Development of a TaqMan qPCR for the Simultaneous Detection of the TuMV and BBWV2 Viruses Responsible for the Viral Disease in Pseudostellaria heterophylla
Abstract
:1. Introduction
2. Material and Methods
2.1. Planting and Management of Experimental Materials
2.2. Construction of Clone and Infectious Plasmids of P. heterophylla BBWV2 Virus
2.3. Design of TaqMan qPCR Primers and Hydrolysis Probes for P. heterophylla TuMV and BBWV2 Viruses
2.4. Optimisation of the Singleplex and Duplex TaqMan qPCR Assays
2.5. Linear Equation of Concentration of Virus Plasmid with Cq Value
2.6. Sensitivity and Reproducibility Test of Singleplex and Duplex qPCR Assays
2.7. Detection of TuMV and BBWV2 in the Main Production Areas of P. heterophylla
2.8. Investigation of BBWV2 Virus Levels During Its Infection Against N. benthamiana and P. heterophylla
3. Results
3.1. Optimisation of TaqMan qPCR for the Detection of Singleplex and Duplex Viruses
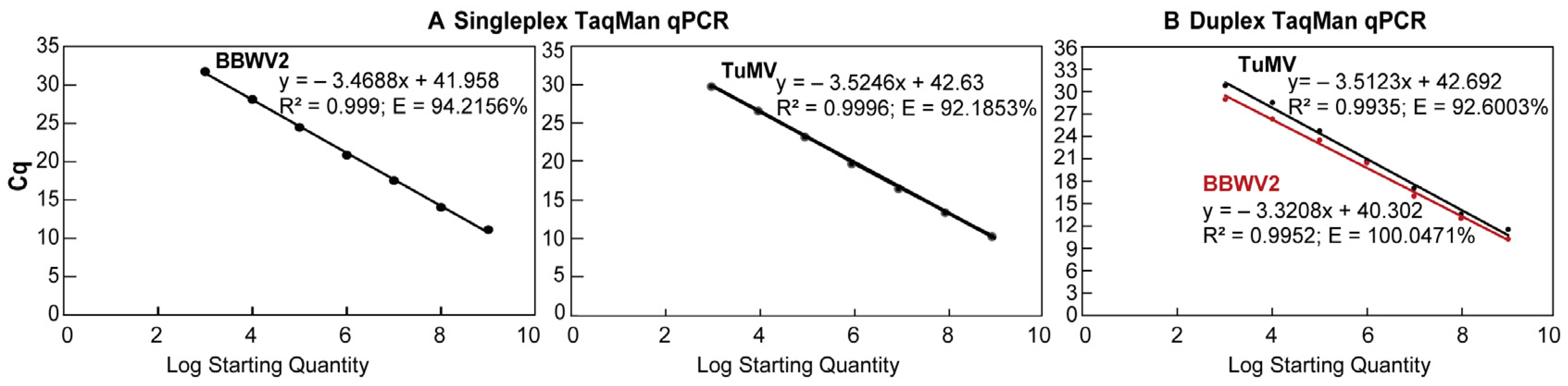
3.2. Establishment of Standard Curve Between Cq and Virus Concentration
3.3. Sensitivity and Reproducibility of Singleplex and Duplex TaqMan qPCR
3.4. The Use of Duplex TaqMan qPCR for the Investigation of the Levels of Viruses Present in P. heterophylla in Major Production Areas
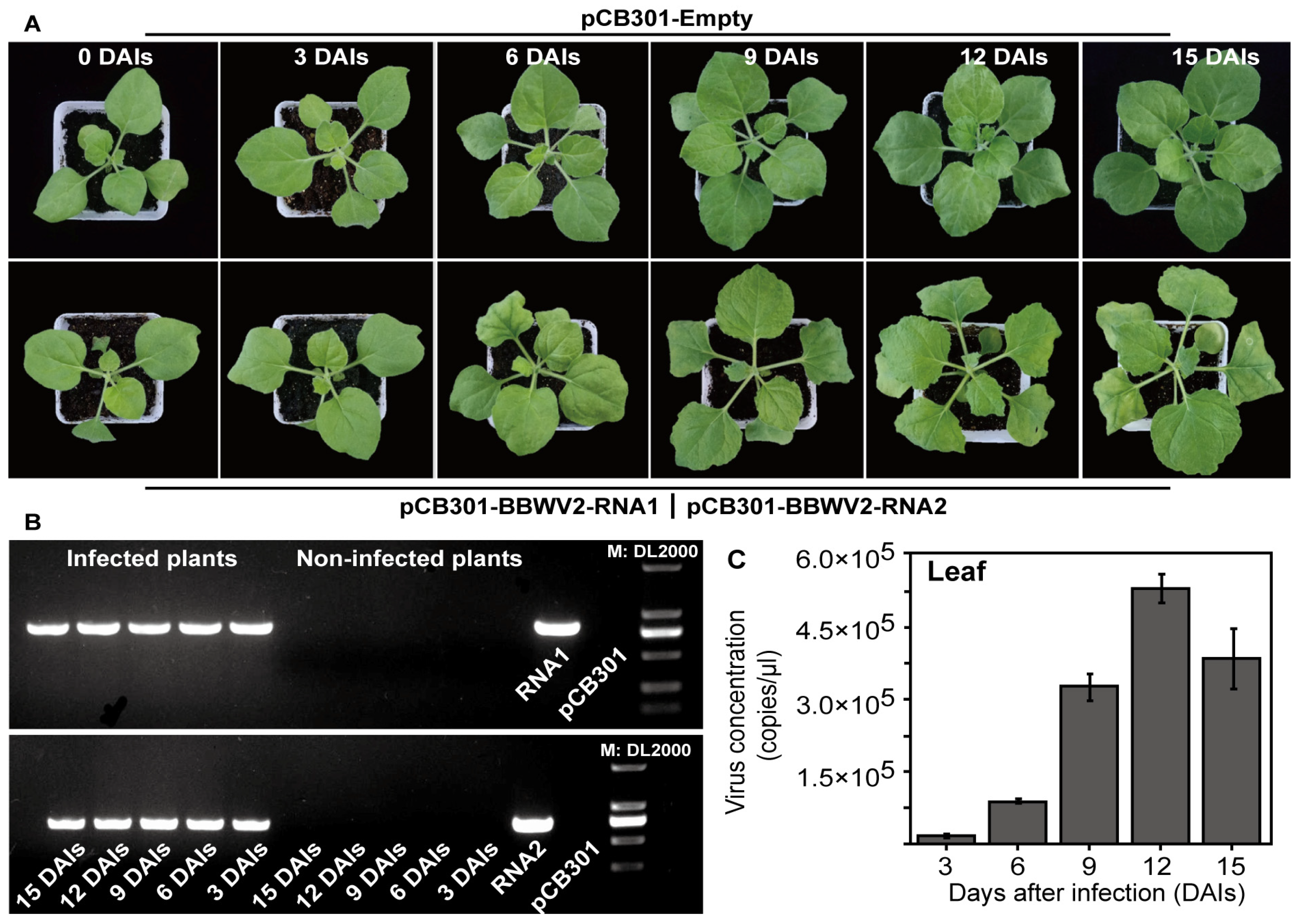
3.5. Monitoring Virus Levels During the Process of BBWV2 Infecting P. heterophylla Through Singleplex TaqMan qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhu, Y.; Zhou, X.H.; Qin, M.J. Advances in research on virus diseases of Pseudostellaria heterophylla. Chin. J. Wild Plant Resour. 2005, 24, 31–32. [Google Scholar] [CrossRef]
- Wu, C.X.; Ma, X.M.; Lin, Y.S. The virus elimination of Pseudostellaria heterophylla by shoot-tip culture and its effect on the yield increasing. J. Fujian Agric. For. Univ. (Nat. Sci.) 2006, 35, 129–133. [Google Scholar] [CrossRef]
- Wu, X.H.; Lou, F.; Li, W.; Li, T.Z.; Liu, H.C. Comparison of different propagation materials of Pseudostellaria heterophylla. J. Zhejiang Agric. Sci. 2021, 62, 530–531. [Google Scholar] [CrossRef]
- Chen, S.L.; Chen, D.H.; Zhang, L.R.; Liu, Q.Q. Discovery of viral pathogens in diseased Pseudostellaria heterophylla plants. Chin. J. Nat. 1982, 5, 238–239. [Google Scholar]
- Song, R.H.; Pu, Z.Q. Studies of Taizisheng (Pseudostellaria heterophylla) virus diseases. Acta Agric. Shanghai 1991, 2, 80–85. [Google Scholar]
- Gao, W.; Zhang, J.S.; Zhang, J.H.; Zhang, L.R. Detection and control of Taizishen mosaic virus. Virol. Sin. 1993, 8, 390–393. [Google Scholar]
- Huang, Y.Y.; Lin, C.F. Occurrence and control of Tizhi jinseng virus diseases in Mindong. J. Ningde Norm. Univ. (Nat. Sci.) 2004, 16, 65–68, 73. [Google Scholar] [CrossRef]
- Kuang, Y.B.; Ye, W.; Li, J.H.; Yu, J.L.; Ye, Z.Y.; Lai, Z.X. Cloning and sequence difference analysis of coat protein genes of turnip mosaic virus isolates from Pseudostellaria heteophylla. Chin. J. Trop. Crop. 2016, 7, 1974–1979. [Google Scholar] [CrossRef]
- Kuang, Y.B.; Chen, M.Z.; Lu, Y.R.; Chen, J.; Ye, Z.Y. Detection of Turnip mosaic virus and Broad bean wilt virus in Pseudostellaria heterophylla by Duplex RT-PCR. Acta Hortic. Sin. 2017, 44, 784–791. [Google Scholar] [CrossRef]
- Wang, R.; Li, Y.; Wei, R.F.; Chen, B.W.; Wang, T.Y.; Ding, W.L. Investigation on the occurrence of virus diseases in the main producing areas of Pseudostellaria heterophylla in China. Plant Prot. 2022, 48, 204–210. [Google Scholar] [CrossRef]
- Liang, S.; Chen, X.R.; Liu, Y.; Feng, W.Z.; Li, C.; Chen, X.S.; Li, Z. Rapid detection of Broad bean wilt virus 2 and Turnip mosaic virus in Pseudostellaria heterophylla by reverse transcription loop-mediated isothermal amplification assay. J. Phytopathol. 2022, 170, 535–545. [Google Scholar] [CrossRef]
- Li, Y.; Liu, S.; Guo, K.; Ding, W.L.; Wang, R. Virome of Pseudostellaria heterophylla: Identification and characterization of three novel carlaviruses and one novel amalgavirus associated with viral diseases of Pseudostellaria heterophylla. Front. Microbiol. 2022, 13, 955089. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Hu, X.P. High-throughput detection of banana bunchy top virus in banana plants and aphids using real-time TaqMan (®) PCR. J. Virol. Methods 2013, 193, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Alsaheli, Z.; Abdallah, A.; Incerti, O.; Shalaby, A.; Youssef, S.; Digiaro, M.; Elbeaino, T. Development of singleplex and multiplex real-time (Taqman®) RT-PCR assays for the detection of viruses associated with fig mosaic disease. J. Virol. Methods 2021, 293, 114145. [Google Scholar] [CrossRef]
- van Gent-Pelzer, M.P.E.; Dullemans, A.M.; Verbeek, M.; Bonants, P.J.M.; van der Lee, T.A.J. Development and evaluation of one-step RT-qPCR TaqMan multiplex panels applied to six viruses occurring in lily and tulip bulbs. J. Virol. Methods 2024, 329, 114987. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Li, M.J.; Yang, X.W.; Wang, J.M.; Sun, R.B.; Xu, Q.M.; Lin, W.M.; Yuan, F.Y.; Zhang, L.D.; Zhang, Z.Y. Rapid Identification and Cloning of Full-Length Genome of Medicinal Pseudostellaria heterophylla virus. CN Patent 202110548653.3, 20 May 2021. [Google Scholar]
- Yang, X.W.; Gu, L.; Liu, H.X.; Liu, C.S.; Yuan, J.D.; Qian, S.; Wang, J.M.; Yuan, F.Y.; Zhang, Z.Y.; Mu, J.; et al. Identification of a TuMV isolate (TuMV-ZR) from Pseudostellaria heterophylla and its development into a viral expression vector. Virus Res. 2023, 332, 199127. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.B.; Han, P.; Lutziger, I.; Wang, K.; Oliver, D.J. A mini binary vector series for plant transformation. Plant Mol. Biol. 1999, 40, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Malandraki, I.; Beris, D.; Isaioglou, I.; Olmos, A.; Varveri, C.; Vassilakos, N. Simultaneous detection of three pome fruit tree viruses by one-step multiplex quantitative RT-PCR. PLoS ONE 2017, 12, e0180877. [Google Scholar] [CrossRef] [PubMed]
- Pallás, V.; Sánchez-Navarro, J.A.; James, D. Recent advances on the multiplex molecular detection of plant viruses and viroids. Front. Microbiol. 2018, 9, 2087. [Google Scholar] [CrossRef] [PubMed]
- Santiago, G.A.; Vázquez, J.; Courtney, S.; Matías, K.Y.; Andersen, L.E.; Colón, C.; Butler, A.E.; Roulo, R.; Bowzard, J.; Villanueva, J.M.; et al. Performance of the Trioplex real-time RT-PCR assay for detection of Zika, dengue, and chikungunya viruses. Nat. Commun. 2018, 9, 1391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, F.; Gao, J.; Zhang, W.M.; Xu, X.G. Development of multiplex TaqMan qPCR for simultaneous detection and differentiation of eight common swine viral and bacterial pathogens. Braz. J. Microbiol. 2022, 53, 359–368. [Google Scholar] [CrossRef]
- Jiang, W.C.; Yue, S.W.; He, S.; Chen, C.; Liu, S.S.; Jiang, H.; Tong, H.L.; Liu, X.T.; Wang, J.N.; Zhang, F.; et al. New design of probe and central-homo primer pairs to improve TaqMan™ PCR accuracy for HBV detection. J. Virol. Methods 2018, 254, 25–30. [Google Scholar] [CrossRef]
- Ugwu, C.C.; Hair-Bejo, M.; Nurulfiza, M.I.; Omar, A.R.; Aini, I. TaqMan probe-based qPCR method for specific detection and quantification of fowl adenovirus 8b challenge from chickens inoculated with live attenuated or inactivated virus. Open Vet. J. 2023, 13, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.X.; Shen, T.R.; Feng, Z.X.; Diao, S.J.; Yan, Y.; Du, Z.K.; Jin, Y.L.; Gu, J.Y.; Zhou, J.Y.; Liao, M.; et al. Development of a highly sensitive TaqMan method based on multi-probe strategy: Its application in ASFV detection. Biol. Methods Protoc. 2024, 9, bpae011. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, S.; Prasad, B.V.L.S.; Selvarajan, R. RNA dependent RNA Polymerases: Insights from structure, function and evolution. Viruses 2018, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.Z.; Maren, N.A.; Kosentka, P.Z.; Liao, Y.Y.; Lu, H.Y.; Duduit, J.R.; Huang, D.B.; Ashrafi, H.; Zhao, T.J.; Huerta, A.I.; et al. An optimized protocol for stepwise optimization of real-time RT-PCR analysis. Hortic. Res. 2021, 8, 179. [Google Scholar] [CrossRef]
- Gangisetty, O.; Reddy, D.S. The optimization of TaqMan real-time RT-PCR assay for transcriptional profiling of GABA-A receptor subunit plasticity. J. Neurosci. Methods 2009, 181, 58–66. [Google Scholar] [CrossRef]
- Pan, Z.Z.; Lu, J.X.; Wang, N.N.; He, W.T.; Zhang, L.T.; Zhao, W.; Su, S. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 2020, 11, 707–718. [Google Scholar] [CrossRef]
- Abbasi, H.; Nikoo, H.R.; Fotouhi, F.; Khosravi, A. Development of a robust TaqMan probe-based one-step multiplex RT-qPCR for simultaneous detection of SARS-CoV-2 and Influenza A/B viruses. BMC Microbiol. 2023, 23, 335. [Google Scholar] [CrossRef]
- Pollari, M.; De, S.; Wang, A.M.; Mäkinen, K. The potyviral silencing suppressor HCPro recruits and employs host ARGONAUTE1 in pro-viral functions. PLoS Pathog. 2020, 16, e1008965. [Google Scholar] [CrossRef]
- Cheng, X.F.; Wang, A.M. The Potyvirus silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J. Virol. 2016, 91, e01478-16. [Google Scholar] [CrossRef] [PubMed]
- Kasschau, K.D.; Xie, Z.X.; Allen, E.; Llave, C.; Chapman, E.J.; Krizan, K.A.; Carrington, J.C. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 2003, 4, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Ogero, K.O.; Kreuze, J.F.; McEwan, M.A.; Luambano, N.D.; Bachwenkizi, H.; Garrett, K.A.; Andersen, K.F.; Thomas-Sharma, S.; van der Vlugt, R.A.A. Efficiency of insect-proof net tunnels in reducing virus-related seed degeneration in sweet potato. Plant Pathol. 2019, 68, 1472–1480. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.; Kwon, S.J.; Kim, M.H.; Choe, S.; Kwak, H.R.; Kim, M.K.; Jung, C.; Seo, J.K. A plant virus-based vector system for gene function studies in Pepper. Plant Physiol. 2019, 181, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Sivalingam, P.N.; Dokka, N.; Mahajan, M.M.; Sahu, B.; Marathe, A.; Kaushal, P.; Ghosh, P.K. Achieving maximum efficiency of Mungbean yellow mosaic India virus infection in mungbean by agroinoculation. 3 Biotech 2022, 12, 29. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.-J.; Wang, X.; Pang, H.-L.; Jia, L.-Y.; Feng, H.Q. Effects of Agrobacterium tumefaciens strain and its infection time and concentration on transient expression of foreign genes based on expression vector of bean yellow dwarf virus. Plant Sci. J. 2021, 39, 297–305. [Google Scholar] [CrossRef]
- Kumari, N.; Aski, M.S.; Mishra, G.P.; Roy, A.; Dikshit, H.K.; Saxena, S.; Kohli, M.; Mandal, B.; Sinha, S.K.; Mishra, D.C.; et al. Development of infectious clones of mungbean yellow mosaic India virus (MYMIV, Begomovirus vignaradiataindiaense) infecting mungbean [Vigna radiata (L.) R. Wilczek] and evaluation of a RIL population for MYMIV resistance. PLoS ONE 2024, 19, e0310003. [Google Scholar] [CrossRef] [PubMed]




| Annealing Temperature (°C) | Standard Plasmid | Cq (Mean ± SD) | Standard Plasmid | Cq (Mean ± SD) |
|---|---|---|---|---|
| 56 | BBWV2 | 10.97 ± 0.225 | TuMV | 11.81 ± 0.076 |
| 58 | BBWV2 | 10.54 ± 0.240 | TuMV | 11.14 ± 0.131 |
| 60 | BBWV2 | 11.60 ± 0.128 | TuMV | 11.86 ± 0.071 |
| 62 | BBWV2 | 11.07 ± 0.273 | TuMV | 10.92 ± 0.375 |
| 64 | BBWV2 | 10.72 ± 0.267 | TuMV | 10.78 ± 0.555 |
| Viruses | Probe Concentration (μM) | Primer Concentration (μM) | |||
|---|---|---|---|---|---|
| 0.1 | 0.2 | 0.3 | 0.4 | ||
| BBWV2 | 0.05 | 9.07 ± 0.044 | 10.14 ± 0.067 | 10.34 ± 0.346 | 10.02 ± 0.072 |
| BBWV2 | 0.10 | 8.71 ± 0.150 | 9.99 ± 0.325 | 10.15 ± 0.139 | 9.53 ± 0.197 |
| BBWV2 | 0.15 | 8.64 ± 0.137 | 9.84 ± 0.014 | 9.87 ± 0.088 | 9.59 ± 0.035 |
| BBWV2 | 0.20 | 8.37 ± 0.027 | 10.01 ± 0.04 | 10.01 ± 0.031 | 11.00 ± 0.107 |
| BBWV2 | 0.25 | 8.12 ± 0.145 | 9.92 ± 0.051 | 9.76 ± 0.108 | 9.48 ± 0.061 |
| TuMV | 0.05 | 9.64 ± 0.296 | 11.19 ± 0.045 | 11.47 ± 0.094 | 11.33 ± 0.037 |
| TuMV | 0.10 | 10.25 ± 0.038 | 11.24 ± 0.097 | 11.02 ± 0.048 | 11.04 ± 0.014 |
| TuMV | 0.15 | 9.56 ± 0.436 | 10.95 ± 0.134 | 10.71 ± 0.473 | 11.00 ± 0.133 |
| TuMV | 0.20 | 9.63 ± 0.142 | 11.19 ± 0.086 | 11.12 ± 0.096 | 10.67 ± 0.035 |
| TuMV | 0.25 | 10.21 ± 0.346 | 11.07 ± 0.053 | 11.18 ± 0.016 | 10.94 ± 0.083 |
| Plasmid Standards | Concentration (Copies/μL) | Intra-Assay | Inter-Assay | ||||
|---|---|---|---|---|---|---|---|
| Average Cq | SD | CV (%) | Average Cq | SD | CV (%) | ||
| Singleplex qPCR | |||||||
| BBWV2 | 2.38 × 106 | 21.57 | 0.09 | 0.40 | 20.85 | 0.06 | 0.26 |
| BBWV2 | 2.38 × 105 | 24.46 | 0.06 | 0.25 | 24.49 | 0.07 | 0.29 |
| BBWV2 | 2.38 × 104 | 27.42 | 0.11 | 0.40 | 28.16 | 0.28 | 1.00 |
| TuMV | 1.12 × 106 | 20.8 | 0.22 | 1.04 | 21.28 | 0.09 | 0.44 |
| TuMV | 1.12 × 105 | 25.05 | 0.14 | 0.54 | 25.04 | 0.09 | 0.38 |
| TuMV | 1.12 × 104 | 29.03 | 0.22 | 0.77 | 28.87 | 0.31 | 1.09 |
| Duplex qPCR | |||||||
| BBWV2 | 2.38 × 106 | 21.01 | 0.09 | 0.41 | 20.98 | 0.1 | 0.48 |
| BBWV2 | 2.38 × 105 | 24.16 | 0.18 | 0.73 | 24.1 | 0.12 | 0.52 |
| BBWV2 | 2.38 × 104 | 27.17 | 0.28 | 1.02 | 27.11 | 0.17 | 0.62 |
| TuMV | 1.12 × 106 | 23.89 | 0.1 | 0.42 | 23.83 | 0.04 | 0.16 |
| TuMV | 1.12 × 105 | 25.57 | 0.22 | 0.86 | 25.46 | 0.18 | 0.71 |
| TuMV | 1.12 × 104 | 29.62 | 0.21 | 0.70 | 29.56 | 0.09 | 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, L.; Liu, C.; Yao, S.; Wu, J.; Wang, L.; Mu, J.; Wang, Y.; Wang, J.; Zhang, Z.; Li, M. Development of a TaqMan qPCR for the Simultaneous Detection of the TuMV and BBWV2 Viruses Responsible for the Viral Disease in Pseudostellaria heterophylla. Microorganisms 2024, 12, 2663. https://doi.org/10.3390/microorganisms12122663
Gu L, Liu C, Yao S, Wu J, Wang L, Mu J, Wang Y, Wang J, Zhang Z, Li M. Development of a TaqMan qPCR for the Simultaneous Detection of the TuMV and BBWV2 Viruses Responsible for the Viral Disease in Pseudostellaria heterophylla. Microorganisms. 2024; 12(12):2663. https://doi.org/10.3390/microorganisms12122663
Chicago/Turabian StyleGu, Li, Chensi Liu, Shuting Yao, Jiaxin Wu, Lianghong Wang, Jing Mu, Yankun Wang, Jianming Wang, Zhongyi Zhang, and Mingjie Li. 2024. "Development of a TaqMan qPCR for the Simultaneous Detection of the TuMV and BBWV2 Viruses Responsible for the Viral Disease in Pseudostellaria heterophylla" Microorganisms 12, no. 12: 2663. https://doi.org/10.3390/microorganisms12122663
APA StyleGu, L., Liu, C., Yao, S., Wu, J., Wang, L., Mu, J., Wang, Y., Wang, J., Zhang, Z., & Li, M. (2024). Development of a TaqMan qPCR for the Simultaneous Detection of the TuMV and BBWV2 Viruses Responsible for the Viral Disease in Pseudostellaria heterophylla. Microorganisms, 12(12), 2663. https://doi.org/10.3390/microorganisms12122663






