UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolation and Cultivation
2.2. Screening of Cd2+ Adsorption Strains
2.3. Determination of Physiological Characterization
2.4. Genome Sequencing, Assembly, and Annotation
2.5. Biological Adsorption Experiment
2.5.1. Preparation of Cell Suspension
2.5.2. Adsorption Efficiency Experiment
2.6. SEM-EDS Analysis
2.7. Untargeted Metabolomics Analysis of Sphingomonas sp. M1-B02
2.8. FTIR Spectrometer Analysis
3. Results
3.1. Screening and Determination of Optimal Adsorption Strain
3.2. Identification of a Novel Species Sphingomonas sp. M1-B02T
3.3. Effect of Cd2+ on Sphingomonas sp. M1-B02
3.4. The Genome Annotation of Sphingomonas sp. M1-B02
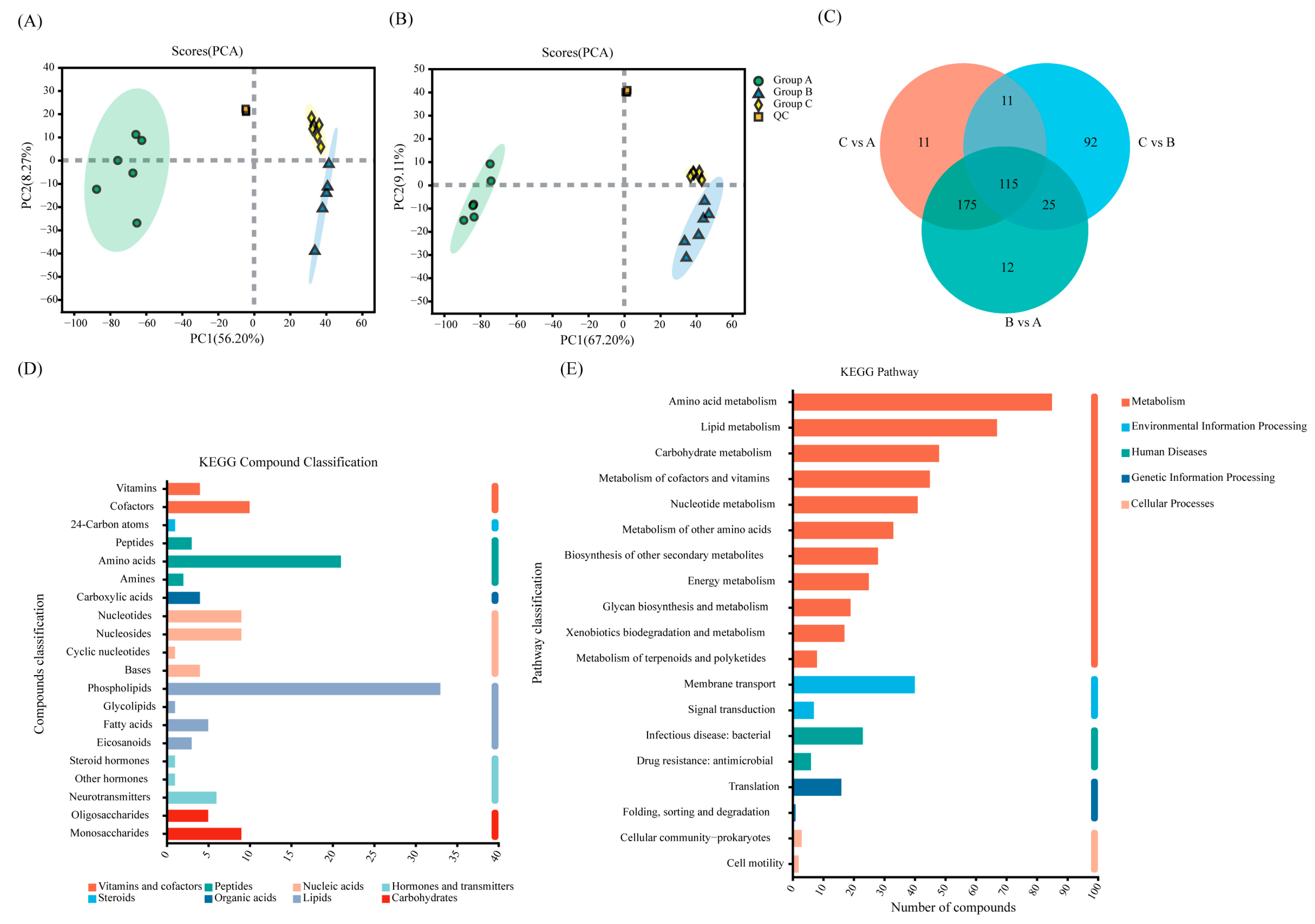
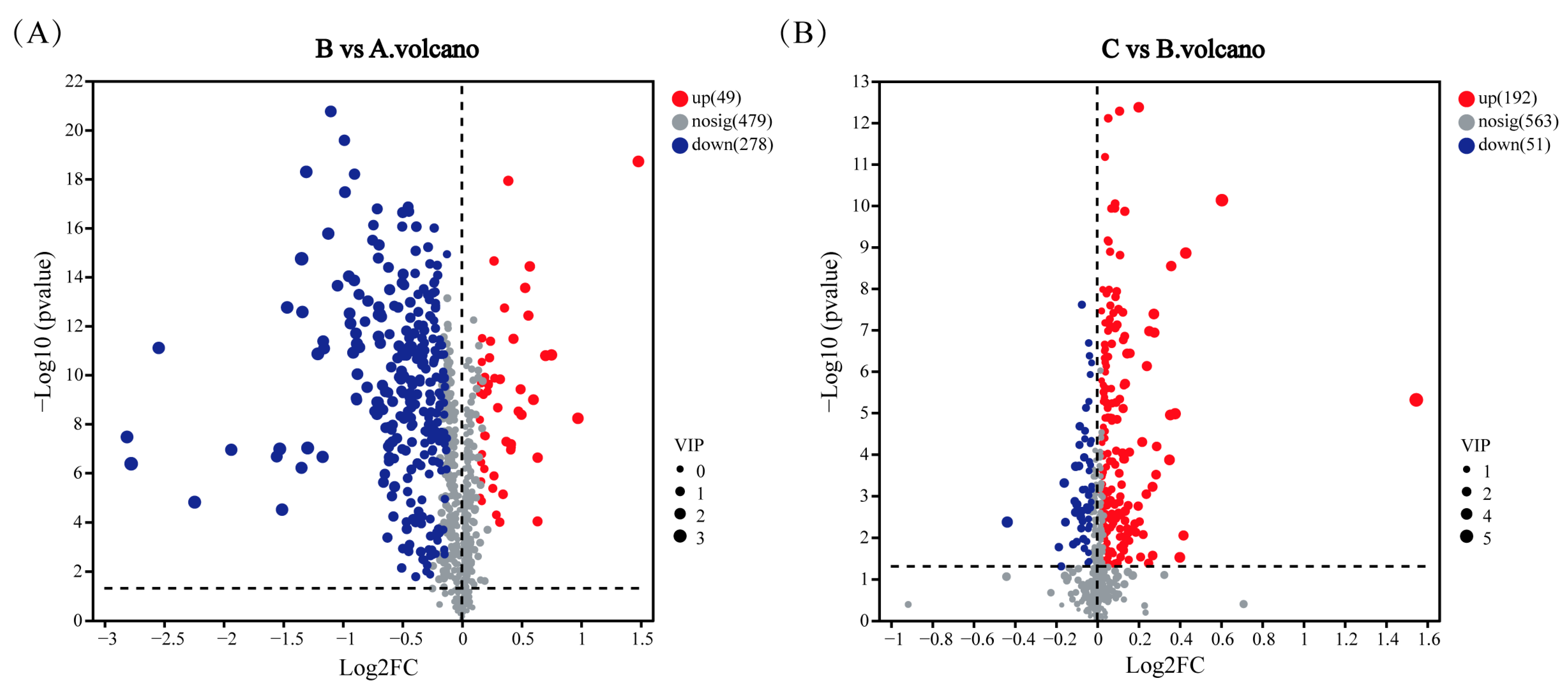
3.5. Analysis of Metabolites of Cd2+ Biosorption by Sphingomonas sp. M1-B02
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Choudhury, S.; Chatterjee, A. Microbial Application in Remediation of Heavy Metals: An Overview. Arch. Microbiol. 2022, 204, 268. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, R.; Binato, G.; Guidotti, M.; Morelli, S.; Pastorelli, A.A.; Sagratella, E.; Ciardullo, S.; Stacchini, P. Cadmium Bioaccumulation in Mediterranean Spider Crab (Maya squinado): Human Consumption and Health Implications for Exposure in Italian Population. Chemosphere 2014, 100, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Hasan, Z.; Zuberi, A. Heavy Metals in Three Commercially Valuable Cyprinids in the River Panjkora, District Lower Dir, Khyber Pakhtunkhwa, Pakistan. Toxicol. Environ. Chem. 2016, 98, 64–76. [Google Scholar] [CrossRef]
- Peng, W.; Li, X.; Song, J.; Jiang, W.; Liu, Y.; Fan, W. Bioremediation of Cadmium- and Zinc-Contaminated Soil Using Rhodobacter sphaeroides. Chemosphere 2018, 197, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Chen, J.; Shim, H.; Bai, Z. New Advances in Plant Growth-Promoting Rhizobacteria for Bioremediation. Environ. Int. 2007, 33, 406–413. [Google Scholar] [CrossRef]
- Li, Y.; Chen, W.; Yang, Y.; Wang, T.; Dai, Y. Quantifying Source-Specific Intake Risks of Wheat Cadmium by Associating Source Contributions of Soil Cadmium with Human Health Risk. Ecotoxicol. Environ. Saf. 2021, 228, 112982. [Google Scholar] [CrossRef] [PubMed]
- Rahman, Z.; Singh, V.P. The Relative Impact of Toxic Heavy Metals (THMs) (Arsenic (As), Cadmium (Cd), Chromium (Cr)(VI), Mercury (Hg), and Lead (Pb)) on the Total Environment: An Overview. Environ. Monit. Assess. 2019, 191, 419. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Shah, K. Evidences for Reduced Metal-Uptake and Membrane Injury upon Application of Nitric Oxide Donor in Cadmium Stressed Rice Seedlings. Plant Physiol. Biochem. 2014, 83, 180–184. [Google Scholar] [CrossRef]
- Yang, L.; Hu, W.; Chang, Z.; Liu, T.; Fang, D.; Shao, P.; Shi, H.; Luo, X. Electrochemical Recovery and High Value-Added Reutilization of Heavy Metal Ions from Wastewater: Recent Advances and Future Trends. Environ. Int. 2021, 152, 106512. [Google Scholar] [CrossRef] [PubMed]
- Rashid, R.; Shafiq, I.; Akhter, P.; Iqbal, M.J.; Hussain, M. A State-of-the-Art Review on Wastewater Treatment Techniques: The Effectiveness of Adsorption Method. Environ. Sci. Pollut. Res. 2021, 28, 9050–9066. [Google Scholar] [CrossRef] [PubMed]
- Nourbakhsh, M.; Sag̃, Y.; Özer, D.; Aksu, Z.; Kutsal, T.; Çag̃lar, A. A Comparative Study of Various Biosorbents for Removal of Chromium(VI) Ions from Industrial Waste Waters. Process Biochem. 1994, 29, 1–5. [Google Scholar] [CrossRef]
- Ahluwalia, S.S.; Goyal, D. Microbial and Plant Derived Biomass for Removal of Heavy Metals from Wastewater. Bioresour. Technol. 2007, 98, 2243–2257. [Google Scholar] [CrossRef] [PubMed]
- Smułek, W.; Zdarta, A.; Łuczak, M.; Krawczyk, P.; Jesionowski, T.; Kaczorek, E. Sapindus Saponins’ Impact on Hydrocarbon Biodegradation by Bacteria Strains after Short- and Long-Term Contact with Pollutant. Colloids Surf. B Biointerfaces 2016, 142, 207–213. [Google Scholar] [CrossRef]
- El Mahdi, A.M.; Aziz, H.A. Hydrocarbon Biodegradation Using Agro-Industrial Wastes as Co-Substrates. In Handbook of Research on Inventive Bioremediation Techniques; Bhakta, J.N., Ed.; IGI Global: Hershey, PA, USA, 2017; pp. 155–185. [Google Scholar]
- Stolz, A. Molecular Characteristics of Xenobiotic-Degrading Sphingomonads. Appl. Microbiol. Biotechnol. 2009, 81, 793–811. [Google Scholar] [CrossRef] [PubMed]
- Colquhoun, D.R.; Hartmann, E.M.; Halden, R.U. Proteomic Profiling of the Dioxin-Degrading Bacterium Sphingomonas wittichii RW1. BioMed Res. Int. 2012, 2012, 408690. [Google Scholar]
- Hesham, A.E.L.; Mawad, A.M.M.; Mostafa, Y.M.; Shoreit, A. Biodegradation Ability and Catabolic Genes of Petroleum-Degrading Sphingomonas koreensis Strain ASU-06 Isolated from Egyptian Oily Soil. BioMed Res. Int. 2014, 2014, 127674. [Google Scholar] [CrossRef] [PubMed]
- Kohler, J.; Hernández, J.A.; Caravaca, F.; Roldán, A. Plant-Growth-Promoting Rhizobacteria and Arbuscular Mycorrhizal Fungi Modify Alleviation Biochemical Mechanisms in Water-Stressed Plants. Funct. Plant Biol. 2008, 35, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.L.; Waqas, M.; Kang, S.M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.-Y.; et al. Bacterial Endophyte Sphingomonas sp. LK11 Produces Gibberellins and IAA and Promotes Tomato Plant Growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From Diversity and Genomics to Functional Role in Environmental Remediation and Plant Growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Pan, F.; Meng, Q.; Wang, Q.; Luo, S.; Chen, B.; Khan, K.Y.; Yang, X.; Feng, Y. Endophytic Bacterium Sphingomonas SaMR12 Promotes Cadmium Accumulation by Increasing Glutathione Biosynthesis in Sedum alfredii Hance. Chemosphere 2016, 154, 358–366. [Google Scholar] [CrossRef]
- Khan, A.L.; Ullah, I.; Hussain, J.; Kang, S.M.; Al-Harrasi, A.; Al-Rawahi, A.; Lee, I.J. Regulations of Essential Amino Acids and Proteomics of Bacterial Endophytes Sphingomonas sp. Lk11 during Cadmium Uptake. Environ. Toxicol. 2016, 31, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Shukla, M.; Chaturvedi, R.; Tamhane, D.; Vyas, P.; Archana, G.; Apte, S.; Bandekar, J.; Desai, A. Multiple-Stress Tolerance of Ionizing Radiation-Resistant Bacterial Isolates Obtained from Various Habitats: Correlation Between Stresses. Curr. Microbiol. 2007, 54, 142–148. [Google Scholar] [CrossRef]
- Reasoner, D.J.; Geldreich, E.E. A New Medium for the Enumeration and Subculture of Bacteria from Potable Water. Appl. Environ. Microbiol. 1985, 49, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Liao, M.; Zhang, Y.; Xu, N.; Xie, X.; Fan, Q. Cadmium Resistance, Microbial Biosorptive Performance and Mechanisms of a Novel Biocontrol Bacterium Paenibacillus sp. LYX-1. Environ. Sci. Pollut. Res. 2022, 29, 68692–68706. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, T.; Cui, X.; Xu, Y.; Hu, S.; Zhao, Y.; Zhang, W.; Liu, G.; Zhang, G. Sphingomonas radiodurans sp. Nov., a Novel Radiation-Resistant Bacterium Isolated from the North Slope of Mount Everest. Int. J. Syst. Evol. Microbiol. 2022, 72, 005312. [Google Scholar] [CrossRef] [PubMed]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An Empirically Improved Memory-Efficient Short-Read de Novo Assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving Bacterial Genome Assemblies from Short and Long Sequencing Reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference Sequence (RefSeq) Database at NCBI: Current Status, Taxonomic Expansion, and Functional Annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam Protein Families Database: Towards a More Sustainable Future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jia, Q.; Jing, W.; Dahms, H.-U.; Wang, L. Screening Strains for Microbial Biosorption Technology of Cadmium. Chemosphere 2020, 251, 126428. [Google Scholar] [CrossRef]
- Ojeda, J.J.; Dittrich, M. Fourier Transform Infrared Spectroscopy for Molecular Analysis of Microbial Cells. In Microbial Systems Biology: Methods and Protocols; Navid, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 187–211. [Google Scholar]
- Ayangbenro, A.S.; Babalola, O.O. A New Strategy for Heavy Metal Polluted Environments: A Review of Microbial Biosorbents. Int. J. Environ. Res. Public Health 2017, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, W.; Li, C.; Zhu, R.; Ge, F.; Zheng, Y.; Tang, Y. Self-Mediated pH Changes in Culture Medium Affecting Biosorption and Biomineralization of Cd2+ by Bacillus Cereus Cd01. J. Hazard. Mater. 2018, 358, 178–186. [Google Scholar] [CrossRef]
- Khadivinia, E.; Sharafi, H.; Hadi, F.; Zahiri, H.S.; Modiri, S.; Tohidi, A.; Mousavi, A.; Salmanian, A.H.; Noghabi, K.A. Cadmium Biosorption by a Glyphosate-Degrading Bacterium, a Novel Biosorbent Isolated from Pesticide-Contaminated Agricultural Soils. J. Ind. Eng. Chem. 2014, 20, 4304–4310. [Google Scholar] [CrossRef]
- Xu, S.; Xing, Y.; Liu, S.; Hao, X.; Chen, W.; Huang, Q. Characterization of Cd2+ Biosorption by Pseudomonas sp. Strain 375, a Novel Biosorbent Isolated from Soil Polluted with Heavy Metals in Southern China. Chemosphere 2020, 240, 124893. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Towards a Genome-Based Taxonomy for Prokaryotes. J. Bacteriol. 2005, 187, 6258–6264. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S rRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a Reference Resource for Gene and Protein Annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The Human Metabolome Database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Guan, N.; Li, J.; Shin, H.; Du, G.; Chen, J.; Liu, L. Microbial Response to Environmental Stresses: From Fundamental Mechanisms to Practical Applications. Appl. Microbiol. Biotechnol. 2017, 101, 3991–4008. [Google Scholar] [CrossRef] [PubMed]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus radiodurans: Functions Necessary to Survive Ionizing Radiation Are Also Necessary to Survive Prolonged Desiccation. J. Bacteriol. 1996, 178, 633–637. [Google Scholar] [CrossRef] [PubMed]
- Battista, J.R. AGAINST ALL ODDS: The Survival Strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 1997, 51, 203–224. [Google Scholar] [CrossRef]
- Potts, M. Desiccation Tolerance of Prokaryotes. Microbiol. Rev. 1994, 58, 755–805. [Google Scholar] [CrossRef]
- Lokshin, M.; Li, Y.; Gaiddon, C.; Prives, C. p53 and p73 Display Common and Distinct Requirements for Sequence Specific Binding to DNA. Nucleic Acids Res. 2007, 35, 340–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gorlatova, N.; Kelman, Z.; Herzberg, O. Structures of p63 DNA Binding Domain in Complexes with Half-Site and with Spacer-Containing Full Response Elements. Proc. Natl. Acad. Sci. USA 2011, 108, 6456–6461. [Google Scholar] [CrossRef] [PubMed]
- Ethayathulla, A.S.; Nguyen, H.T.; Viadiu, H. Crystal Structures of the DNA-Binding Domain Tetramer of the p53 Tumor Suppressor Family Member p73 Bound to Different Full-Site Response Elements. J. Biol. Chem. 2013, 288, 4744–4754. [Google Scholar] [CrossRef] [PubMed]
- Tichý, V.; Navrátilová, L.; Adámik, M.; Fojta, M.; Brázdová, M. Redox State of p63 and p73 Core Domains Regulates Sequence-Specific DNA Binding. Biochem. Biophys. Res. Commun. 2013, 433, 445–449. [Google Scholar] [CrossRef]
- Gobrecht, J.; McDyre, C.; Comotto, J.; Reynolds, M. Induction of Cytotoxic and Genotoxic Damage Following Exposure of V79 Cells to Cadmium Chloride. Mutat. Res. Toxicol. Environ. Mutagen. 2017, 816–817, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Wei, C.; Liao, C.; Wu, H. Damage to DNA of Effective Microorganisms by Heavy Metals: Impact on Wastewater Treatment. J. Environ. Sci. 2008, 20, 1514–1518. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free Radicals, Metals and Antioxidants in Oxidative Stress-Induced Cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Qu, W.; Ke, H.; Pi, J.; Broderick, D.; French, J.E.; Webber, M.M.; Waalkes, M.P. Acquisition of apoptotic resistance in cadmium-transformed human prostate epithelial cells: Bcl-2 overexpression blocks the activation of JNK signal transduction pathway. Environ. Health. Perspect. 2007, 115, 1094–1100. [Google Scholar] [CrossRef]
- Tapisso, J.T.; Marques, C.C.; Mathias, M.L.; Ramalhinho, M.G. Induction of micronuclei and sister chromatid exchange in bone-marrow cells and abnormalities in sperm of Algerian mice (Mus. spretus) exposed to cadmium, lead and zinc. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2009, 678, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Waisberg, M.; Joseph, P.; Hale, B.; Beyersmann, D. Molecular and cellular mechanisms of cadmium carcinogenesis. Toxicology 2003, 192, 95–117. [Google Scholar] [CrossRef]
- Fernandes, A.S.; Mazzei, J.L.; Evangelista, H.; Marques, M.R.C.; Ferraz, E.R.A.; Felzenszwalb, I. Protection against UV-induced oxidative stress and DNA damage by Amazon moss extracts. J. Photochem. Photobiol. B. 2018, 183, 331–341. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, M.; Bo, L.; Li, S.; Hu, L.; Zhao, X.; Sun, C. Metabolomic analysis of the toxic effect of chronic exposure of cadmium on rat urine. Environ. Sci. Pollut. Res. Int. 2018, 25, 3765–3774. [Google Scholar] [CrossRef]
- Ortega, J.; Lee, G.S.; Gu, L.; Yang, W.; Li, G.M. Mispair-bound human MutS-MutL complex triggers DNA incisions and activates mismatch repair. Cell Res. 2021, 31, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, J.; Hanawalt, P.C. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli. Mol. Gen. Genet. 1999, 262, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Courcelle, J.; Hanawalt, P.C. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl. Acad. Sci. USA 2001, 98, 8196–8202. [Google Scholar] [CrossRef] [PubMed]
- Welker, D.L.; Deering, R.A. Interactions between radiation-sensitive mutations in double-mutant haploids of Dictyostelium discoideum. Mol. Gen. Genet. 1979, 167, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Amundsen, S.K.; Fero, J.; Salama, N.R.; Smith, G.R. Dual nuclease and helicase activities of Helicobacter pylori AddAB are required for DNA repair, recombination, and mouse infectivity. J. Biol. Chem. 2009, 284, 16759–16766. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Wang, H.; Hu, H.; Pang, L.; Huang, B.; Zhu, R. Isolation, identification and heavy metals biosorption of a lead and cadmium-tolerant strain. Sheng Wu Gong Cheng Xue Bao 2020, 36, 1600–1609. [Google Scholar] [PubMed]
- Alonso, J.C.; Stiege, A.C.; Lüder, G. Genetic recombination in Bacillus subtilis 168: Effect of recN, recF, recH and addAB mutations on DNA repair and recombination. Mol. Gen. Genet. 1993, 239, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.; Silver, S. Bacterial resistance mechanisms for heavy metals of environmental concern. J. Ind. Microbiol. 1995, 14, 61–75. [Google Scholar] [CrossRef]
- Petris, M.J.; Mercer, J.F.; Culvenor, J.G.; Lockhart, P.; Gleeson, P.A.; Camakaris, J. Ligand-regulated transport of the Menkes copper P-type ATPase efflux pump from the Golgi apparatus to the plasma membrane: A novel mechanism of regulated trafficking. EMBO. J. 1996, 15, 6084–6095. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, A.I.; Loukidou, M.X.; Matis, K.A. Biosorption of toxic metals from aqueous solutions by bacteria strains isolated from metal-polluted soils. Process Biochem. 2004, 39, 909–916. [Google Scholar] [CrossRef]
- Haq, F.; Butt, M.; Hazrat, A.; Chaudhary, H.J. Biosorption of cadmium and chromium from water by endophytic Kocuria rhizophila: Equilibrium and kinetic studies. Desalination Water Treat. 2015, 57, 19946–19958. [Google Scholar] [CrossRef]
- Huang, F.; Dang, Z.; Guo, C.; Lu, G.; Gu, R.R.; Liu, H.; Zhang, H. Biosorption of Cd(II) by live and dead cells of Bacillus cereus RC-1 isolated from cadmium-contaminated soil. Colloids. Surf. B. Biointerfaces. 2013, 107, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Özdemir, S.; Kılınç, E.; Poli, A.; Nicolaus, B.; Güven, K. Biosorption of Cd, Cu, Ni, Mn and Zn from aqueous solutions by thermophilic bacteria, Geobacillus toebii sub. sp. decanicus and Geobacillus thermoleovorans sub. sp. stromboliensis: Equilibrium, kinetic and thermodynamic studies. Chem. Eng. J. 2009, 152, 195–206. [Google Scholar] [CrossRef]
- Sarı, A.; Tuzen, M. Kinetic and equilibrium studies of biosorption of Pb(II) and Cd(II) from aqueous solution by macrofungus (Amanita rubescens) biomass. J. Hazard. Mater. 2009, 164, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, Y.; Zhao, F. Kinetic and equilibrium studies of chromium (VI) biosorption by spent macroalgae Polysiphonia urceolata and Chondrus ocellatus. Biotechnol. Biotechnol. Equip. 2015, 29, 498–505. [Google Scholar] [CrossRef]
- Wu, S.C.; Peng, X.L.; Cheung, K.C.; Liu, S.L.; Wong, M.H. Adsorption kinetics of Pb and Cd by two plant growth promoting rhizobacteria. Bioresour. Technol. 2009, 100, 4559–4563. [Google Scholar] [CrossRef]
- Blokker, P.; Schouten, S.; Ende, H.; Leeuw, J.W.; Hatcher, P.G.; Damsté, J.S.S. Chemical structure of algaenans from the fresh water algae Tetraedron minimum, Scenedesmus communis and Pediastrum boryanum. Org. Geochem. 1998, 29, 1453–1468. [Google Scholar] [CrossRef]
- Sigee, D.C.; Dean, A.; Levado, E.; Tobin, M.J. Fourier-transform infrared spectroscopy of Pediastrum duplex: Characterization of a micro-population isolated from a eutrophic lake. J. Eur. J. Phycol. Tobin 2002, 37, 19–26. [Google Scholar] [CrossRef]
- Fischer, G.; Braun, S.; Thissen, R.; Dott, W. FT-IR spectroscopy as a tool for rapid identification and intra-species characterization of airborne filamentous fungi. J. Microbiol. Methods 2006, 64, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Fourest, E.; Canal, C.; Roux, J.C. Improvement of heavy metal biosorption by mycelial dead biomasses (Rhizopus arrhizus, Mucor miehei and Penicillium chrysogenum): pH control and cationic activation. FEMS Microbiol. Rev. 1994, 14, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Puranik, P.R.; Modak, J.M.; Paknikar, K.M. A Comparative Study of the Mass Transfer Kinetics of Metal Biosorption by Microbial Biomass. Hydrometallurgy 1999, 52, 189–197. [Google Scholar] [CrossRef]
- Forsberg, A.; Söderlund, S.; Frank, A.; Petersson, L.R.; Pedersén, M. Studies on metal content in the brown seaweed, Fucus vesiculosus, from the Archipelago of Stockholm. Environ. Pollut. 1988, 49, 245–263. [Google Scholar] [CrossRef] [PubMed]
- Dumas, P.; Miller, L. The use of synchrotron infrared microspectroscopy in biological and biomedical investigations. Vib. Spectrosc. 2003, 32, 3–21. [Google Scholar] [CrossRef]
- Wolkers, W.F.; Oliver, A.E.; Tablin, F.; Crowe, J.H. A Fourier-transform infrared spectroscopy study of sugar glasses. Carbohydr. Res. 2004, 339, 1077–1085. [Google Scholar] [CrossRef]
- Yee, N.; Benning, L.G.; Phoenix, V.R.; Ferris, F.G. Characterization of metal-cyanobacteria sorption reactions: A combined macroscopic and infrared spectroscopic investigation. Environ. Sci. Technol. 2004, 38, 775–782. [Google Scholar] [CrossRef]
- Fry, F.H.; Jensen, P.; Kepert, C.M.; Spiccia, L. Macrocyclic Copper(II) and Zinc(II) Complexes Incorporating Phosphate Esters. Inorg. Chem. 2003, 42, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, L.; Devi, P.; Shridhar, D.M.; Naik, C.G. Use of Fourier Transform Infrared (FTIR) spectroscopy to study cadmium-induced changes in Padina tetrastromatica (Hauck). Anal. Chem. Insights. 2008, 3, 135–143. [Google Scholar] [CrossRef]
- Jameson, E.; Rowe, O.F.; Hallberg, K.B.; Johnson, D.B. Sulfidogenesis and selective precipitation of metals at low pH mediated by Acidithiobacillus spp. and acidophilic sulfate-reducing bacteria. Hydrometallurgy 2010, 104, 488–493. [Google Scholar] [CrossRef]
- Mohammed Fayaz, A.; Girilal, M.; Rahman, M.; Venkatesan, R.; Kalaichelvan, P.T. Biosynthesis of silver and gold nanoparticles using thermophilic bacterium Geobacillus stearothermophilus. Process Biochem. 2011, 46, 1958–1962. [Google Scholar] [CrossRef]
- Andrejević, T.P.; Ašanin, D.P.; Pantović, B.V.; Stevanović, N.L.; Marković, V.R.; Djuran, M.I.; Glišić, B.Đ. Metal complexes with valuable biomolecules produced by Pseudomonas aeruginosa: A review of the coordination properties of pyocyanin, pyochelin and pyoverdines. Dalton Trans. 2023, 52, 4276–4289. [Google Scholar] [CrossRef] [PubMed]
- Bao, T.; Sun, T.; Sun, L. Effect of cadmium hyperaccumulation on antioxidative defense and proline accumulation of Solanum nigrum. Afr. J. Biotechnol. 2011, 10, 7198–7206. [Google Scholar]
- Nourimand, M.; Todd, C.D. Allantoin Increases Cadmium Tolerance in Arabidopsis via Activation of Antioxidant Mechanisms. Plant. Cell Physiol. 2016, 57, 2485–2496. [Google Scholar] [CrossRef]
- Carballo, R.; Castiñeiras, A.; Domínguez-Martín, A.; García-Santos, I.; Niclós-Gutiérrez, J. Solid state structures of cadmium complexes with relevance for biological systems. Met. Ions. Life. Sci. 2013, 11, 145–189. [Google Scholar] [PubMed]
- Wei, Y.; Shen, H.; Gao, C.; Du, Y.; Zhao, Y.; Wang, Y.; Zhou, S.; Li, J.; Zhao, B.; Wu, D. Electrochemical detection mechanism of estrogen effect induced by cadmium: The regulation of purine metabolism by the estrogen effect of cadmium. Chemosphere 2022, 311, 136970. [Google Scholar] [CrossRef]
- Macaskie, L.E.; Dean, A.C. Cadmium accumulation by a Citrobacter sp. J. Gen. Microbiol. 1984, 130, 53–62. [Google Scholar] [CrossRef]
- Michel, L.J.; Macaskie, L.E.; Dean, A.C. Cadmium accumulation by immobilized cells of a Citrobacter sp. using various phosphate donors. Biotechnol. Bioeng. 1986, 28, 1358–1365. [Google Scholar] [CrossRef]
- Macaskie, L.E.; Wates, J.M.; Dean, A.C. Cadmium accumulation by a Citrobacter sp. immobilized on gel and solid supports: Applicability to the treatment of liquid wastes containing heavy metal cations. Biotechnol. Bioeng. 1987, 30, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xia, W.; Liu, H.; Liu, F.; Li, H.; Chang, H.; Sun, J.; Liu, W.; Sun, X.; Jiang, Y.; et al. Urinary metabolomics reveals novel interactions between metal exposure and amino acid metabolic stress during pregnancy. Toxicol. Res. 2018, 7, 1164–1172. [Google Scholar] [CrossRef]
- Gratão, P.L.; Polle, A.; Lea, P.J.; Azevedo, R.A. Making the life of heavy metal-stressed plants a little easier. Funct. Plant Biol. 2005, 32, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Das, S. Oxidative stress-induced DNA damage and DNA repair mechanisms in mangrove bacteria exposed to climatic and heavy metal stressors. Environ. Pollut. 2023, 339, 122722. [Google Scholar] [CrossRef]
- Trevors, J.T.; Stratton, G.W.; Gadd, G.M. Cadmium transport, resistance, and toxicity in bacteria, algae, and fungi. Can. J. Microbiol. 1986, 32, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, H.; Song, Y.; Zhang, F.; Yang, Z.; Yang, Y.; Grohmann, T. Response of glutathione pools to cadmium stress and the strategy to translocate cadmium from roots to leaves (Daucus carota L.). Sci. Total. Environ. 2022, 823, 153575. [Google Scholar] [CrossRef]
- Guney, Y.; Turkcu, U.O.; Hicsonmez, A.; Andrieu, M.N.; Guney, H.Z.; Bilgihan, A.; Kurtman, C. Carnosine may reduce lung injury caused by radiation therapy. Med. Hypotheses 2006, 66, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Hipkiss, A.R. Carnosine and its possible roles in nutrition and health. Adv. Food. Nutr. Res. 2009, 57, 87–154. [Google Scholar] [PubMed]
- Ghodsi, R.; Kheirouri, S. Carnosine and advanced glycation end products: A systematic review. Amino. Acids. 2018, 50, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Zhong, Y.; Wu, Y.; Luo, Z.; Sun, Y.; Wang, G.; Kurihara, H.; Li, Y.; He, R. Carnosine attenuates cyclophosphamide-induced bone marrow suppression by reducing oxidative DNA damage. Redox. Biol. 2018, 14, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Vistoli, G.; Straniero, V.; Pedretti, A.; Fumagalli, L.; Bolchi, C.; Pallavicini, M.; Valoti, E.; Testa, B. Predicting the physicochemical profile of diastereoisomeric histidine-containing dipeptides by property space analysis. Chirality 2012, 24, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Torreggiani, A.; Bonora, S.; Fini, G. Raman and IR spectroscopic investigation of zinc(II)-carnosine complexes. Biopolymers 2000, 57, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Torreggiani, A.; Tamba, M.; Fini, G. Binding of copper(II) to carnosine: Raman and IR spectroscopic study. Biopolymers 2000, 57, 149–159. [Google Scholar] [CrossRef]






| Type | Dynamic Parameters | |||
|---|---|---|---|---|
| Group B | First-order | 34.63 | 1.83 | 0.97 |
| Second-order | 37.07 | 0.07 | 0.93 | |
| Group C | First-order | 40.18 | 1.47 | 0.98 |
| Second-order | 43.05 | 0.05 | 0.95 | |
| Wave Number (cm−1) | Group Type (v) | Peak Intensity |
|---|---|---|
| 3500–3300 | Multimolecular association vO-H | S |
| Carboxyl vO-H | VS | |
| Amide vN-H | Variable | |
| 1615–1510 | -NO2 | S |
| 1380 | -CH3 | - |
| 1275–1210 | Aromatic ether | S |
| 1000–650 | σC-H | Variable |
| 1400–500 | C-X |
| Bacteria | pH | Temperature (°C) | Concentration (mg/L) | Sorption Capacity (mg/g) | Reference |
|---|---|---|---|---|---|
| Bacillus laterosporus | 7 | 25 | 1000 | 159.5 | [70] |
| Kocuria rhizophila | 8 | 35 | 150 | 9.07 | [71] |
| Sphingomonas sp. LK11 | - | 28 | - | 44 | [22] |
| Paenibacillus sp. LYX-1 | 8 | 30 | 100 | 30.68 | [25] |
| Cedecea sp. SC19 | 7 | 37 | 500 | 126.19 | [65] |
| Bacillus cereus | 5 | 28 | - | 31.95 | [72] |
| Geobacillus toebii sub.sp. decanicus | - | 25 | 280 | 38.8 | [73] |
| Amanita rubescens | 5 | 20 | - | 27.3 | [74] |
| Sphingomonas sp. M1-B02 | 7 | 30 | 100 | 34.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Niu, H.; Li, S.; Yue, M.; Zhang, G. UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02. Microorganisms 2024, 12, 2620. https://doi.org/10.3390/microorganisms12122620
Li Y, Niu H, Li S, Yue M, Zhang G. UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02. Microorganisms. 2024; 12(12):2620. https://doi.org/10.3390/microorganisms12122620
Chicago/Turabian StyleLi, Yunshi, Haoyuan Niu, Shuang Li, Ming Yue, and Gaosen Zhang. 2024. "UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02" Microorganisms 12, no. 12: 2620. https://doi.org/10.3390/microorganisms12122620
APA StyleLi, Y., Niu, H., Li, S., Yue, M., & Zhang, G. (2024). UV-C Exposure Enhanced the Cd2+ Adsorption Capability of the Radiation-Resistant Strain Sphingomonas sp. M1-B02. Microorganisms, 12(12), 2620. https://doi.org/10.3390/microorganisms12122620






