Identification, Characterization, and Ultrastructure Analysis of the Phenol-Degrading Rhodococcus erythropolis 7Ba and Its Viable but Nonculturable Forms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Organisms, Methods of Cultivation
2.2. Biochemical Tests
2.3. Determination of Bacterial Degradation Ability
2.4. Microscopic Investigations
2.4.1. Light Microscopy
2.4.2. Transmission Electron Microscopy
2.5. Preparation of Cell-Free Extract
2.6. Enzyme Activity
2.7. Obtaining VBNC Cells
2.8. Determination of Cell Viability
2.9. DNA Sequencing, Assembly, and Annotation of Complete Genomes
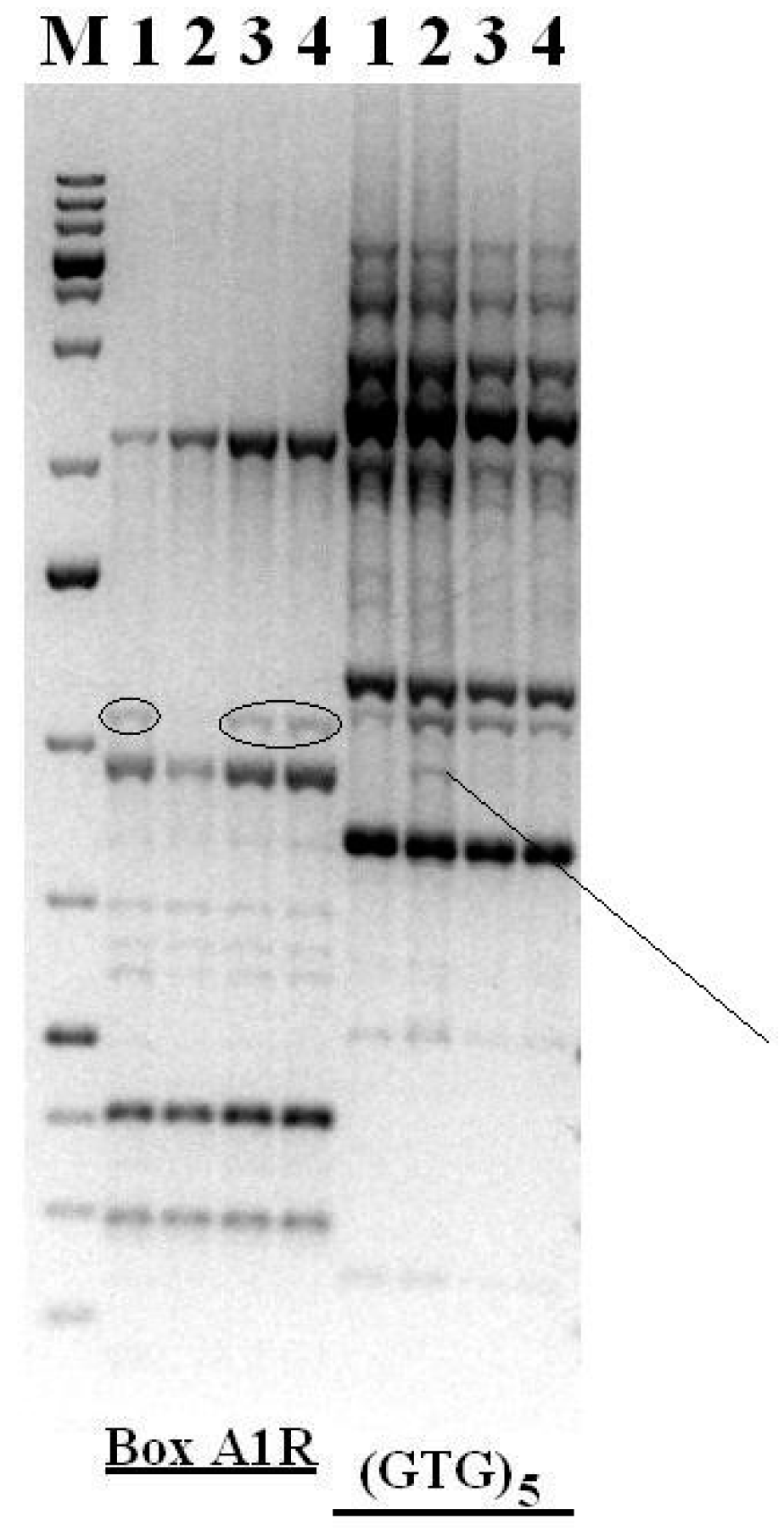
2.10. Genomic Fingerprinting
2.11. Statistical Analysis
3. Results
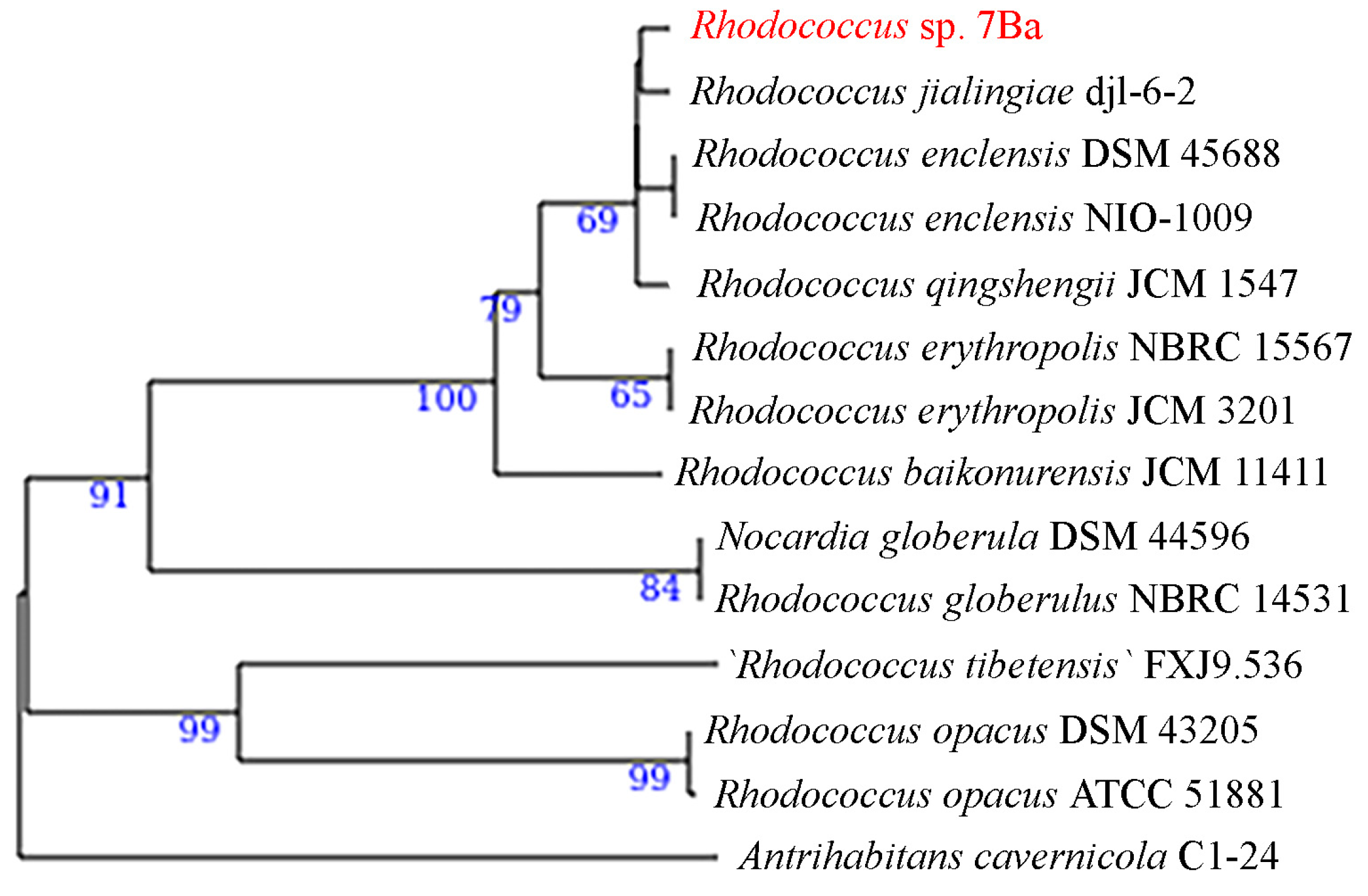
3.1. Isolation, Morphology, and Biochemical Properties
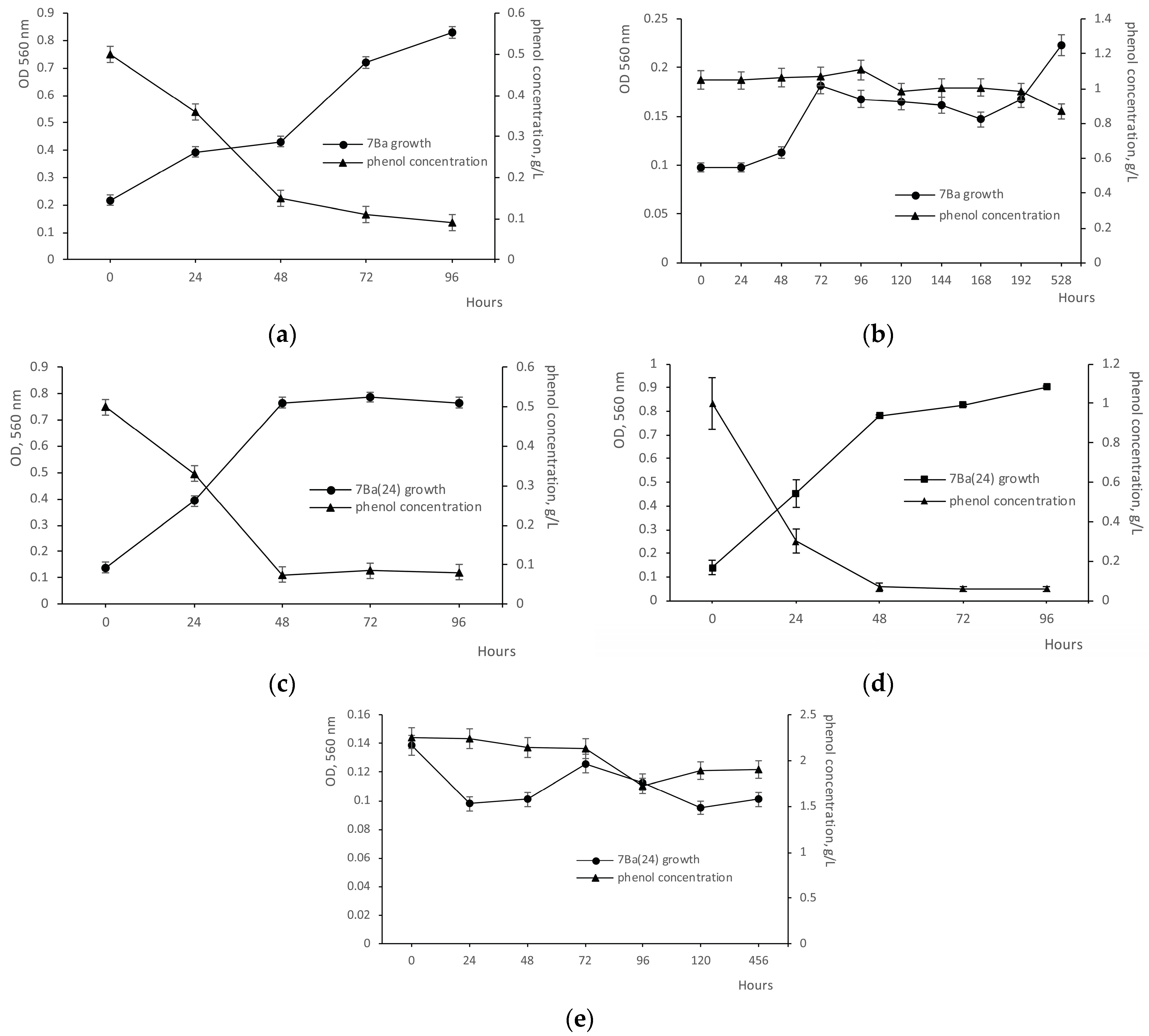
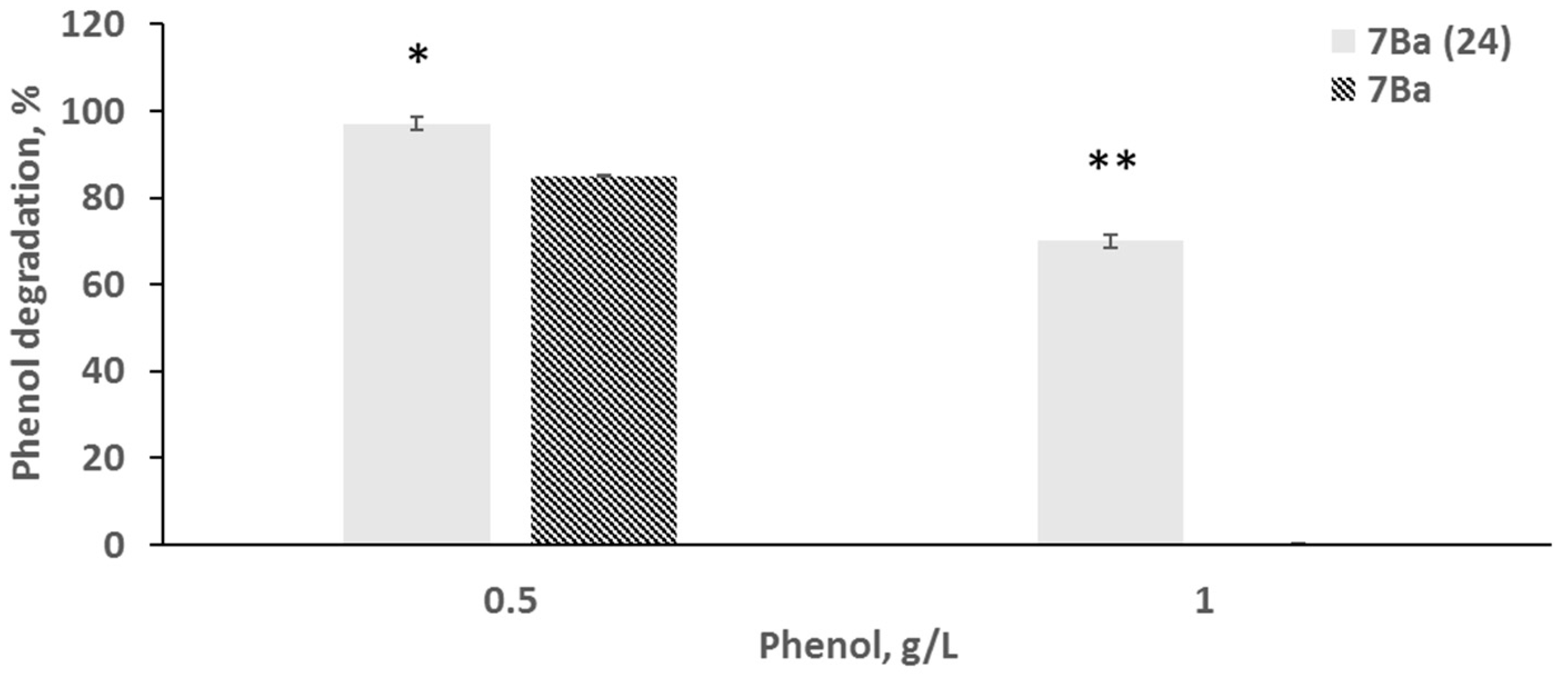
3.2. Phenol Degradation Ability Before and After Recovery from the VBNC State
3.3. Growth on Toxic Substrates Before and After Dormancy
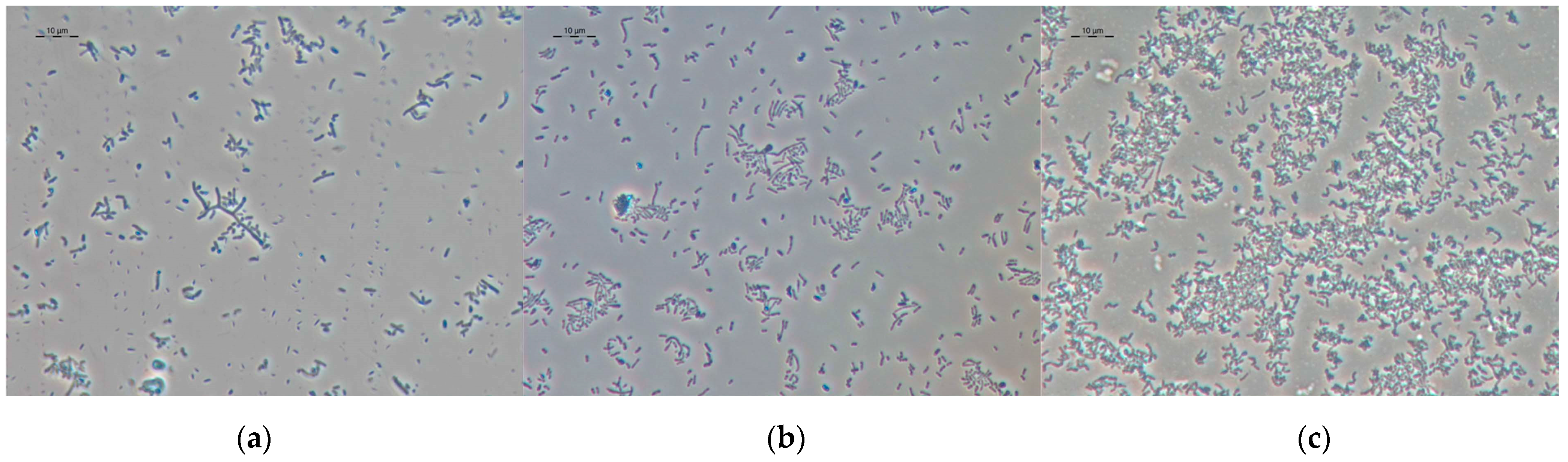
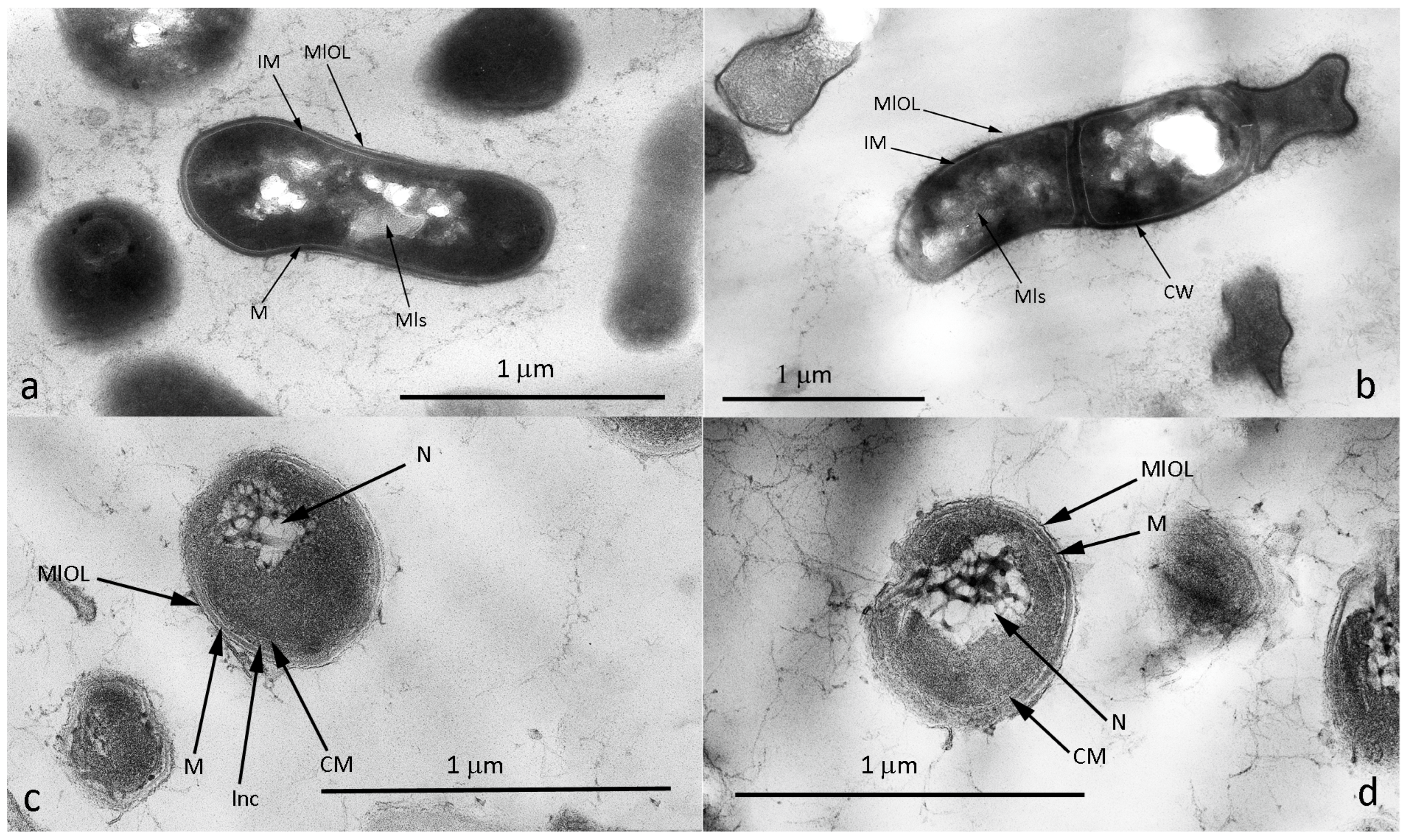
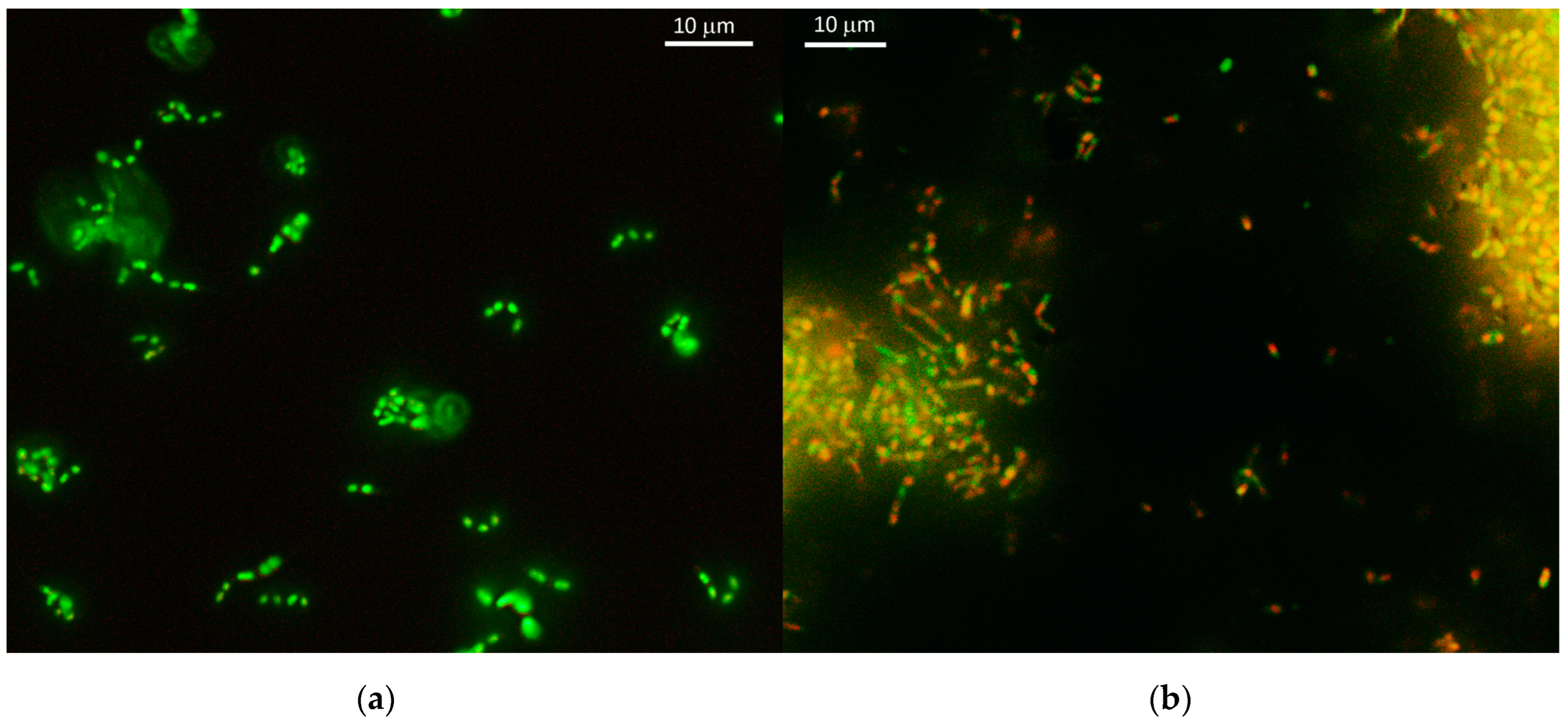
3.4. Microscopy of Vegetative and VBNC Cells
3.5. Determination of Cell Viability
3.6. Whole Genome, Analysis and Gene Search of Major Degradation Enzymes and Rpf Protein
3.7. Genomic Fingerprinting
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kuppan, N.; Padman, M.; Mahadeva, M.; Srinivasan, S.; Devarajan, R. A comprehensive review of sustainable bioremediation techniques: Eco friendly solutions for waste and pollution management. Waste Manag. Bull. 2024, 2, 154–171. [Google Scholar] [CrossRef]
- Alori, E.T.; Gabasawa, A.I.; Elenwo, C.E.; Agbeyegbe, O.O. Bioremediation techniques as affected by limiting factors in soil environment. Front. Soil Sci. 2022, 2, 937186. [Google Scholar] [CrossRef]
- Sun, J.; Mu, Q.; Kimura, H.; Murugadoss, V.; He, M.; Du, W.; Hou, C. Oxidative degradation of phenols and substituted phenols in the water and atmosphere: A review. Adv. Compos. Hybrid Mater. 2022, 5, 627–640. [Google Scholar] [CrossRef]
- Duan, W.; Meng, F.; Cui, H.; Lin, Y.; Wang, G.; Wu, J. Ecotoxicity of phenol and cresols to aquatic organisms: A review. Ecotoxicol. Environ. Saf. 2018, 157, 441–456. [Google Scholar] [CrossRef] [PubMed]
- Mohamad Said, K.A.; Ismail, A.F.; Abdul Karim, Z.; Abdullah, M.S.; Hafeez, A. A review of technologies for the phenolic compounds recovery and phenol removal from wastewater. Process Saf. Environ. Prot. 2021, 151, 257–289. [Google Scholar] [CrossRef]
- Motamedi, M.; Yerushalmi, L.; Haghighat, F.; Chen, Z. Recent developments in photocatalysis of industrial effluents ։ A review and example of phenolic compounds degradation. Chemosphere 2022, 296, 133688. [Google Scholar] [CrossRef]
- Wu, P.; Zhang, Z.; Luo, Y.; Bai, Y.; Fan, J. Bioremediation of phenolic pollutants by algae—Current status and challenges. Bioresour. Technol. 2022, 350, 126930. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Ding, P.; Xu, M.J.; Zhang, C.M.; Xing, K.; Qin, S. Biodegradation of phenol by a halotolerant versatile yeast Candida tropicalis SDP-1 in wastewater and soil under high salinity conditions. J. Environ. Manag. 2021, 289, 112525. [Google Scholar] [CrossRef]
- He, Y.; Wang, Z.; Li, T.; Peng, X.; Tang, Y.; Jia, X. Biodegradation of phenol by Candida tropicalis sp.: Kinetics, identification of putative genes and reconstruction of catabolic pathways by genomic and transcriptomic characteristics. Chemosphere 2022, 308, 136443. [Google Scholar] [CrossRef] [PubMed]
- Fernández, M.; Morales, G.M.; Agostini, E.; González, P.S. An approach to study ultrastructural changes and adaptive strategies displayed by Acinetobacter guillouiae SFC 500-1A under simultaneous Cr(VI) and phenol treatment. Environ. Sci. Pollut. Res. 2017, 24, 20390–20400. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, N.; Priyadarshini, A.; Sahoo, M.M.; Verma, A.K.; Daverey, A.; Sahoo, N.K. A comprehensive review on eco-toxicity and biodegradation of phenolics: Recent progress and future outlook. Environ. Technol. Innov. 2022, 27, 102423. [Google Scholar] [CrossRef]
- Chiesa, S.; Scarpato, A.; Lucentini, L.; Maltese, S.; Aliko, V.; Multisanti, C.R.; Turani, B.; Faggio, C. Get Rid of Marine Pollution: Bioremediation an Innovative, Attractive, and Successful Cleaning Strategy. Sustainability 2022, 14, 11784. [Google Scholar] [CrossRef]
- Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M.; Sridhar, K.; Inbaraj, B.S.; Tripathi, M. Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment. Toxics 2022, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- Baskaran, D.; Byun, H.S. Current trend of polycyclic aromatic hydrocarbon bioremediation: Mechanism, artificial mixed microbial strategy, machine learning, ground application, cost and policy implications. Chem. Eng. J. 2024, 498, 155334. [Google Scholar] [CrossRef]
- Ayrapetyan, M.; Williams, T.; Oliver, J.D. Relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol. 2018, 200, e00249-18. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol. Rev. 2010, 34, 415–425. [Google Scholar] [CrossRef]
- Oliver, J.D. The Viable but Nonculturable State in Bacteria. J. Microbiol. 2005, 43, 93–100. [Google Scholar] [PubMed]
- Shleeva, M.O.; Bagramyan, K.; Telkov, M.V.; Mukamolova, G.V.; Young, M.; Kell, D.B.; Kaprelyants, A.S. Formation and resuscitation of “non-culturable” cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology 2002, 148, 1581–1591. [Google Scholar] [CrossRef]
- Jia, Y.; Yu, C.; Fan, J.; Fu, Y.; Ye, Z.; Guo, X.; Xu, Y.; Shen, C. Alterations in the Cell Wall of Rhodococcus biphenylivorans Under Norfloxacin Stress. Front. Microbiol. 2020, 11, 554957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Z.; Pang, S.; Jiang, B.; Yang, Y.; Duan, Q.; Zhu, G. The impact of cell structure, metabolism and group behavior for the survival of bacteria under stress conditions. Arch. Microbiol. 2020, 203, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Nowakowska, J.; Oliver, J.D. Resistance to environmental stresses by Vibrio vulnificus in the viable but nonculturable state. FEMS Microbiol. Ecol. 2013, 84, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wang, Y.; Xue, B.; Zhang, Y.; Mei, R.; Zhang, Y.; Hashmi, M.Z.; Lin, H.; Chen, J.; Sun, F. Resuscitation of functional bacterial community for enhancing biodegradation of phenol under high salinity conditions based on Rpf. Bioresour. Technol. 2018, 261, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Fida, T.T.; Moreno-Forero, S.K.; Heipieper, H.J.; Springael, D. Physiology and transcriptome of the polycyclic aromatic hydrocarbon-degrading Sphingomonas sp. LH128 after long-term starvation. Microbiology 2013, 159, 1807–1817. [Google Scholar] [CrossRef]
- Pinto, D.; Santos, M.A.; Chambel, L. Thirty years of viable but nonculturable state research: Unsolved molecular mechanisms. Crit. Rev. Microbiol. 2015, 41, 61–76. [Google Scholar] [CrossRef]
- Maalej, S.; Denis, M.; Dukan, S. Temperature and growth-phase effects on Aeromonas hydrophila survival in natural seawater microcosms: Role of protein synthesis and nucleic acid content on viable but temporarily nonculturable response. Microbiology 2004, 150, 181–187. [Google Scholar] [CrossRef]
- Pinto, D.; Almeida, V.; Almeida Santos, M.; Chambel, L.M.M. Resuscitation of Escherichia coli VBNC cells depends on a variety of environmental or chemical stimuli. J. Appl. Microbiol. 2011, 110, 1601–1611. [Google Scholar] [CrossRef]
- Kaprelyants, A.S.; Gottschal, J.C.; Kell, D.B. Dormancy in non-sporulating bacteria. FEMS Microbiol. Lett. 1993, 104, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Mukamolova, G.V.; Yanopolskaya, N.D.; Kell, D.B.; Kaprelyants, A.S. On resuscitation from the dormant state of Micrococcus luteus. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 1998, 73, 237–243. [Google Scholar] [CrossRef]
- Telkov, M.V.; Demina, G.R.; Voloshin, S.A.; Salina, E.G.; Dudik, T.V.; Stekhanova, T.N.; Mukamolova, G.V.; Kazaryan, K.A.; Goncharenko, A.V.; Young, M.; et al. Proteins of the Rpf (resuscitation promoting factor) family are peptidoglycan hydrolases. Biochemistry 2006, 71, 414–422. [Google Scholar] [CrossRef]
- Nikitushkin, V.D.; Demina, G.R.; Shleeva, M.O.; Kaprelyants, A.S. Peptidoglycan fragments stimulate resuscitation of “non- culturable” mycobacteria. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2013, 103, 37–46. [Google Scholar] [CrossRef]
- Ruggiero, A.; Squeglia, F.; Romano, M.; Vitagliano, L.; De Simone, A.; Berisio, R. The structure of Resuscitation promoting factor B from M. tuberculosis reveals unexpected ubiquitin-like domains. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2016, 1860, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Nikitushkin, V.D.; Demina, G.R.; Kaprelyants, A.S. Rpf proteins are the factors of reactivation of the dormant forms of actinobacteria. Biochemistry 2016, 81, 1719–1734. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Chen, X.; Hu, J.; Shen, C.; Ding, L. Exploring the potential environmental functions of viable but non-culturable bacteria. World J. Microbiol. Biotechnol. 2013, 29, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, J.; Wang, Y.; Li, Y.C.; Chen, S.Y.; Zhang, Q. Promoting Resuscitation of the VBNC Bacteria and Enrichment of Naphthalene-degrading Bacteria from Oil-contaminated Soils with the Rpf4 from Rhodococcus erythropolis KB1. Geomicrobiol. J. 2022, 39, 916–924. [Google Scholar] [CrossRef]
- Shi, J.; Zhou, X.; Zhang, S.; Sun, F.; Shen, C.; Su, X. Unveiling the distribution characteristics of rpf-like genes and indigenous resuscitation promoting factor production in PCB-contaminated soils. J. Environ. Manag. 2024, 357, 120803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Ahmad, W.; Zhu, X.Y.; Chen, J.; Austin, B. Viable but nonculturable bacteria and their resuscitation: Implications for cultivating uncultured marine microorganisms. Mar. Life Sci. Technol. 2021, 3, 189–203. [Google Scholar] [CrossRef]
- Su, X.M.; Bamba, A.M.; Zhang, S.; Zhang, Y.G.; Hashmi, M.Z.; Lin, H.J.; Ding, L.X. Revealing potential functions of VBNC bacteria in polycyclic aromatic hydrocarbons biodegradation. Lett. Appl. Microbiol. 2018, 66, 277–283. [Google Scholar] [CrossRef]
- Solyanikova, I.P.; Mulyukin, A.L.; Suzina, N.E.; El-Registan, G.I.; Golovleva, L.A. Improved xenobiotic-degrading activity of Rhodococcus opacus strain 1CP after dormancy. J. Environ. Sci. Health Part B 2011, 46, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Li, H.; Jia, Y.; Fan, J.; Wan, J.; Guo, L.; Su, X.; Zhang, Y.; Wu, W.M.; Shen, C. Supplementing resuscitation-promoting factor (Rpf) enhanced biodegradation of polychlorinated biphenyls (PCBs) by Rhodococcus biphenylivorans strain TG9T. Environ. Pollut. 2020, 263, 114488. [Google Scholar] [CrossRef]
- Su, X.; Xie, M.; Han, Z.; Xiao, Y.; Wang, R.; Shen, C.; Hashmi, M.Z.; Sun, F. Resuscitation-Promoting Factor Accelerates Enrichment of Highly Active Tetrachloroethene/Polychlorinated Biphenyl-Dechlorinating Cultures. Appl. Environ. Microbiol. 2023, 89, e01951-22. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, S.; Wang, R.; An, Z.; Sun, F.; Shen, C.; Lin, H.; Su, X. A novel strategy for enhancing bioremediation of polychlorinated biphenyl-contaminated soil with resuscitation promoting factor and resuscitated strain. J. Hazard. Mater. 2023, 447, 130781. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Chen, J.; Wang, Y.; Meng, T.; Yue, L.; Luo, D.; Wang, X. Promoting effect of the recombinant resuscitation promoting factors-2 of Rhodococcus erythropolis on petroleum degradation and cultivable bacterial diversities of the oil-contaminated soils. Lett. Appl. Microbiol. 2022, 74, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E.S. The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef]
- Polivtseva, V.N.; Anokhina, T.O.; Iminova, L.R.; Borzova, O.V.; Esikova, T.Z.; Solyanikova, I.P. Evaluation of the Biotechnological Potential of New Bacterial Strains Capable of Phenol Degradation. Appl. Biochem. Microbiol. 2020, 56, 298–305. [Google Scholar] [CrossRef]
- Egozarian, N.S.; Emelyanova, E.V.; Suzina, N.E.; Sazonova, O.I.; Polivtseva, V.N.; Anokhina, T.O.; Wu, Y.; Solyanikova, I.P. Removal of Phenol by Rhodococcus opacus 1CP after Dormancy: Insight into Enzymes’ Induction, Specificity, and Cells Viability. Microorganisms 2024, 12, 597. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Brown, C.L.; Mullet, J.; Hindi, F.; Stoll, J.E.; Gupta, S.; Choi, M.; Keenum, I.; Vikesland, P.; Pruden, A.; Zhang, L. mobileOG-db: A Manually Curated Database of Protein Families Mediating the Life Cycle of Bacterial Mobile Genetic Elements. Appl. Environ. Microbiol. 2022, 88, e00991-22. [Google Scholar] [CrossRef] [PubMed]
- Vernikos, G.S.; Parkhill, J. Interpolated variable order motifs for identification of horizontally acquired DNA: Revisiting the Salmonella pathogenicity islands. Bioinformatics 2006, 22, 2196–2203. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Schneider, M.; de Bruijn, F.; Lupski, J. Genomic fingerprinting of bacteria with repetitive sequencebased polymerase chain reaction. Methods Mol. Cell Biol. 1994, 5, 25–40. [Google Scholar]
- Mohapatra, B.R.; Broersma, K.; Mazumder, A. Comparison of five rep-PCR genomic fingerprinting methods for differentiation of fecal Escherichia coli from humans, poultry and wild birds. FEMS Microbiol. Lett. 2007, 277, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; Volume 33, pp. 75–78. [Google Scholar]
- Jones, A.L.; Goodfellow, M. Rhodococcus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Bergey’s Manual Trust: Glasgow, UK, 2015; pp. 1–50. [Google Scholar] [CrossRef]
- Solyanikova, I.P.; Suzina, N.E.; Emelyanova, E.V.; Polivtseva, V.N.; Pshenichnikova, A.B.; Lobanok, A.G.; Golovleva, L.A. Morphological, physiological, and biochemical characteristics of a benzoate-degrading strain Rhodococcus opacus 1CP under stress conditions. Microbiology 2017, 86, 202–212. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef]
- Whyte, L.G.; Smits, T.H.M.; Labbé, D.; Witholt, B.; Greer, C.W.; Van Beilen, J.B. Gene cloning and characterization of multiple alkane hydroxylase systems in Rhodococcus strains Q15 and NRRL B-16531. Appl. Environ. Microbiol. 2002, 68, 5933–5942. [Google Scholar] [CrossRef] [PubMed]
- Markova, Y.A.; Petrushin, I.S.; Belovezhets, L.A. Detection of gene clusters for biodegradation of alkanes and aromatic compounds in the Rhodococcus qingshengii VKM Ac-2784D genome. Vavilov J. Genet. Breed. 2023, 27, 276–282. [Google Scholar] [CrossRef]
- Benning, S.; Pritsch, K.; Radl, V.; Siani, R.; Wang, Z.; Schloter, M. (Pan)genomic analysis of two Rhodococcus isolates and their role in phenolic compound degradation. Microbiol. Spectr. 2024, 12, e03783-23. [Google Scholar] [CrossRef] [PubMed]
- Delegan, Y.; Petrikov, K.; Frantsuzova, E.; Rudenko, N.; Solomentsev, V.; Suzina, N.; Travkin, V.; Solyanikova, I.P. Complete Genome Analysis of Rhodococcus opacus S8 Capable of Degrading Alkanes and Producing Biosurfactant Reveals Its Genetic Adaptation for Crude Oil Decomposition. Microorganisms 2022, 10, 1172. [Google Scholar] [CrossRef] [PubMed]
- Shumkova, E.S.; Egorova, D.O.; Boronnikova, S.V.; Plotnikova, E.G. Polymorphism of the bphA genes in bacteria destructing biphenyl/chlorinated biphenils. Mol. Biol. 2015, 49, 569–580. [Google Scholar] [CrossRef]
- Yu, C.; Wang, H.; Blaustein, R.A.; Guo, L.; Ye, Q.; Fu, Y.; Fan, J.; Su, X.; Hartmann, E.M.; Shen, C. Pangenomic and functional investigations for dormancy and biodegradation features of an organic pollutant-degrading bacterium Rhodococcus biphenylivorans TG9. Sci. Total Environ. 2022, 809, 151141. [Google Scholar] [CrossRef] [PubMed]
- Signoretto, C.; Lleò, M.D.M.; Tafi, M.C.; Canepari, P. Cell wall chemical composition of Enterococcus faecalis in the viable but nonculturable state. Appl. Environ. Microbiol. 2000, 66, 1953–1959. [Google Scholar] [CrossRef]
- Ivshina, I.B.; Mukhutdinova, A.N.; Tyumina, H.A.; Vikhareva, H.V.; Suzina, N.E.; El’-Registan, G.I.; Mulyukin, A.L. Drotaverine Hydrochloride Degradation Using Cyst-like Dormant Cells of Rhodococcus ruber. Curr. Microbiol. 2015, 70, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Bodor, A.; Bounedjoum, N.; Vincze, G.E.; Erdeiné Kis, Á.; Laczi, K.; Bende, G.; Szilágyi, Á.; Kovács, T.; Perei, K.; Rákhely, G. Challenges of unculturable bacteria: Environmental perspectives. Rev. Environ. Sci. Bio/Technol. 2020, 19, 1–22. [Google Scholar] [CrossRef]
- West, S.A.; Cooper, G.A. Division of labour in microorganisms: An evolutionary perspective. Nat. Rev. Microbiol. 2016, 14, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.N.; Turkarslan, S.; Beer, K.D.; Yin Lo, F.; Baliga, N.S. Adaptation of cells to new environments. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 3, 544–561. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, A.G. Stress Responses of Bacterial Cells as Mechanism of Development of Antibiotic Tolerance (Review). Appl. Biochem. Microbiol. 2018, 54, 108–127. [Google Scholar] [CrossRef]
- da Cruz Nizer, W.S.; Inkovskiy, V.; Overhage, J. Surviving Reactive Chlorine Stress: Responses of Gram-Negative Bacteria to Hypochlorous Acid. Microorganisms 2020, 8, 1220. [Google Scholar] [CrossRef]
- Ye, C.; Lin, H.; Zhang, M.; Chen, S.; Yu, X. Characterization and potential mechanisms of highly antibiotic tolerant VBNC Escherichia coli induced by low level chlorination. Sci. Rep. 2020, 10, 1957. [Google Scholar] [CrossRef]
- Jin, Y.; Gan, G.; Yu, X.; Wu, D.; Zhang, L.; Yang, N.; Hu, J.; Liu, Z.; Zhang, L.; Hong, H.; et al. Isolation of Viable but Non-culturable Bacteria from Printing and Dyeing Wastewater Bioreactor Based on Resuscitation Promoting Factor. Curr. Microbiol. 2017, 74, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, Y.; Wang, Y.; Mei, R.; Zhang, Y.; Hashmi, M.Z.; Lin, H.; Su, X. A New Approach of Rpf Addition to Explore Bacterial Consortium for Enhanced Phenol Degradation Under High Salinity Conditions. Curr. Microbiol. 2018, 75, 1046–1054. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Wang, Y.; Xue, B.; Hashmi, M.Z.; Lin, H.; Chen, J.; Wang, Z.; Mei, R.; Sun, F. Impact of resuscitation promoting factor (Rpf) in membrane bioreactor treating high-saline phenolic wastewater: Performance robustness and Rpf-responsive bacterial populations. Chem. Eng. J. 2019, 357, 715–723. [Google Scholar] [CrossRef]
- Liu, Y.; Su, X.; Lu, L.; Ding, L.; Shen, C. A novel approach to enhance biological nutrient removal using a culture supernatant from Micrococcus luteus containing resuscitation-promoting factor (Rpf) in SBR process. Environ. Sci. Pollut. Res. 2016, 23, 4498–4508. [Google Scholar] [CrossRef]
- Wang, S.; Li, C.; Zhang, W.; Lin, M.; Li, J. Degradation of phenol from water by Rhodococcus ruber promoted by MgO nanoparticles. J. Environ. Chem. Eng. 2024, 12, 113946. [Google Scholar] [CrossRef]
- Tengku-Mazuki, T.A.; Subramaniam, K.; Zakaria, N.N.; Convey, P.; Abdul Khalil, K.; Lee, G.L.Y.; Zulkharnain, A.; Shaharuddin, N.A.; Ahmad, S.A. Optimization of phenol degradation by Antarctic bacterium Rhodococcus sp. Antarct. Sci. 2020, 32, 486–495. [Google Scholar] [CrossRef]
- Zhao, T.; Gao, Y.; Yu, T.; Zhang, Y.; Zhang, Z.; Zhang, L.; Zhang, L. Biodegradation of phenol by a highly tolerant strain Rhodococcus ruber C1: Biochemical characterization and comparative genome analysis. Ecotoxicol. Environ. Saf. 2021, 208, 111709. [Google Scholar] [CrossRef]
- Grechishnikova, E.G.; Shemyakina, A.O.; Novikov, A.D.; Lavrov, K.V.; Yanenko, A.S. Rhodococcus: Sequences of genetic parts, analysis of their functionality, and development prospects as a molecular biology platform. Crit. Rev. Biotechnol. 2023, 43, 835–850. [Google Scholar] [CrossRef] [PubMed]
- Grechishnikova, E.G.; Shemyakina, A.O.; Novikov, A.D.; Kalinina, T.I.; Lavrov, K.V.; Yanenko, A.S. Structure of the regulatory region of nitrile hydratase genes in Rhodococcus rhodochrous M8, a biocatalyst for production of acrylic heteropolymers. Microbiology 2024, 93, 432–437. [Google Scholar] [CrossRef]
- Spence, E.M.; Calvo-Bado, L.; Mines, P.; Bugg, T.D.H. Metabolic engineering of Rhodococcus jostii RHA1 for production of pyridine-dicarboxylic acids from lignin. Microb. Cell Factories 2021, 20, 15. [Google Scholar] [CrossRef]
- Jansen, Z.; Alameri, A.; Wei, Q.; Kulhanek, D.L.; Gilmour, A.R.; Halper, S.; Schwalm, N.D.; Thyer, R. A modular toolkit for environmental Rhodococcus, Gordonia, and Nocardia enables complex metabolic manipulation. Appl. Environ. Microbiol. 2024, 90, e00340-24. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Value |
|---|---|
| Input Read Pairs | 1,349,672 |
| Both Surviving | 1,173,100 (86.92%) |
| Forward Only Surviving | 163,148 (12.09%) |
| Reverse Only Surviving | 6314 (0.47%) |
| Dropped | 7110 (0.53%) |
| Substrate | 7Ba 1 | 7Ba(24) 2 | 7Ba(4) 3 |
|---|---|---|---|
| Benzoate (0.5 g/L) | + | + | + |
| Phenol (0.2 g/L) | + | + | + |
| Phenol (0.5 g/L) | + | + | + |
| Phenol (1 g/L) | − | + | − |
| 4-chlorophenol (0.1 g/L) | − | + | − |
| 2,4-dichlorophenol (0.1 g/L) | − | + | − |
| 2,4,6-trichlorophenol (0.1 g/L) | − | + | − |
| Hexadecane (0.1 g/L) | + | + | + |
| Parameter | Value |
|---|---|
| Number of contigs | 102 |
| Total length of assembly, bp | 6,767,064 |
| Longest contig, bp | 947,547 |
| Shortest contig, bp | 501 |
| N50, bp | 310,791 |
| N75, bp | 226,191 |
| N90, bp | 87,567 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polivtseva, V.N.; Zvonarev, A.N.; Sazonova, O.I.; Delegan, Y.A.; Kocharovskaya, Y.N.; Bogun, A.G.; Suzina, N.E. Identification, Characterization, and Ultrastructure Analysis of the Phenol-Degrading Rhodococcus erythropolis 7Ba and Its Viable but Nonculturable Forms. Microorganisms 2024, 12, 2662. https://doi.org/10.3390/microorganisms12122662
Polivtseva VN, Zvonarev AN, Sazonova OI, Delegan YA, Kocharovskaya YN, Bogun AG, Suzina NE. Identification, Characterization, and Ultrastructure Analysis of the Phenol-Degrading Rhodococcus erythropolis 7Ba and Its Viable but Nonculturable Forms. Microorganisms. 2024; 12(12):2662. https://doi.org/10.3390/microorganisms12122662
Chicago/Turabian StylePolivtseva, Valentina N., Anton N. Zvonarev, Olesya I. Sazonova, Yanina A. Delegan, Yulia N. Kocharovskaya, Alexander G. Bogun, and Nataliya E. Suzina. 2024. "Identification, Characterization, and Ultrastructure Analysis of the Phenol-Degrading Rhodococcus erythropolis 7Ba and Its Viable but Nonculturable Forms" Microorganisms 12, no. 12: 2662. https://doi.org/10.3390/microorganisms12122662
APA StylePolivtseva, V. N., Zvonarev, A. N., Sazonova, O. I., Delegan, Y. A., Kocharovskaya, Y. N., Bogun, A. G., & Suzina, N. E. (2024). Identification, Characterization, and Ultrastructure Analysis of the Phenol-Degrading Rhodococcus erythropolis 7Ba and Its Viable but Nonculturable Forms. Microorganisms, 12(12), 2662. https://doi.org/10.3390/microorganisms12122662






