. 2015, 196, 3–14. [Google Scholar] [CrossRef]
- Rocha, L.F.; Bond, J.P.; Fakhoury, A.M. Wheat Production Alters Soil Microbial Profiles and Enhances Beneficial Microbes in Double-Cropping Soybean. Front. Agron. 2022, 3, 807112. [Google Scholar] [CrossRef]
- Tkalec, V.; Mahnic, A.; Gselman, P.; Rupnik, M. Analysis of seed-associated bacteria and fungi on staple crops using the cultivation and metagenomic approaches. Folia Microbiol. 2022, 67, 351–361. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Chen, S. Diazotroph paenibacillus triticisoli bj-18 drives the variation in bacterial, diazotrophic and fungal communities in the rhizosphere and root/shoot endosphere of maize. Int. J. Mol. Sci. 2021, 22, 1460. [Google Scholar] [CrossRef]
- Ke, X.; Feng, S.; Wang, J.; Lu, W.; Zhang, W.; Chen, M.; Lin, M. Effect of inoculation with nitrogen-fixing bacterium Pseudomonas stutzeri A1501 on maize plant growth and the microbiome indigenous to the rhizosphere. Syst. Appl. Microbiol. 2019, 42, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Manriquez, B.; Muller, D.; Prigent-Combaret, C. Experimental Evolution in Plant-Microbe Systems: A Tool for Deciphering the Functioning and Evolution of Plant-Associated Microbial Communities. Front. Microbiol. 2021, 12, 619122. [Google Scholar] [CrossRef] [PubMed]
- Henry, L.P.; Bergelson, J. Evolutionary implications of host genetic control for engineering beneficial microbiomes. Curr. Opin. Syst. Biol. 2023, 34, 100455. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Pereira, P.J.; Teixeira, L.; Dangl, J.L. The Plant Microbiome: From Ecology to Reductionism and Beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e14. [Google Scholar] [CrossRef] [PubMed]
- Stringlis, I.A.; Yu, K.; Feussner, K.; De Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.A.H.M.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef] [PubMed]
- Morcillo, R.J.; Singh, S.K.; He, D.; An, G.; Vílchez, J.I.; Tang, K.; Yuan, F.; Sun, Y.; Shao, C.; Zhang, S.; et al. Rhizobacterium-derived diacetyl modulates plant immunity in a phosphate-dependent manner. EMBO J. 2020, 39, e102602. [Google Scholar] [CrossRef] [PubMed]
- Dowen, R.H.; Pelizzola, M.; Schmitz, R.J.; Lister, R.; Dowen, J.M.; Nery, J.R.; Dixon, J.E.; Ecker, J.R. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 2012, 109, E2183–E2191. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, R.; Peng, L.; Singh, S.K.; Zhang, M.; Zhang, X.; Vílchez, J.I.; Wang, Z.; He, D.; Yang, Y.; Lv, S. Dicer-like proteins influence Arabidopsis root microbiota independent of RNA-directed DNA methylation. Microbiome 2021, 9, 57. [Google Scholar] [CrossRef]
- Baubec, T.; Dinh, H.Q.; Pecinka, A.; Rakic, B.; Rozhon, W.; Wohlrab, B.; von Haeseler, A.; Scheid, O.M. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell 2010, 22, 34–47. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, X.; Miki, D.; Cutler, S.; La, H.; Hou, Y.J.; Oh, J.E.; Zhu, J.K. Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 2012, 24, 1230–1241. [Google Scholar] [CrossRef]
- Wei, L.; Gu, L.; Song, X.; Cui, X.; Lu, Z.; Zhou, M.; Wang, L.; Hu, F.; Zhai, J.; Meyers, B.C.; et al. Dicer-like 3 produces transposable element-associated 24-nt siRNAs that control agricultural traits in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 3877–3882. [Google Scholar] [CrossRef]
- Halter, T.; Wang, J.; Amesefe, D.; Lastrucci, E.; Charvin, M.; Rastogi, M.S.; Navarro, L. The arabidopsis active demethylase ros1 cis-regulates defense genes by erasing dna methylation at promoter-regulatory regions. Elife 2021, 10, e62994. [Google Scholar] [CrossRef]
- Le, T.N.; Schumann, U.; Smith, N.A.; Tiwari, S.; Khang Au, P.C.; Zhu, Q.H.; Taylor, J.M.; Kazan, K.; Llewellyn, D.J.; Zhang, R.; et al. DNA demethylases target promoter transposable elements to positively regulate stress responsive genes in Arabidopsis. Genome Biol. 2014, 15, 458. [Google Scholar] [CrossRef]
- Schumann, U.; Lee, J.M.; Smith, N.A.; Zhong, C.; Zhu, J.K.; Dennis, E.S.; Millar, A.A.; Wang, M.B. DEMETER plays a role in DNA demethylation and disease response in somatic tissues of Arabidopsis. Epigenetics 2019, 14, 1074–1087. [Google Scholar] [CrossRef]
- Hüther, P.; Hagmann, J.; Nunn, A.; Kakoulidou, I.; Pisupati, R.; Langenberger, D.; Weigel, D.; Johannes, F.; Schultheiss, S.J.; Becker, C. MethylScore, a pipeline for accurate and context-aware identification of differentially methylated regions from population-scale plant whole-genome bisulfite sequencing data. Quant. Plant Biol. 2022, 3, e19. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Satgé, C.; Moreau, S.; Sallet, E.; Lefort, G.; Auriac, M.C.; Remblière, C.; Cottret, L.; Gallardo, K.; Noirot, C.; Jardinaud, M.F.; et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat. Plants 2016, 2, 16166. [Google Scholar] [CrossRef] [PubMed]
- Vigneaud, J.; Kohler, A.; Sow, M.D.; Delaunay, A.; Fauchery, L.; Guinet, F.; Daviaud, C.; Barry, K.W.; Keymanesh, K.; Johnson, J.; et al. DNA hypomethylation of the host tree impairs interaction with mutualistic ectomycorrhizal fungus. New Phytol. 2023, 238, 2561–2577. [Google Scholar] [CrossRef] [PubMed]
- Varga, S.; Soulsbury, C.D. Paternal arbuscular mycorrhizal fungal status affects DNA methylation in seeds. Biol. Lett. 2017, 13, 20170407. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Wang, M.; Huang, Y.; Xu, Z.; Xu, P.; Nie, Y.; Tang, H. Guided by the principles of microbiome engineering: Accomplishments and perspectives for environmental use. mLife 2022, 1, 382–398. [Google Scholar] [CrossRef]
- Prakash, O.; Nimonkar, Y.; Desai, D. A Recent Overview of Microbes and Microbiome Preservation. Indian J. Microbiol. 2020, 60, 297–309. [Google Scholar] [CrossRef]
- Head, S.R.; Kiyomi Komori, H.; LaMere, S.A.; Whisenant, T.; Van Nieuwerburgh, F.; Salomon, D.R.; Ordoukhanian, P. Library construction for next-generation sequencing: Overviews and challenges. Biotechniques 2014, 56, 61–77. [Google Scholar] [CrossRef]
- Hess, J.F.; Kohl, T.A.; Kotrová, M.; Rönsch, K.; Paprotka, T.; Mohr, V.; Hutzenlaub, T.; Brüggemann, M.; Zengerle, R.; Niemann, S.; et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol. Adv. 2020, 41, 107537. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Zhang, C.; Zhou, W.; Li, W.; Chu, L.; Yan, J.; Li, H. Diversity and plant growth-promoting ability of endophytic fungi from the five flower plant species collected from Yunnan, Southwest China. J. Plant Interact. 2014, 9, 585–591. [Google Scholar] [CrossRef]
- Lu, J.; Salzberg, S.L. Ultrafast and accurate 16S rRNA microbial community analysis using Kraken 2. Microbiome 2020, 8, 124. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nature Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Albrecht, B.; Bağci, C.; Bessarab, I.; Górska, A.; Jolic, D.; Williams, R.B.H. MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biol. Direct 2018, 13, 6. [Google Scholar] [CrossRef]
- Schmitz-Hübsch, D.J.; Kurtz, S. MetaGenomeThreader: A software tool for predicting genes in DNA-sequences of metagenome projects. Methods Mol. Biol. 2010, 668, 325–338. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2, High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Gweon, H.S.; Oliver, A.; Taylor, J.; Booth, T.; Gibbs, M.; Read, D.S.; Griffiths, R.I.; Schonrogge, K. PIPITS: An automated pipeline for analyses of fungal internal transcribed spacer sequences from the Illumina sequencing platform. Methods Ecol. Evol. 2015, 6, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Zhang, K.; Lin, R.; Chang, Y.; Zhou, Q.; Zhang, Z. 16S-FASAS: An integrated pipeline for synthetic full-length 16S rRNA gene sequencing data analysis. PeerJ 2022, 10, e14043. [Google Scholar] [CrossRef] [PubMed]
- Fuks, G.; Elgart, M.; Amir, A.; Zeisel, A.; Turnbaugh, P.J.; Soen, Y.; Shental, N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 2018, 6, 17. [Google Scholar] [CrossRef]
- Krivonos, D.V.; Konanov, D.N.; Ilina, E.N. FunFun: ITS-based functional annotator of fungal communities. Ecol. Evol. 2023, 13, e9874. [Google Scholar] [CrossRef]
- Sikolenko, M.A.; Valentovich, L.N. RiboGrove: A database of full-length prokaryotic 16S rRNA genes derived from completely assembled genomes. Res. Microbiol. 2022, 173, 103936. [Google Scholar] [CrossRef] [PubMed]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 2023. [Google Scholar] [CrossRef]
- Kõljalg, U.; Abarenkov, K.; Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S. UNITE: A database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol. 2005, 166, 1063–1068. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Keegan, K.P.; Glass, E.M.; Meyer, F. MG-RAST, a metagenomics service for analysis of microbial community structure and function. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; pp. 207–233. [Google Scholar]
- Huson, D.H.; Albrecht, B.; Bağci, C.; Bessarab, I.; Górska, A.; Jolic, D.; Williams, R.B.H. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Milanese, A.; Mende, D.R.; Paoli, L.; Salazar, G.; Ruscheweyh, H.J.; Cuenca, M.; Hingamp, P.; Alves, R.; Costea, P.I.; Coelho, L.P.; et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019, 10, 1014. [Google Scholar] [CrossRef] [PubMed]
- Pierce, N.T.; Irber, L.; Reiter, T.; Brooks, P.; Brown, C.T. Large-scale sequence comparisons with sourmash. F1000Research 2019, 8, 1006. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Menzel, P.; Ng, K.L.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed]
- Ounit, R.; Wanamaker, S.; Close, T.J.; Lonardi, S. CLARK: Fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genom. 2015, 16, 236. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.; Manimaran, S.; Shen, Y.; Perez-Rogers, J.F.; Byrd, A.L.; Castro-Nallar, E.; Crandall, K.A.; Johnson, W.E. PathoScope 2.0, A complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome 2014, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Van der Jeugt, F.; Maertens, R.; Steyaert, A.; Verschaffelt, P.; De Tender, C.; Dawyndt, P.; Mesuere, B. UMGAP: The Unipept MetaGenomics Analysis Pipeline. BMC Genom. 2022, 23, 433. [Google Scholar] [CrossRef] [PubMed]
- Uritskiy, G.V.; Diruggiero, J.; Taylor, J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis 08 Information and Computing Sciences 0803 Computer Software 08 Information and Computing Sciences 0806 Information Systems. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef]
- Sultana Tithi Corresp, S.; Aylward, F.O.; Jensen, R.V.; Zhang, L.; Author, C.; Sultana Tithi, S. FastViromeExplorer: A pipeline for virus and phage identification and abundance profiling in metagenomics data. PeerJ 2018, 6, e4227. Available online: http://bench.cs.vt.edu/FastViromeExplorer/ (accessed on 15 January 2024).
- Ru, J.; Khan Mirzaei, M.; Xue, J.; Peng, X.; Deng, L. ViroProfiler: A containerized bioinformatics pipeline for viral metagenomic data analysis. Gut Microbes 2023, 15, 2192522. [Google Scholar] [CrossRef]
- Plyusnin, I.; Vapalahti, O.; Sironen, T.; Kant, R.; Smura, T. Enhanced Viral Metagenomics with Lazypipe 2. Viruses 2023, 15, 431. [Google Scholar] [CrossRef]
- Ji, Y.; Huotari, T.; Roslin, T.; Schmidt, N.M.; Wang, J.; Yu, D.W.; Ovaskainen, O. SPIKEPIPE: A metagenomic pipeline for the accurate quantification of eukaryotic species occurrences and intraspecific abundance change using DNA barcodes or mitogenomes. Mol. Ecol. Resour. 2020, 20, 256–267. [Google Scholar] [CrossRef]
- Zhao, Y.; Tang, H.; Ye, Y. RAPSearch2, A fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 2012, 28, 125–126. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Zhang, J.; Kim, Y.; Ho, J.; Liu, K.; Keenum, I.; Gupta, S.; Davis, B.; Hepp, S.L.; Zhang, L.; et al. ARGem: A new metagenomics pipeline for antibiotic resistance genes: Metadata, analysis, and visualization. Front. Genet. 2023, 14, 1219297. [Google Scholar] [CrossRef] [PubMed]
- Apweiler, R. UniProt: The Universal Protein knowledgebase. Nucleic Acids Res. 2004, 32, 115D–119D. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Bik, H.M. Phinch: An interactive, exploratory data visualization framework for-Omic datasets. bioRxiv 2014. [Google Scholar] [CrossRef]
- Breitwieser, F.P.; Salzberg, S.L. Pavian: Interactive analysis of metagenomics data for microbiome studies and pathogen identification. Bioinformatics 2020, 36, 1303–1304. [Google Scholar] [CrossRef] [PubMed]
- Mclaren, M.R.; Willis, A.D.; Callahan, B.J. Consistent and correctable bias in metagenomic sequencing measurements. Elife 2019, 8, e46923. [Google Scholar] [CrossRef]
- Moinard, S.; Piau, D.; Laporte, F.; Taberlet, P.; Coissac, E. PCR Bias in metabarcoding Towards quantitative DNA Metabarcoding: A method to overcome PCR amplification bias. bioRxiv 2023. [Google Scholar] [CrossRef]
- Piñol, J.; Mir, G.; Gomez-Polo, P.; Agustí, N. Universal and blocking primer mismatches limit the use of high-throughput DNA sequencing for the quantitative metabarcoding of arthropods. Mol. Ecol. Resour. 2015, 15, 819–830. [Google Scholar] [CrossRef]
- Piper, A.M.; Batovska, J.; Cogan, N.O.I.; Weiss, J.; Cunningham, J.P.; Rodoni, B.C.; Blacket, M.J. Prospects and challenges of implementing DNA metabarcoding for high-throughput insect surveillance. GigaScience 2019, 8, giz092. [Google Scholar] [CrossRef]
- Smith, R.H.; Glendinning, L.; Walker, A.W.; Watson, M. Investigating the impact of database choice on the accuracy of metagenomic read classification for the rumen microbiome. Anim. Microbiome 2022, 4, 57. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Zhao, S.; Guo, W.; Peng, L.; Zhao, F.; Wang, L.; Fan, G.; Zhu, Y.; Xu, D.; Liu, G.; et al. Sequencing introduced false positive rare taxa lead to biased microbial community diversity, assembly, and interaction interpretation in amplicon studies. Environ. Microbiomes 2022, 17, 43. [Google Scholar] [CrossRef] [PubMed]
- Hiergeist, A.; Ruelle, J.; Emler, S.; Gessner, A. Reliability of species detection in 16S microbiome analysis: Comparison of five widely used pipelines and recommendations for a more standardized approach. PLoS ONE 2023, 18, e0280870. [Google Scholar] [CrossRef] [PubMed]
- Jeske, J.T.; Gallert, C. Microbiome Analysis via OTU and ASV-Based Pipelines—A Comparative Interpretation of Ecological Data in WWTP Systems. Bioengineering 2022, 9, 146. [Google Scholar] [CrossRef] [PubMed]
- Mallawaarachchi, V.; Roach, M.J.; Decewicz, P.; Papudeshi, B.; Giles, S.K.; Grigson, S.R.; Bouras, G.; Hesse, R.D.; Inglis, L.K.; Hutton, A.L.K.; et al. Phables: From fragmented assemblies to high-quality bacteriophage genomes. Bioinformatics 2023, 39, btad586. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Moon, C.D.; Zheng, N.; Huws, S.; Zhao, S.; Wang, J. Opportunities and challenges of using metagenomic data to bring uncultured microbes into cultivation. Microbiome 2022, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, S.; Albaum, S.P.; Blumenkamp, P.; Linke, B.; Stoye, J.; Goesmann, A. Flexible metagenome analysis using the MGX framework. Microbiome 2018, 6, 76. [Google Scholar] [CrossRef]
- Deng, Z.; Zhang, J.; Li, J.; Zhang, X. Application of Deep Learning in Plant–Microbiota Association Analysis. Front. Genet. 2021, 12, 697090. [Google Scholar] [CrossRef]
- Castrillo, G.; Pereira, P.J.; Teixeira, L.; Paredes, S.H.; Law, T.F.; De Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; et al. Root Microbiota Drive Direct Integration of Phosphate Stress and Immunity. Available online: http://www.nature.com/authors/editorial_policies/license.html#terms(2017) (accessed on 27 December 2023).
- van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; Da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I.; et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef]
- Nichols, R.V.; Vollmers, C.; Newsom, L.A.; Wang, Y.; Heintzman, P.D.; Leighton, M.; Green, R.E.; Shapiro, B. Minimizing polymerase biases in metabarcoding. Mol. Ecol. Resour. 2018, 18, 927–939. [Google Scholar] [CrossRef]
- Pinto, A.J.; Raskin, L. PCR biases distort bacterial and archaeal community structure in pyrosequencing datasets. PLoS ONE 2012, 7, e43093. [Google Scholar] [CrossRef]
- Schloss, P.D. identifying and overcoming threats to reproducibility, replicability, robustness, and generalizability in microbiome research. mBio 2018, 9, e00525-18. [Google Scholar] [CrossRef]
- Vecherskii, M.V.; Semenov, M.V.; Lisenkova, A.A.; Stepankov, A.A. Metagenomics: A New Direction in Ecology. Biol. Bull. 2021, 48, S107–S117. [Google Scholar] [CrossRef]
- Allali, I.; Arnold, J.W.; Roach, J.; Cadenas, M.B.; Butz, N.; Hassan, H.M.; Koci, M.; Ballou, A.; Mendoza, M.; Ali, R. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- Sul, W.J.; Cole, J.R.; Jesus Eda, C.; Wang, Q.; Farris, R.J.; Fish, J.A.; Tiedje, J.M. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc. Natl. Acad. Sci. USA 2011, 108, 14637–14642. [Google Scholar] [CrossRef] [PubMed]
- Kimotho, R.N.; Maina, S. Unraveling plant-microbe interactions using integrated omics approaches. J. Exp. Bot. 2023, 75, 1289–1313. [Google Scholar] [CrossRef] [PubMed]
- Bukhat, S.; Imran, A.; Javaid, S.; Shahid, M.; Majeed, A.; Naqqash, T. Communication of plants with microbial world: Exploring the regulatory networks for PGPR mediated defense signaling. Microbiol. Res. 2020, 238, 126486. [Google Scholar] [CrossRef] [PubMed]
- Abdullaeva, Y.; Ambika Manirajan, B.; Honermeier, B.; Schnell, S.; Cardinale, M. Domestication affects the composition, diversity, and co-occurrence of the cereal seed microbiota. J. Adv. Res. 2021, 31, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Lui, L.M.; Nielsen, T.N.; Arkin, A.P. A method for achieving complete microbial genomes and improving bins from metagenomics data. PLoS Comput. Biol. 2021, 17, e1008972. [Google Scholar] [CrossRef] [PubMed]
- Sereika, M.; Kirkegaard, R.H.; Karst, S.M.; Michaelsen, T.Y.; Sørensen, E.A.; Wollenberg, R.D.; Albertsen, M. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat. Methods 2022, 19, 823–826. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Beaulaurier, J.; Zhu, S.; Deikus, G.; Mogno, I.; Zhang, X.S.; Davis-Richardson, A.; Canepa, R.; Triplett, E.W.; Faith, J.J.; Sebra, R.; et al. Metagenomic binning and association of plasmids with bacterial host genomes using DNA methylation. Nat. Biotechnol. 2018, 36, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Elbers, J.P.; Rogers, M.F.; Perelman, P.L.; Proskuryakova, A.A.; Serdyukova, N.A.; Johnson, W.E.; Horin, P.; Corander, J.; Murphy, D.; Burger, P.A. Improving Illumina assemblies with Hi-C and long reads: An example with the North African dromedary. Mol. Ecol. Resour. 2019, 19, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, M.; Kolmogorov, M.; Tseng, E.; Portik, D.M.; Tolstoganov, I.; Uritskiy, G.; Liachko, I.; Sullivan, S.T.; Shin, B.; Zorea, A. Generation of lineage-resolved complete metagenome-assembled genomes by precision phasing. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wu, R.; Davison, M.R.; Nelson, W.C.; Smith, M.L.; Lipton, M.S.; Jansson, J.K. Hi-C metagenome sequencing reveals soil phage–host interactions. Nat. Commun. 2023, 14, 7666. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.X.; Haudenshield, J.S.; Bowen, C.R.; Hartman, G.L. Metagenome-wide association study and machine learning prediction of bulk soil microbiome and crop productivity. Front. Microbiol. 2017, 8, 519. [Google Scholar] [CrossRef]
- Aguiar-Pulido, V.; Huang, W.; Suarez-Ulloa, V.; Cickovski, T.; Mathee, K.; Narasimhan, G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis. Evol. Bioinform. 2016, 12, 5–16. [Google Scholar] [CrossRef] [PubMed]

| Cereal Diseases | Bacteria/Fungi | Symptoms | Cereal Crops | References |
|---|---|---|---|---|
| Fusarium head blight | Fusarium graminearum | Bleached or discolored spikelets, premature ripening, and pink or orange fungal spore masses on infected heads. | Wheat, rice, barley | [65] |
| Bacterial leaf blight | Xanthomonas campestris | Symptoms include water-soaked lesions with yellow halos on leaves. Lesions may expand and coalesce, leading to leaf wilting and plant death. | Wheat, sorghum, barley crops | [66] |
| Common charcoal root rot | Cochliobolus sativus, Macrophomina phaseolina | Symptoms include dark brown to black lesions on the roots and lower stem. Infected plants may exhibit stunted growth, reduced tillering, and wilting. | Sorghum, barley, wheat | [67] |
| Tan spot | Pyrenophora tritici-repentis | Symptoms include tan or brown necrotic lesions with yellow halos on leaves. Lesions may coalesce, leading to extensive leaf damage and reduced grain yield. | Wheat, maize, sorghum | [68] |
| Fungal leaf blight | Exserhilum turcicum | Large cigar-shaped lesion oriented lengthwise along the leaf. | Sorghum, wheat, maize | [69] |
| Bacterial leaf spot | Pseudomonas syringae | Water-soaked spot lesions on leaves. | Sorghum, wheat | [70] |
| Bacterial leaf stripe | Burkholderia andropogonis, Pseudomonas andropogonis, Pseudomonas sorghicola | Characterized by long, narrow stripes that can vary from red to black. | Maize, wheat, oats, sorghum | [71] |
| Anthracnose | Colletotrichum sublineolum | Small, circular, elliptical, or elongated spots. | Sorghum, maize, Barley, rye, oats | [72] |
| Leaf Scald | Rhynchosporium secalis | Elongated, brown lesions with yellow halos on leaves. Severe infections can lead to premature leaf death and reduced grain yield. | Barley | [73] |
| Grain molds | Fusarium spp., Curvularia lunata, Alternaria alternata, Phoma sorghina and other fungi | Pink, orange, or white seeds found on the infected heads. | Sorghum, maize, Wheat, oats | [74] |
| Powdery mildew | Blumeria graminis | White or gray powdery fungal growth on leaves, stems, and panicles. Infected plants may exhibit stunted growth, reduced photosynthesis, and premature senescence. | Sorghum, maize, Barley, oats | [75] |
| Rust | Puccinia purpurea | Reddish-brown pustules on stems, leaves, and spikelets. Infected plants may exhibit stunted growth, chlorosis, and reduced grain yield. | Sorghum, maize, Barley, oats | [76] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masenya, K.; Manganyi, M.C.; Dikobe, T.B. Exploring Cereal Metagenomics: Unravelling Microbial Communities for Improved Food Security. Microorganisms 2024, 12, 510. https://doi.org/10.3390/microorganisms12030510
Masenya K, Manganyi MC, Dikobe TB. Exploring Cereal Metagenomics: Unravelling Microbial Communities for Improved Food Security. Microorganisms. 2024; 12(3):510. https://doi.org/10.3390/microorganisms12030510
Chicago/Turabian StyleMasenya, Kedibone, Madira Coutlyne Manganyi, and Tshegofatso Bridget Dikobe. 2024. "Exploring Cereal Metagenomics: Unravelling Microbial Communities for Improved Food Security" Microorganisms 12, no. 3: 510. https://doi.org/10.3390/microorganisms12030510
APA StyleMasenya, K., Manganyi, M. C., & Dikobe, T. B. (2024). Exploring Cereal Metagenomics: Unravelling Microbial Communities for Improved Food Security. Microorganisms, 12(3), 510. https://doi.org/10.3390/microorganisms12030510






