Characterization and Transcriptional Regulation of the 2-Ketogluconate Utilization Operon in Pseudomonas plecoglossicida
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Primers
2.2. lacZ Reporter Gene Fusions
2.3. Comparison of 2KGA Utilization Abilities
2.4. 2KGA Fermentation
2.5. Primer Extension
2.6. Electrophoretic Mobility Shift Assay (EMSA)
2.7. DNase I Footprinting
2.8. Real-Time Fluorescence Quantitative PCR (RT-qPCR)
2.9. Statistical Analysis
3. Results and Discussion
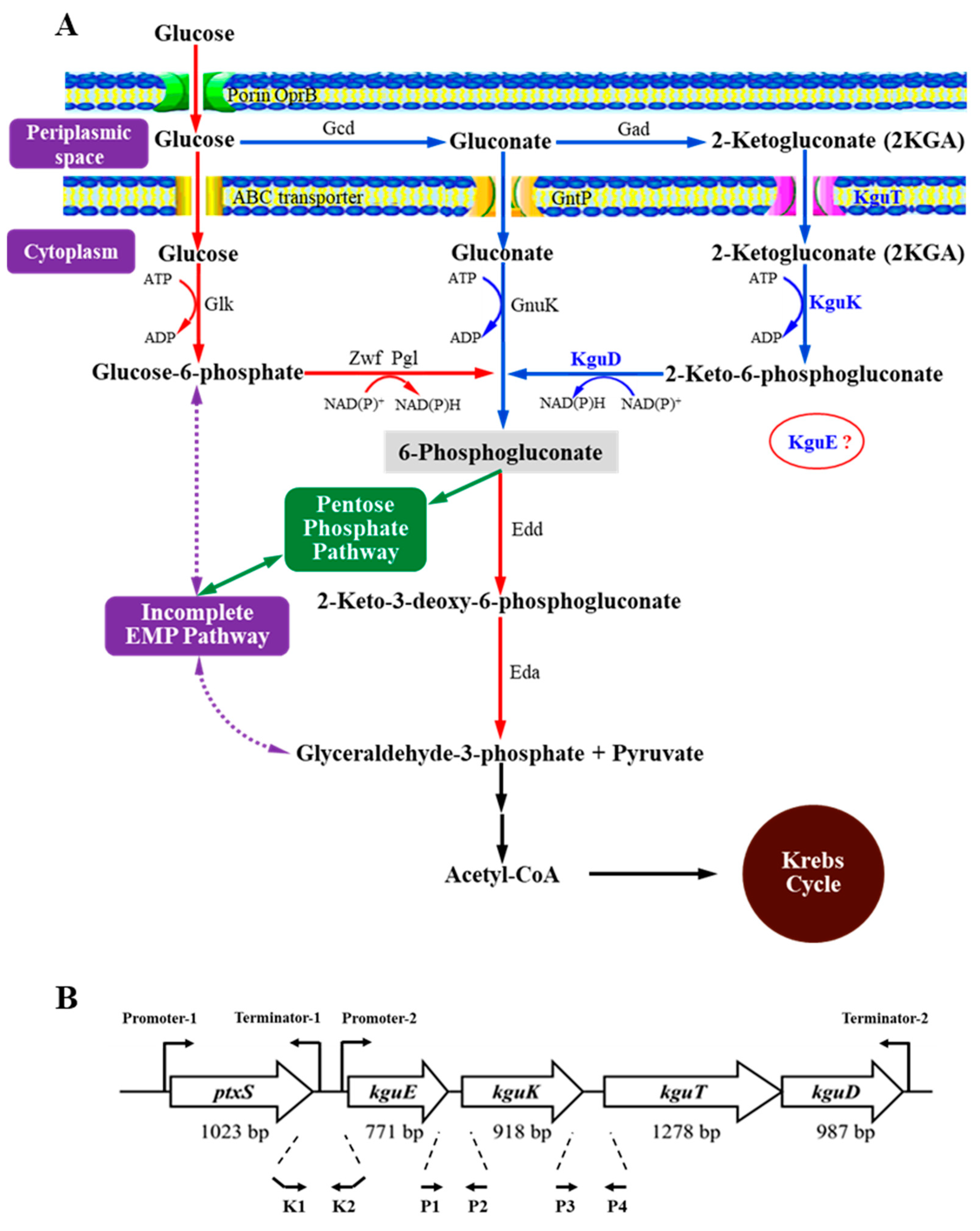
3.1. Identification of the Structural Genes of the kgu Operon in Pseudomonas plecoglossicida
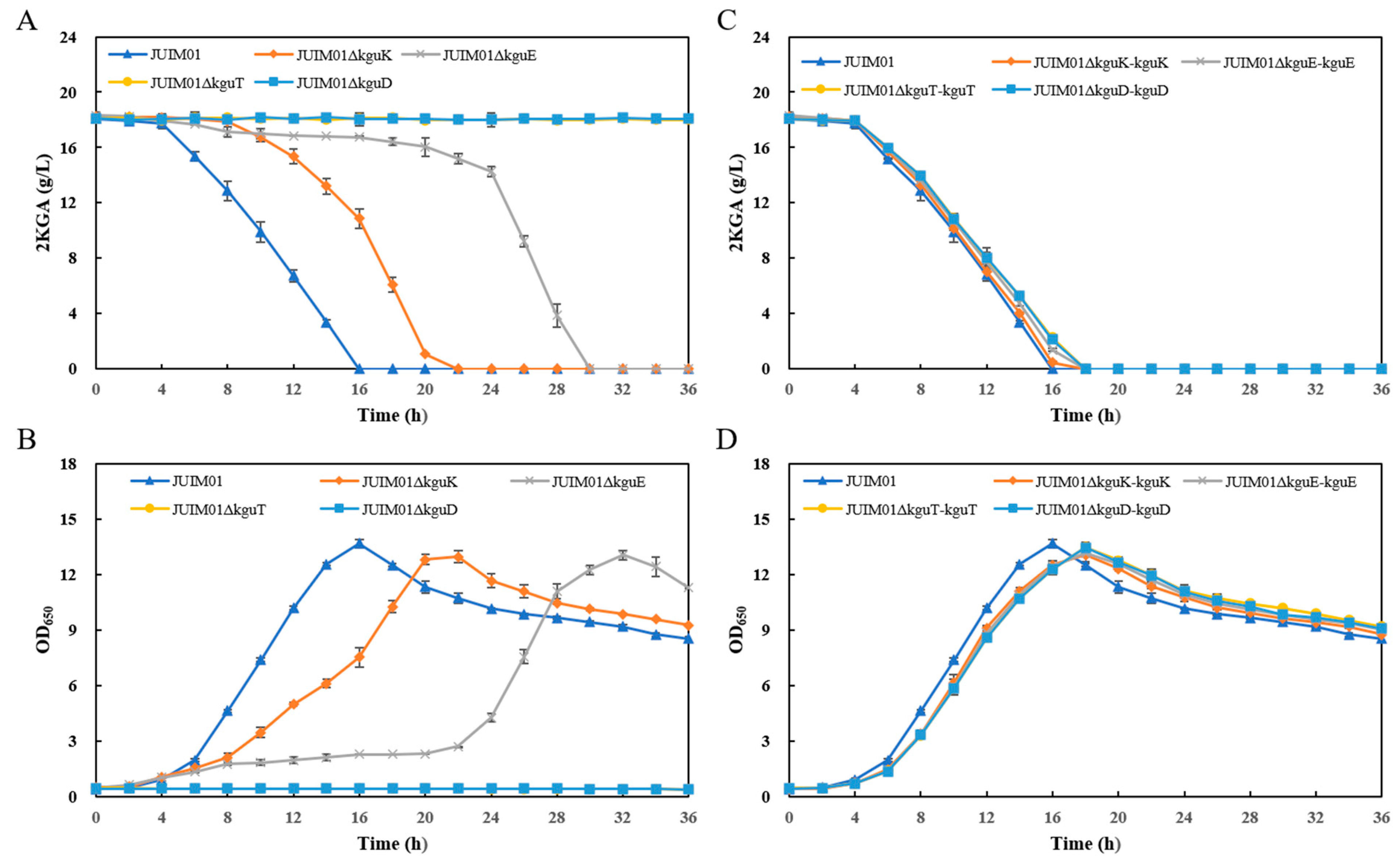
3.2. Effects of the Structural Genes of the kgu Operon on 2KGA Catabolism in P. plecoglossicida
3.3. 2KGA Fermentation Using the kguK/kguE/kguT/kguD-Knockout and Complementation Recombinants Derived from P. plecoglossicida JUIM01
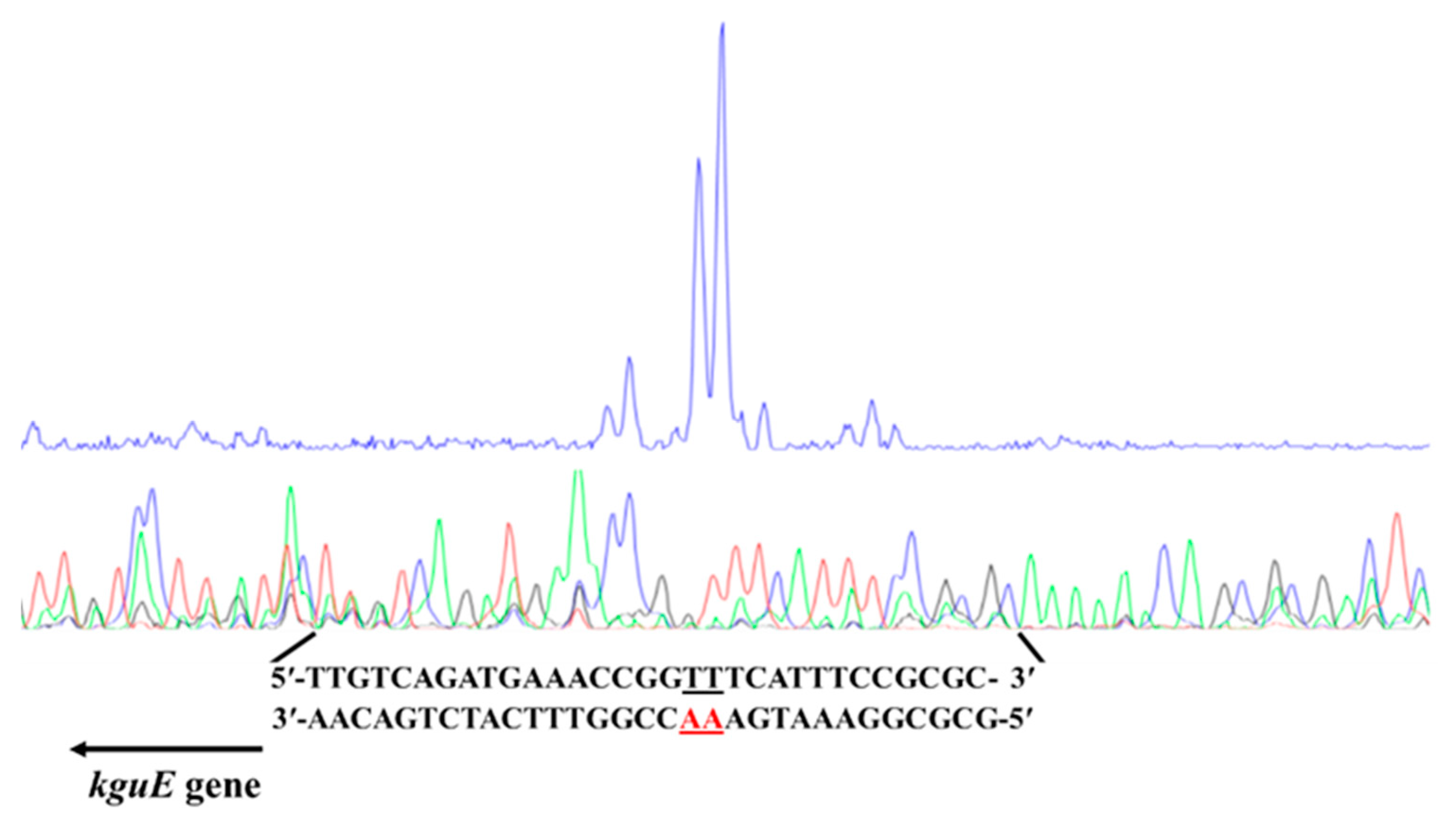
3.4. Transcriptional Start Site of the kgu Operon in P. plecoglossicida
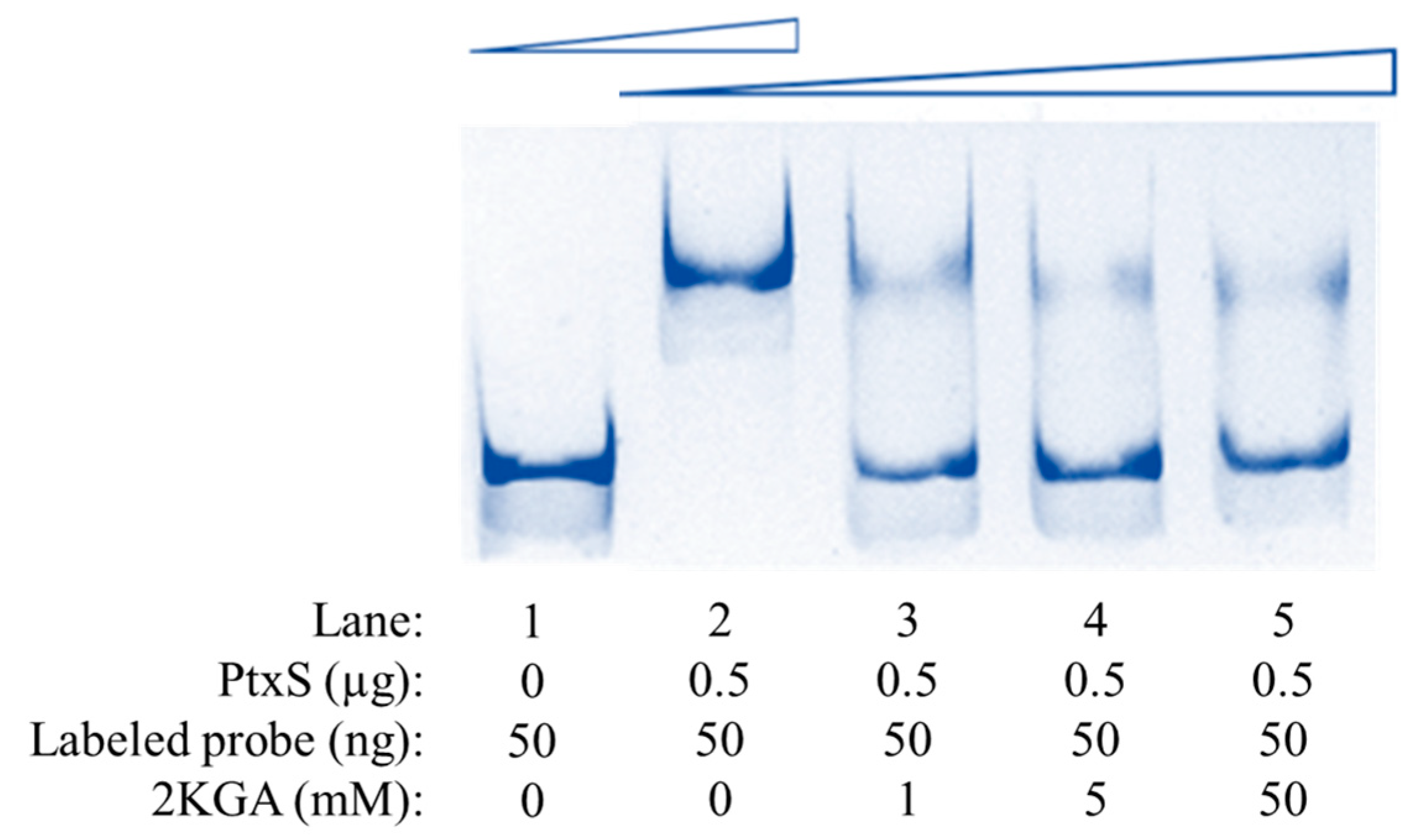
3.5. Specific Binding Between PtxS and the kgu Operon Promoter Region in P. plecoglossicida
3.6. Transcriptional Regulation of PtxS on the kgu Operon in P. plecoglossicida
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stottmeister, U.; Aurich, A.; Wilde, H.; Andersch, J.; Schmidt, S.; Sicker, D. White biotechnology for green chemistry: Fermentative 2-oxocarboxylic acids as novel building blocks for subsequent chemical syntheses. J. Ind. Microbiol. Biot. 2005, 32, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Pappenberger, G.; Hohmann, H.P. Industrial production of l-ascorbic acid (vitamin C) and d-isoascorbic acid. In Biotechnology of Food and Feed Additives; Advances in Biochemical Engineering/Biotechnology; Zorn, H., Czermak, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; Volume 143, pp. 143–188. [Google Scholar] [CrossRef]
- EFSA Panel on Food Additives and Nutrient Sources Added to Food (ANS). Scientific Opinion on the re-evaluation of erythorbic acid (E 315) and sodium erythorbate (E 316) as food additives. EFSA J. 2016, 14, 4360. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, X.J.; Zhou, Y.Z.; Peng, Z.; Cui, F.J.; Zhou, Q.; Man, Z.W.; Guo, J.; Sun, W.J. Can cadmium-contaminated rice be used to produce food additive sodium erythorbate? Food Chem. 2025, 462, 140923. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.J.; Wang, Q.H.; Luan, F.; Man, Z.W.; Cui, F.J.; Qi, X.H. The role of kguT gene in 2-ketogluconate-producing Pseudomonas plecoglossicida JUIM01. Appl. Biochem. Biotech. 2019, 187, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, D.M.; Sun, W.J.; Cui, F.J.; Gong, J.S.; Zhang, X.M.; Shi, J.S.; Xu, Z.H. Two-stage semi-continuous 2-keto-gluconic acid (2KGA) production by Pseudomonas plecoglossicida JUIM01 from rice starch hydrolyzate. Front. Bioeng. Biotech. 2020, 8, 120. [Google Scholar] [CrossRef]
- Wang, D.M.; Chen, X.; Guo, H.; Wang, Q.H.; Sun, L.; Sun, W.J. Exploring the response mechanism of Pseudomonas plecoglossicida to high-temperature stress by transcriptomic analyses for 2-keto gluconic acid production. Food Biosci. 2024, 62, 105063. [Google Scholar] [CrossRef]
- Sun, W.J.; Zhang, Q.N.; Li, L.L.; Qu, M.X.; Zan, X.Y.; Cui, F.J.; Zhou, Q.; Wang, D.M.; Sun, L. The functional characterization of the 6-phosphogluconate dehydratase operon in 2-ketogluconic acid industrial producing strain Pseudomonas plecoglossicida JUIM01. Foods 2024, 13, 3444. [Google Scholar] [CrossRef]
- del Castillo, T.; Ramos, J.L.; Rodríguez-Herva, J.J.; Fuhrer, T.; Sauer, U.; Duque, E. Convergent peripheral pathways catalyze initial glucose catabolism in Pseudomonas putida: Genomic and flux analysis. J. Bacteriol. 2007, 189, 5142–5152. [Google Scholar] [CrossRef]
- del Castillo, T.; Duque, E.; Ramos, J.L. A set of activators and repressors control peripheral glucose pathways in Pseudomonas putida to yield a common central intermediate. J. Bacteriol. 2008, 190, 2331–2339. [Google Scholar] [CrossRef]
- Daddaoua, A.; Krell, T.; Alfonso, C.; Morel, B.; Ramos, J.L. Compartmentalized glucose metabolism in Pseudomonas putida is controlled by the PtxS repressor. J. Bacteriol. 2010, 192, 4357–4366. [Google Scholar] [CrossRef]
- Daddaoua, A.; Fillet, S.; Fernández, M.; Udaondo, Z.; Krell, T.; Ramos, J.L. Genes for carbon metabolism and the ToxA virulence factor in Pseudomonas aeruginosa are regulated through molecular interactions of PtxR and PtxS. PLoS ONE 2012, 7, e39390. [Google Scholar] [CrossRef] [PubMed]
- Udaondo, Z.; Ramos, J.L.; Segura, A.; Krell, T.; Daddaoua, A. Regulation of carbohydrate degradation pathways in Pseudomonas involves a versatile set of transcriptional regulators. Microb. Biotechnol. 2018, 11, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.M.; Sun, L.; Sun, W.J.; Cui, F.J.; Gong, J.S.; Zhang, X.M.; Shi, J.S.; Xu, Z.H. Purification, characterization and gene identification of a membrane-bound glucose dehydrogenase from 2-keto-d-gluconic acid industrial producing strain Pseudomonas plecoglossicida JUIM01. Int. J. Biol. Macromol. 2018, 118, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.M.; Sun, L.; Sun, W.J.; Cui, F.J.; Gong, J.S.; Zhang, X.M.; Shi, J.S.; Xu, Z.H. A membrane-bound gluconate dehydrogenase from 2-keto-d-gluconic acid industrial producing strain Pseudomonas plecoglossicida JUIM01: Purification, characterization, and gene identification. Appl. Biochem. Biotech. 2019, 188, 897–913. [Google Scholar] [CrossRef] [PubMed]
- Swanson, B.L.; Hager, P.; Phibbs, P., Jr.; Ochsner, U.; Vasil, M.L.; Hamood, A.N. Characterization of the 2-ketogluconate utilization operon in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 2000, 37, 561–573. [Google Scholar] [CrossRef]
- Luan, F.; Wang, D.M.; Sun, W.J.; Cui, F.J.; Qi, X.H.; Guo, Q.; Zhang, X.F.; Li, Y.Z. Cloning and bioinformatics analysis of 2-ketogluconate utilization operon from Pseudomonas plecoglossicida. J. Chin. Inst. Food Sci. Technol. 2018, 18, 292–298. (In Chinese) [Google Scholar] [CrossRef]
- Colmer, J.A.; Hamood, A.N. Characterization of ptxS, a Pseudomonas aeruginosa gene which interferes with the effect of the exotoxin A positive regulatory gene, ptxR. Mol. Gen. Genet. MGG 1998, 258, 250–259. [Google Scholar] [CrossRef]
- Swanson, B.L.; Colmer, J.A.; Hamood, A.N. The Pseudomonas aeruginosa exotoxin A regulatory gene, ptxS: Evidence for negative autoregulation. J. Bacteriol. 1999, 181, 4890–4895. [Google Scholar] [CrossRef]
- Swanson, B.L.; Hamood, A.N. Autoregulation of the Pseudomonas aeruginosa protein PtxS occurs through a specific operator site within the ptxS upstream region. J. Bacteriol. 2000, 182, 4366–4371. [Google Scholar] [CrossRef]
- Daddaoua, A.; Krell, T.; Ramos, J.L. Transcriptional control by two interacting regulatory proteins: Identification of the PtxS binding site at PtxR. Nucleic Acids Res. 2013, 41, 10150–10156. [Google Scholar] [CrossRef]
- Sun, L.; Wang, D.M.; Sun, W.J.; Zhang, X.F.; Cui, F.J.; Su, C.; Zhang, X.M.; Xu, G.Q.; Shi, J.S.; Xu, Z.H. Characterization of a transcriptional regulator PtxS from Pseudomonas plecoglossicida for regulating 2-ketogluconic acid metabolism. Int. J. Biol. Macromol. 2021, 174, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, D.M.; Sun, W.J.; He, X.Y.; Cui, F.J.; Zhang, X.M.; Gong, J.S.; Shi, J.S.; Xu, Z.H. A 2-ketogluconate kinase KguK in Pseudomonas plecoglossicida JUIM01: Enzymatic characterization and its role in 2-keto-d-gluconic acid metabolism. Int. J. Biol. Macromol. 2020, 165 Pt B, 2640–2648. [Google Scholar] [CrossRef]
- Lu, J.S.; Huang, X.Q.; Li, K.; Li, S.N.; Zhang, M.Y.; Wang, Y.; Jiang, H.X.; Xu, Y.Q. LysR family transcriptional regulator PqsR as repressor of pyoluteorin biosynthesis and activator of phenazine-1-carboxylic acid biosynthesis in Pseudomonas sp. M18. J. Biotechnol. 2009, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fekete, R.A.; Miller, M.J.; Chattoraj, D.K. Fluorescently labeled oligonucleotide extension: A rapid and quantitative protocol for primer extension. Biotechniques 2003, 35, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef]
- de Bruijn, I.; de Kock, M.J.D.; de Waard, P.; van Beek, T.A.; Raaijmakers, J.M. Massetolide A biosynthesis in Pseudomonas fluorescens. J. Bacteriol. 2008, 190, 2777–2789. [Google Scholar] [CrossRef]
- Hwangbo, H.; Park, R.D.; Kim, Y.W.; Rim, Y.S.; Park, K.H.; Kim, T.H.; Suh, J.S.; Kim, K.Y. 2-Ketogluconic acid production and phosphate solubilization by Enterobacter intermedium. Curr. Microbiol. 2003, 47, 0087–0092. [Google Scholar] [CrossRef]
- Archana, G.; Buch, A.; Kumar, G.N. Pivotal role of organic acid secretion by rhizobacteria in plant growth promotion. In Microorganisms in Sustainable Agriculture and Biotechnology; Satyanarayana, T., Johri, B., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 35–53. [Google Scholar] [CrossRef]
- Kumar, C.; Yadav, K.; Archana, G.; Naresh Kumar, G. 2-Ketogluconic acid secretion by incorporation of Pseudomonas putida KT 2440 gluconate dehydrogenase (gad) operon in Enterobacter asburiae PSI3 improves mineral phosphate solubilization. Curr. Microbiol. 2013, 67, 388–394. [Google Scholar] [CrossRef]
- Wagh, J.; Chanchal, K.; Sonal, S.; Praveena, B.; Archana, G.; Kumar, G.N. Inoculation of genetically modified endophytic Herbaspirillum seropedicae Z67 endowed with gluconic and 2-ketogluconic acid secretion, confers beneficial effects on rice (Oriza sativa) plants. Plant Soil 2016, 409, 51–64. [Google Scholar] [CrossRef]
- Yu, K.; Liu, Y.; Tichelaar, R.; Savant, N.; Lagendijk, E.; van Kuijk, S.J.L.; Stringlis, I.A.; van Dijken, A.J.H.; Pieterse, C.M.J.; Bakker, P.A.H.M.; et al. Rhizosphere-associated Pseudomonas suppress local root immune responses by gluconic acid-mediated lowering of environmental pH. Curr. Biol. 2019, 29, 3913–3920. [Google Scholar] [CrossRef]
- Choi, W.J.; Lee, E.Y.; Choi, C.Y. Effect of dissolved oxygen concentration on the metabolism of glucose in Pseudomonas putida BM014. Biotechnol. Bioprocess Eng. 1998, 3, 109–111. [Google Scholar] [CrossRef]
- Latrach Tlemçani, L.; Corroler, D.; Barillier, D.; Mosrati, R. Physiological states and energetic adaptation during growth of Pseudomonas putida mt-2 on glucose. Arch. Microbiol. 2008, 190, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Nikel, P.I.; Chavarría, M.; Fuhrer, T.; Sauer, U.; de Lorenzo, V. Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 2015, 290, 25920–25932. [Google Scholar] [CrossRef] [PubMed]
- Kohlstedt, M.; Wittmann, C. GC-MS-based 13C metabolic flux analysis resolves the parallel and cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. Metab. Eng. 2019, 54, 35–53. [Google Scholar] [CrossRef]
- Volke, D.C.; Gurdo, N.; Milanesi, R.; Nikel, P.I. Time-resolved, deuterium-based fluxomics uncovers the hierarchy and dynamics of sugar processing by Pseudomonas putida. Metab. Eng. 2023, 79, 159–172. [Google Scholar] [CrossRef]



| Strains and Plasmids | Description | Sources |
|---|---|---|
| Strains | ||
| P. plecoglossicida JUIM01 | Industrial 2KGA-producing strain | Our lab [4,5,6,7,8] |
| JUIM01ΔkguE | Derived from JUIM01 with kguE knockout | This work |
| JUIM01ΔkguK | Derived from JUIM01 with kguK knockout | Our lab [23] |
| JUIM01ΔkguT | Derived from JUIM01 with kguT knockout | Our lab [5] |
| JUIM01ΔkguD | Derived from JUIM01 with kguD knockout | This work |
| JUIM01ΔptxS | Derived from JUIM01 with ptxS knockout | This work |
| JUIM01ΔkguE-kguE | Derived from JUIM01ΔkguE with kguE complemented | This work |
| JUIM01ΔkguK-kguK | Derived from JUIM01ΔkguK with kguK complemented | Our lab [23] |
| JUIM01ΔkguT-kguT | Derived from JUIM01ΔkguT with kguT complemented | Our lab [5] |
| JUIM01ΔkguD-kguD | Derived from JUIM01ΔkguD with kguD complemented | This work |
| JUIM01/pME6522 | Negative control for lacZ fusion analysis, TcR | This work |
| JUIM01/pME6522-kgu | Recombinant strain for lacZ fusion analysis, TcR | This work |
| JUIM01ΔptxS/pME6522-kgu | Recombinant strain for lacZ fusion analysis, TcR | This work |
| Escherichia coli BL21(DE3)/pET-28a-ptxS | PtxS heterologous-expression strain, KanR | Our lab [22] |
| Plasmids | ||
| pME6522 | E. coli-Pseudomonas shuttle vector for transcriptional lacZ fusions and promoter probing, TcR | [24] |
| pME6522-kgu | Derived from pME6522, containing a 132 bp ptxS-kguE intergenic sequence, TcR | This work |
| pK18mobSacB | Suicide vector for in-frame deletions, KanR | Our lab [23] |
| pK18mobSacB-ΔkguE | Derived from pK18mobSacB, containing an incomplete kguE gene, for in-frame deletion of kguE in JUIM01, KanR | This work |
| pK18mobSacB-ΔkguD | Derived from pK18mobSacB, containing an incomplete kguD gene, for in-frame deletion of kguD in JUIM01, KanR | This work |
| pK18mobSacB-ΔptxS | Derived from pK18mobSacB, containing an incomplete ptxS gene, for in-frame deletion of ptxS in JUIM01, KanR | This work |
| pBBR1MCS-2 | E. coli-Pseudomonas shuttle vector for expression, KanR | Our lab [23] |
| pBBRkguE | Derived from pBBR1MCS-2, for constitutive expression of kguE, KanR | This work |
| pBBRkguD | Derived from pBBR1MCS-2, for constitutive expression of kguD, KanR | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Yang, W.; Li, L.; Wang, D.; Zan, X.; Cui, F.; Qi, X.; Sun, L.; Sun, W. Characterization and Transcriptional Regulation of the 2-Ketogluconate Utilization Operon in Pseudomonas plecoglossicida. Microorganisms 2024, 12, 2530. https://doi.org/10.3390/microorganisms12122530
Sun L, Yang W, Li L, Wang D, Zan X, Cui F, Qi X, Sun L, Sun W. Characterization and Transcriptional Regulation of the 2-Ketogluconate Utilization Operon in Pseudomonas plecoglossicida. Microorganisms. 2024; 12(12):2530. https://doi.org/10.3390/microorganisms12122530
Chicago/Turabian StyleSun, Lei, Wenqi Yang, Lulu Li, Daming Wang, Xinyi Zan, Fengjie Cui, Xianghui Qi, Ling Sun, and Wenjing Sun. 2024. "Characterization and Transcriptional Regulation of the 2-Ketogluconate Utilization Operon in Pseudomonas plecoglossicida" Microorganisms 12, no. 12: 2530. https://doi.org/10.3390/microorganisms12122530
APA StyleSun, L., Yang, W., Li, L., Wang, D., Zan, X., Cui, F., Qi, X., Sun, L., & Sun, W. (2024). Characterization and Transcriptional Regulation of the 2-Ketogluconate Utilization Operon in Pseudomonas plecoglossicida. Microorganisms, 12(12), 2530. https://doi.org/10.3390/microorganisms12122530









