E2 Ubiquitin-Conjugating Enzymes Regulates Dengue Virus-2 Replication in Aedes albopictus
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Virus Enrichment
2.2. Experimental Infection of Mosquitoes with DENV-2
2.3. Plasmid Construction
2.4. Plasmid Transfection and Virus Infection
2.5. RNA Extraction and Reverse Transcription
2.6. Quantitative Real-Time PCR
2.7. Double-Stranded RNA Preparation
2.8. RNAi Interference of Ubc9 Gene Expression in C6/36 Cells
2.9. RNAi Interference of Ubc9 Gene Expression in the Midgut of Mosquitoes
2.10. Western Blot Analysis
2.10.1. Cellular Proteins
2.10.2. Tissue Proteins
2.11. Immunofluorescence Assay
2.12. Co-IP Assay
2.13. Statistical Analysis
2.14. Ethical Approval
3. Results
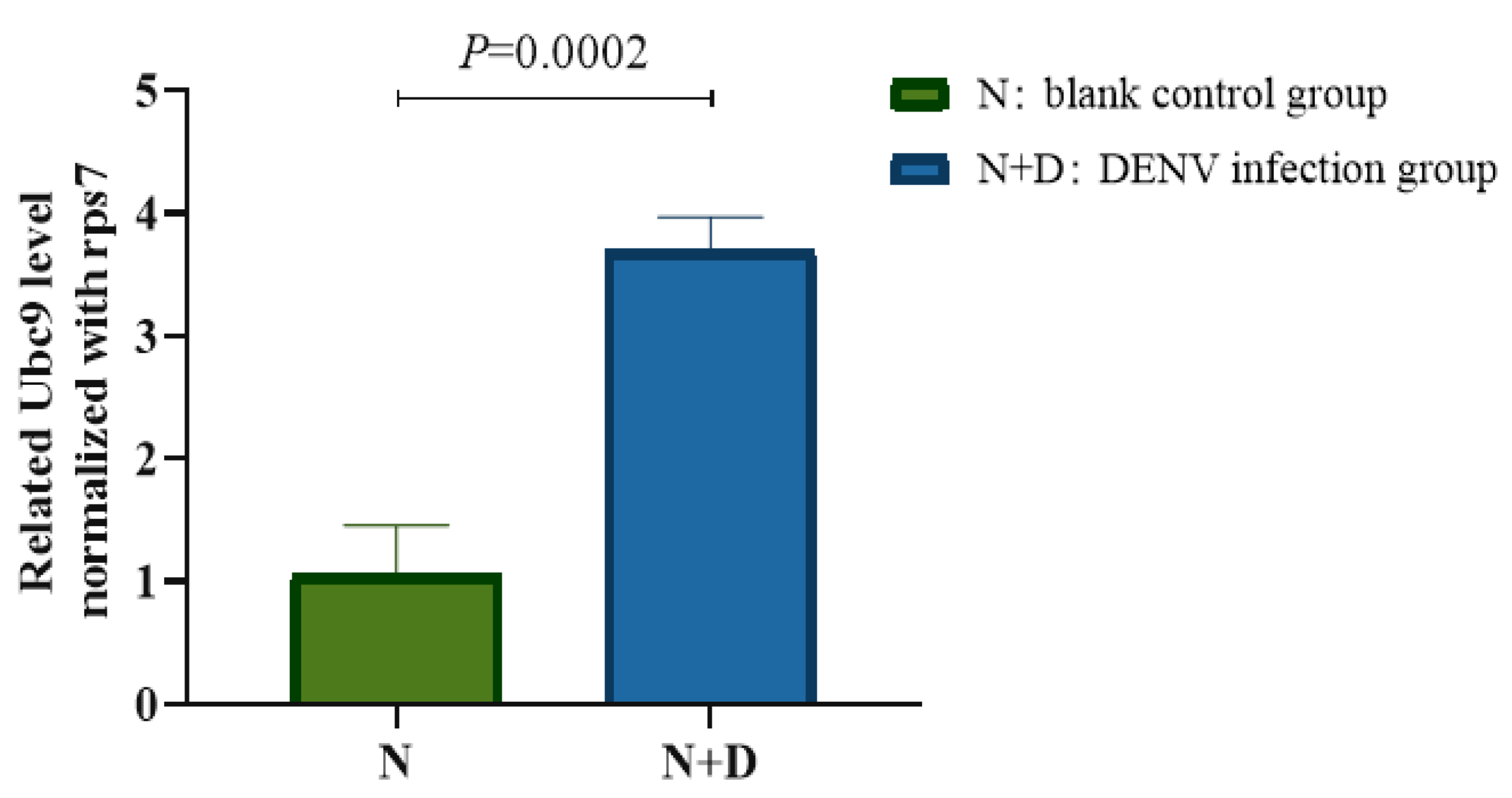
3.1. Upregulation of Ubc9 Expression in the Midgut of A. albopictus After DENV-2 Infection
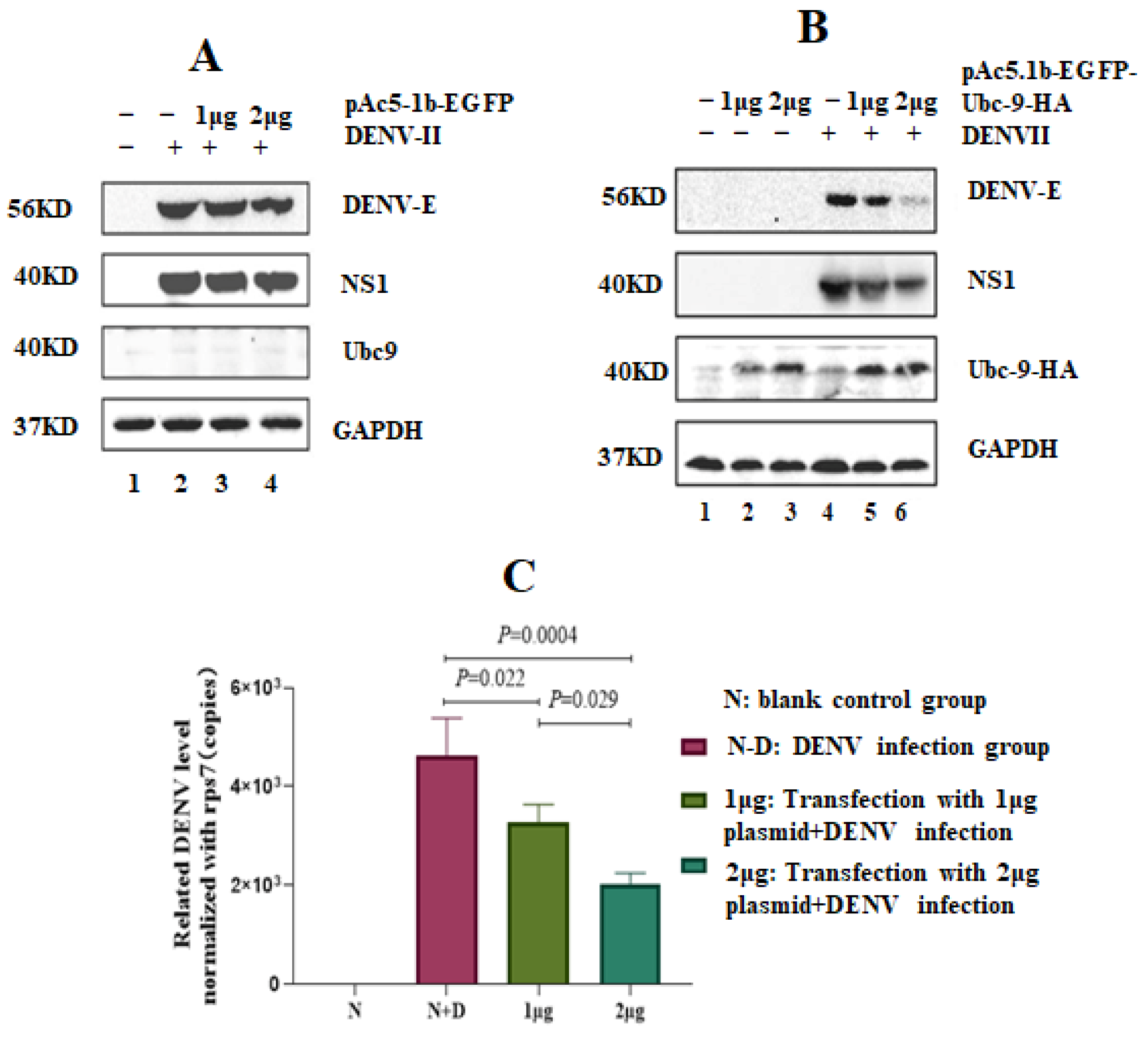
3.2. High Expression of Ubc9 Protein Inhibits the Expression of DENV-2 NS1 and E
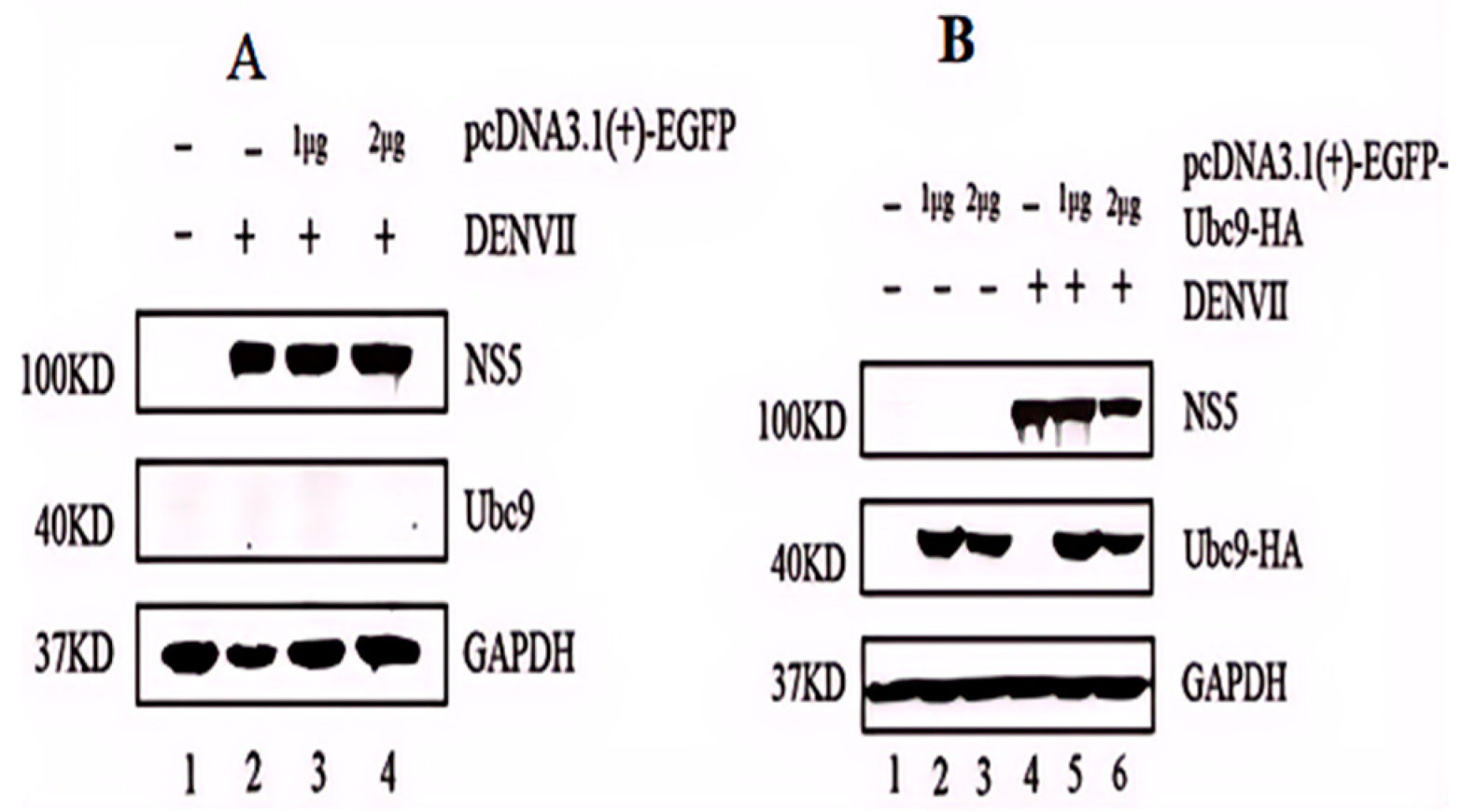
3.3. High Expression of Ubc9 Protein Inhibits the Expression of DENV-2 NS5
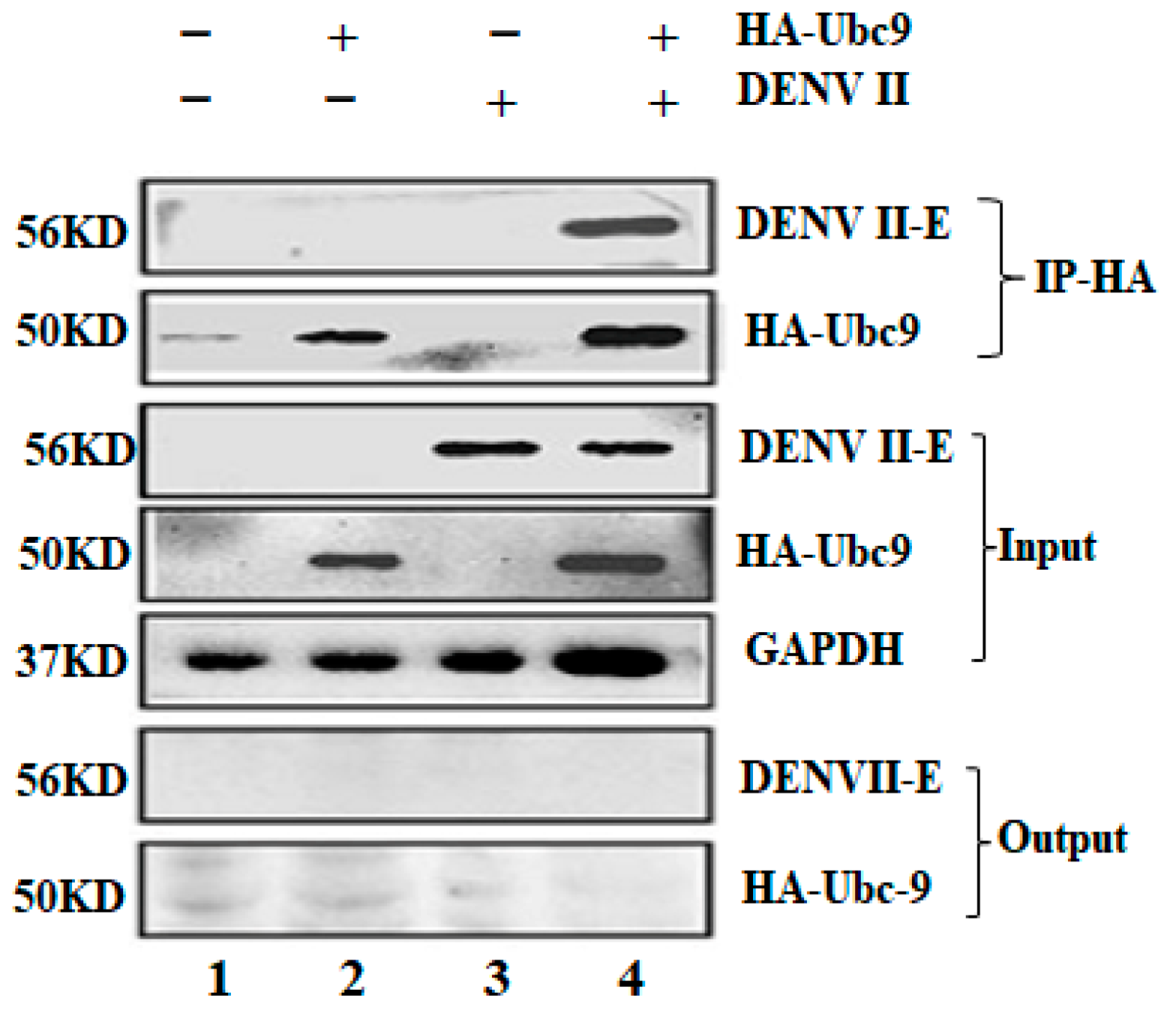
3.4. Co-Immunoprecipitation (CO-IP) Test DENV Env Protein E Interacts with Ubc9 in an Overexpressed System
3.5. Immunoconfocal Technique Confirmed the Colocalization of Ubc9 and Dengue E Protein
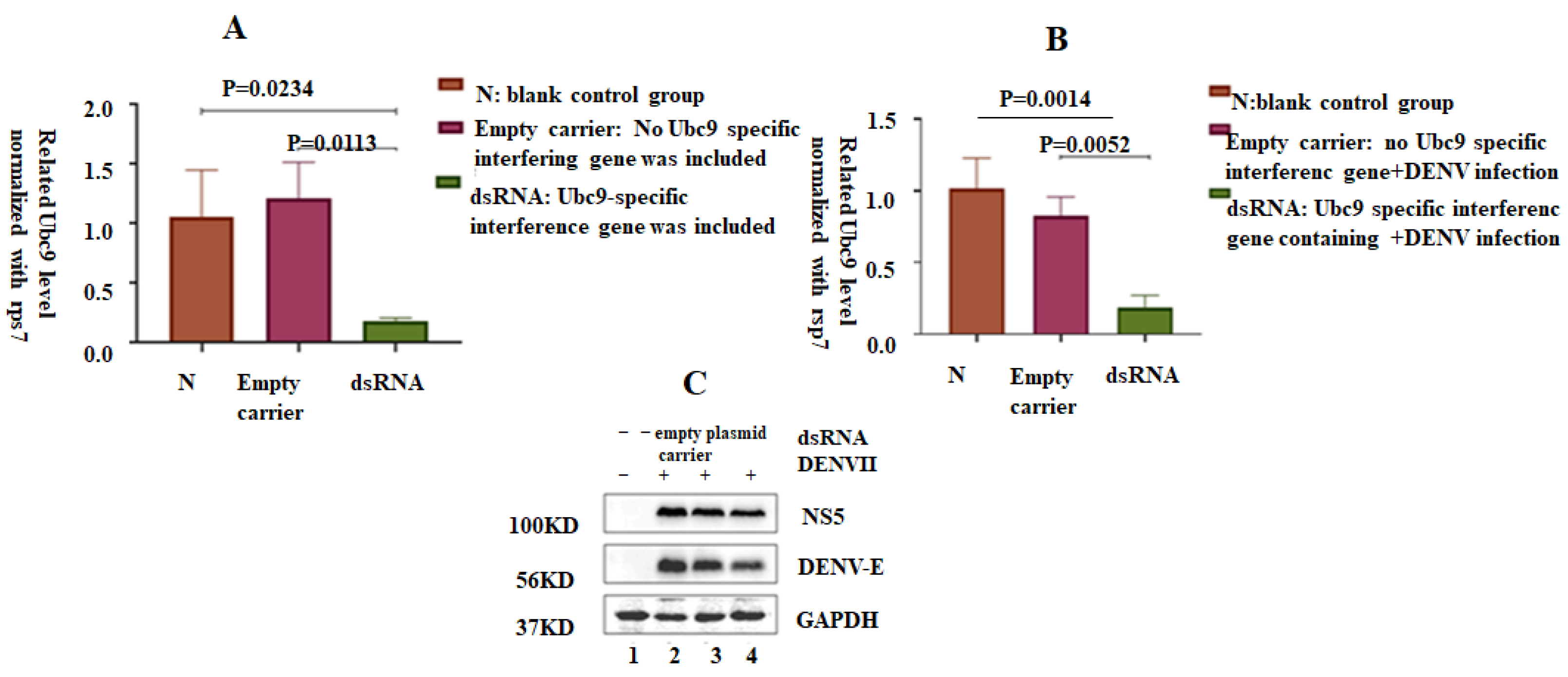
3.6. shRNA-Induced Silencing of Ubc9 in C6/36 Cells Decreased the Expression of NS5 and E Proteins in DENV-2
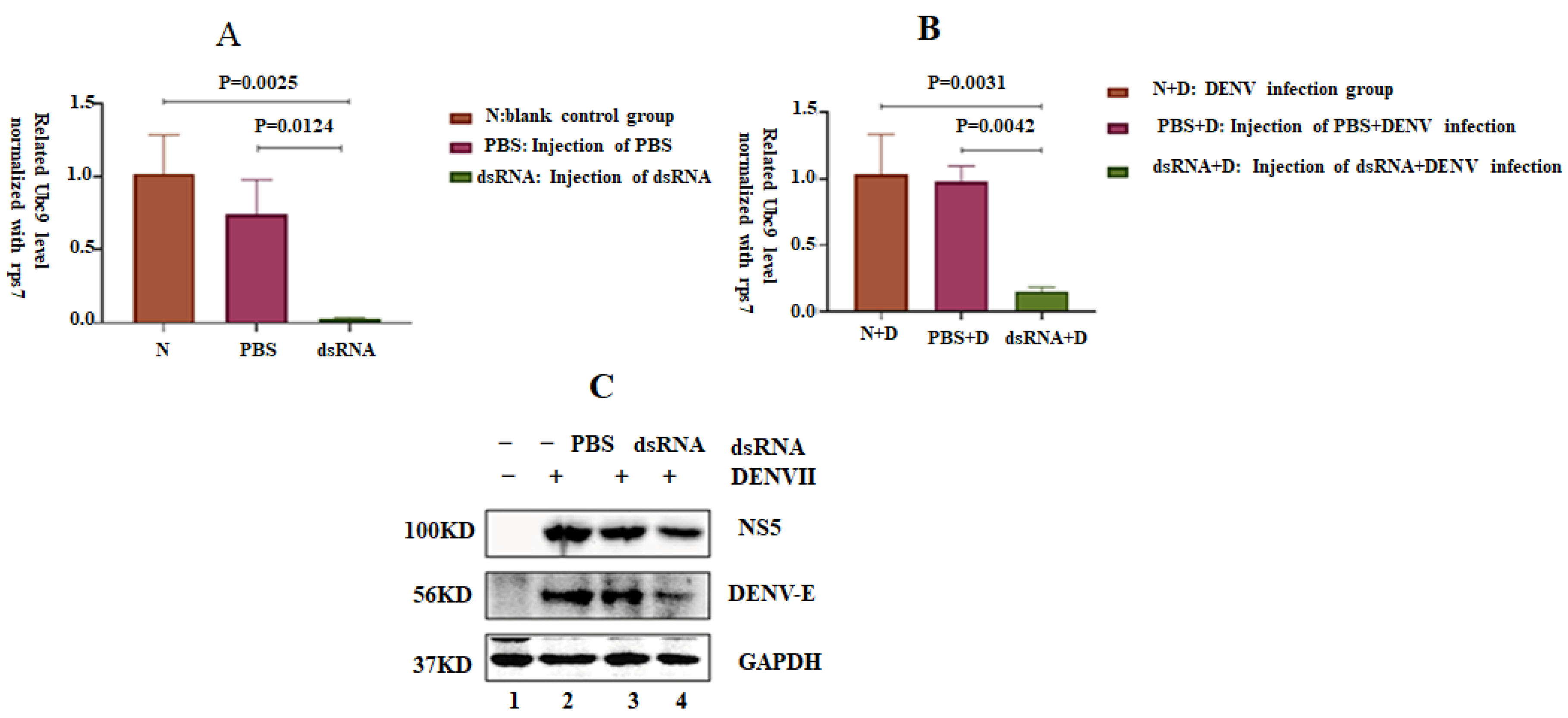
3.7. Silencing the Ubc9 Gene in A. albopictus Decreased the Replication Ability of DENV-2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soo, K.M.; Khalid, B.; Ching, S.M.; Chee, H.Y. Meta-Analysis of Dengue Severity during Infection by Different Dengue Virus Serotypes in Primary and Secondary Infections. PLoS ONE 2016, 11, e0154760. [Google Scholar] [CrossRef] [PubMed]
- Leitner, M.; Etebari, K.; Asgari, S. Transcriptional response of Wolbachia-transinfected Aedes aegypti mosquito cells to dengue virus at early stages of infection. J. Virol. 2018, 92, e00224-18. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, B.; Liu, P.; Li, J.; Chen, X.; Gu, J. piRNA Profiling of Dengue Virus Type 2-Infected Asian Tiger Mosquito and Midgut Tissues. Viruses 2018, 10, 213. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Fan, P.Y.; Wei, Y.; Wang, J.Q.; Zou, W.H.; Zhou, G.F.; Zhong, D.B.; Zheng, X.L. Isobaric tags for relative and absolute quantification-based proteomic analysis of host-pathogen protein interactions in the midgut of Aedes albopictus during dengue virus infection. Front. Microbiol. 2022, 13, 990978. [Google Scholar] [CrossRef]
- Tan, M.J.A.; Chan, K.W.K.; Ng, I.H.W.; Kong, S.Y.Z.; Gwee, C.P.; Vasudevan, W.S. The Potential Role of the ZIKV NS5 Nuclear Spherical-Shell Structures in Cell Type-Specific Host Immune Modulation during ZIKV Infection. Cells 2019, 8, 1519. [Google Scholar] [CrossRef]
- Guzman, M.G.; Halstead, S.B.; Artsob, H.; Buchy, P.; Farrar, J.; Gubler, D.J.; Hunsperger, E.; Kroeger, A.; Margolis, H.S.; Martínez, E.; et al. Dengue: A continuing global threat. Nat. Rev. Microbiol. 2010, 8, S7–S16. [Google Scholar] [CrossRef]
- Lee, E.; Bujalowski, P.J.; Teramoto, T.; Gottipati, K.; Scott, S.D.; Padmanabhan, R.; Chor, K.H. Structures of flavivirus RNA promoters suggest two binding modes with NS5 polymerase. Nat. Commun. 2021, 12, 2530. [Google Scholar] [CrossRef]
- Jones, M.; Davidson, A.; Hibbert, L.; Gruenwald, P.; Schlaak, J.; Ball, S.; Foster, G.R.; Jacobs, M. Dengue virus inhibits alpha interferon signaling by reducing STAT2 expression. J. Virol. 2005, 79, 5414–5420. [Google Scholar] [CrossRef]
- Veit, E.C.; Salim, M.S.; Jung, M.J.; Richardson, R.B.; Boys, L.N.; Meghan, Q.; Barrall, E.A.; Bednarski, E.; Hamilton, R.E.; Kikawa, C. Evolution of STAT2 resistance to flavivirus NS5 occurred multiple times despite genetic constraints. Nat. Commun. 2024, 15, 5426. [Google Scholar] [CrossRef]
- Conde, J.N.; Schutt, W.R.; Mladinich, M.; Sohn, S.Y.; Hearing, P.; Mackow, E.R. NS5 Sumoylation Directs Nuclear Responses That Permit Zika Virus To Persistently Infect Human Brain Microvascular Endothelial Cells. J. Virol. 2020, 94, e01086-20. [Google Scholar] [CrossRef]
- Wei, Y.; He, S.; Wang, J.T.; Fan, P.Y.; He, Y.L.; Hu, K.; Chen, Y.; Zhou, G.F.; Zhong, D.B.; Zheng, X.L. Genome-wide SNPs reveal novel patterns of spatial genetic structure in Aedes albopictus (Diptera Culicidae) population in China. Front. Public Health 2022, 10, 1028026. [Google Scholar] [CrossRef] [PubMed]
- Stokes, S.; Almire, F.; Tatham, M.H.; McFarlane, S.; Mertens, P.; Pondeville, E.; Boutell, C. The SUMOylation pathway suppresses arbovirus replication in Aedes aegypti cells. PLoS Pathog. 2020, 16, e1009134. [Google Scholar] [CrossRef] [PubMed]
- Ruckert, C.; Ebel, G.D. How Do Virus-Mosquito Interactions Lead to Viral Emergence? Trends Parasitol. 2018, 34, 310–321. [Google Scholar] [CrossRef]
- Weng, S.C.; Shiao, S.H. SUMOylation Is Essential for Dengue Virus Replication and Transmission in the Mosquito Aedes aegypti. Front. Microbiol. 2022, 13, 801284. [Google Scholar] [CrossRef]
- Wei, Y.; Wang, J.; Wei, Y.H.; Song, Z.; Hu, K.; Chen, Y.; Zhou, G.F.; Zhong, D.B.; Zheng, X.L. Vector Competence for DENV-2 Among Aedes albopictus (Diptera: Culicidae) Populations in China. Front. Cell. Infect. Microbiol. 2021, 11, 649975. [Google Scholar] [CrossRef] [PubMed]
- Chathuranga, K.; Weerawardhana, A.; Dodantenna, N.; Lee, J.S. Regulation of antiviral innate immune signaling and viral evasion following viral genome sensing. Exp. Mol. Med. 2021, 53, 1647–1668. [Google Scholar] [CrossRef]
- Angleró-Rodríguez, Y.I.; MacLeod, H.J.; Kang, S.; Carlson, J.S.; Jupatanakul, N.; Dimopoulos, G. Aedes aegypti molecular responses to Zika virus: Modulation of infection by the Toll and Jak/Stat immune pathways and virus host factors. Front. Microbiol. 2017, 8, 2050. [Google Scholar] [CrossRef]
- Kean, J.; Rainey, S.M.; McFarlane, M.; Donald, C.L.; Schnettler, E.; Kohl, A.; Pondeville, E. Fighting Arbovirus Transmission: Natural and Engineered Control of Vector Competence in Aedes Mosquitoes. Insects 2015, 6, 236–278. [Google Scholar] [CrossRef]
- Cheng, G.; Liu, Y.; Wang, P.; Xiao, X. Mosquito Defense Strategies against Viral Infection. Trends Parasitol. 2016, 32, 177–186. [Google Scholar] [CrossRef]
- Fan, Y.; Li, X.; Zhang, L.; Zong, Z.; Wang, F.W.; Huang, J.; Zeng, L.H.; Zhang, C.; Yan, H.; Zhang, L.; et al. SUMOylation in Viral Replication and Antiviral Defense. Adv. Sci. 2022, 9, 2104126. [Google Scholar] [CrossRef]
- Cartier, E.; Garcia-Olivares, J.; Janezic, E.; Viana, J.; Moore, M.; Lin, M.L.; Caplan, J.L.; Gonzalo Torres, G.; Kim, Y.H. The SUMO-Conjugase Ubc9 Prevents the Degradation of the Dopamine Transporter, Enhancing Its Cell Surface Level and Dopamine Uptake. Front. Cell. Neurosci. 2019, 13, 35. [Google Scholar] [CrossRef] [PubMed]
- Martinat, C.; Cormier, A.; Tobaly-Tapiero, J.; Palmic, N.; Casartelli, N.; Mahboubi, B.; Coggins, S.A.; Buchrieser, J.; Persaud, M.; Diaz-Griffero, F. SUMOylation of SAMHD1 at Lysine 595 is required for HIV-1 restriction in non-cycling cells. Nat. Commun. 2021, 12, 4582. [Google Scholar] [CrossRef]
- Dang, R.R.; Rodgers, V.G.J.; García-Sastre, A.; Liao, J.Y. Human SUMOylation Pathway Is Critical for Influenza B Virus. Viruses 2022, 14, 314. [Google Scholar] [CrossRef]
- Everett, R.D.; Boutell, C.; Hale, B.G. Interplay between viruses and host sumoylation pathways. Nat. Rev. Microbiol. 2013, 11, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, P.; Schreiner, S.; Dobner, T. Human pathogens and the host cell SUMOylation system. J. Virol. 2012, 86, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Mattoscio, D.; Segré, C.V.; Chiocca, S. Viral manipulation of cellular protein conjugation pathways: The SUMO lesson. World J. Virol. 2013, 2, 79–90. [Google Scholar] [CrossRef]
- Bernier-Villamor, V.; Sampson, D.A.; Matunis, M.J.; Lima, C.D. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 2002, 108, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Goffinont, S.; Coste, F.; Prieu-Serandon, P.; Mance, L.; Gaudon, V.; Garnier, N.; Castaing, B.; Suskiewicz, M.J. Structural insights into the regulation of the human E2∼SUMO conjugate through analysis of its stable mimetic. J. Biol. Chem. 2023, 299, 104870. [Google Scholar] [CrossRef]
- Rodriguez, M.S.; Dargemont, C.; Hay, R.T. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001, 276, 12654–12659. [Google Scholar] [CrossRef]
- Wimmer, P.; Schreiner, S. Viral Mimicry to Usurp Ubiquitin and SUMO Host Pathways. Viruses 2015, 7, 4854–4872. [Google Scholar] [CrossRef]
- Wilson, V.G. Sumoylation at the host-pathogen interface. Biomolecules 2012, 2, 203–227. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.W.; Shih, H.M.; Yang, T.H.; Yang, Y.L. The type 2 dengue virus envelope protein interacts with small ubiquitin-like modifier-1 (SUMO-1) conjugating enzyme 9 (Ubc9). J. Biomed. Sci. 2007, 14, 429–444. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Chu, H.; Wen, L.; Yuan, S.; Chik, K.K.; Yuen, T.T.; Yip, C.C.Y.; Wang, D.; Zhou, J.; Yin, F.F.; et al. Targeting SUMO Modification of the Non-Structural Protein 5 of Zika Virus as a Host-Targeting Antiviral Strategy. Int. J. Mol. Sci. 2019, 20, 392. [Google Scholar] [CrossRef] [PubMed]
- Su, C.I.; Tseng, C.H.; Yu, C.Y.; Lai, M.M.C. SUMO Modification Stabilizes Dengue Virus Nonstructural Protein 5 to Support Virus Replication. J. Virol. 2016, 90, 4308–4319. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zeng, N.; Liu, S.; Miao, Q.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H.C. Interaction of porcine reproductive and respiratory syndrome virus proteins with SUMO-conjugating enzyme reveals the SUMOylation of nucleocapsid protein. PLoS ONE 2017, 12, e0189191. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer. Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef]
- Frachon, N.; Demaretz, S.; Seaayfan, E.; Chelbi, L.; Bakhos-Douaihy, D.; Laghmani, K. AUP1 Regulates the Endoplasmic Reticulum-Associated Degradation and Polyubiquitination of NKCC2. Cells 2024, 13, 389. [Google Scholar] [CrossRef]
- Ting-Chun Yeh and Shawn, B. Bratton. Caspase-dependent regulation of the ubiquitin–proteasome system through direct substrate targeting. Proc. Natl. Acad. Sci. USA 2013, 110, 4284–14289. [Google Scholar] [CrossRef]
- Lewis, T.W.; Barthelemy, J.R.; Virts, E.L.; Kennedy, F.M.; Gadgil, R.Y.; Wiek, C.; Linka, R.M.; Zhang, F.; Andreassen, P.R.; Hanenberg, H.; et al. Deficiency of the Fanconi anemia E2 ubiqitin conjugase UBE2T only partially abrogatesAlu-mediated recombination in a new model of homology dependent recombination. Nucleic Acids Res. 2019, 7, 3503–3520. [Google Scholar] [CrossRef]
- Ye, Y.S.; Tyndall, E.R.; Bui, V.; Bewley, M.C.; Wang, G.F.; Hong, X.P.; Shen, Y.; Flanagan, J.M.; Wang, H.G.; Tian, T. Multifaceted membrane interactions of human Atg3 promote LC3- phosphatidylethanolamine conjugation during autophagy. Nat. Commun. 2023, 14, 5503. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Li, F.H.; Wang, X.J. Regulation of Tripartite Motif Containing Proteins on Immune Response and Viral Evasion. Front. Microbiol. 2021, 12, 794882. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Thurmond, S.; Hai, R.; Song, J. Structure and function of Zika virus NS5 protein: Perspectives for drug design. Cell Mol Life Sci. 2018, 75, 1723–1736. [Google Scholar] [CrossRef] [PubMed]
- Jablunovsky, A.; Jose, J. The Dynamic Landscape of Capsid Proteins and Viral RNA Interactions in Flavivirus Genome Packaging and Virus Assembly. Pathogens 2024, 13, 120. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, M.J.G.; Sanchez-Garrido, J.; Hoben, G.F.; Goddard, P.J.; Shenoy, A.R. The Atypical Ubiquitin E2 Conjugase UBE2L3 Is an Indirect Caspase-1 Target and Controls IL-1β Secretion by Inflammasomes. Cell Rep. 2017, 18, 1285–1297. [Google Scholar] [CrossRef]
- Ureña, E.; Pirone, L.; Chafino, S.; Pérez, C.; Sutherland, J.D.; Lang, V.; Rodriguez, M.S.; Lopitz-Otsoa, F.; Blanco, F.J.; Barrio, R.; et al. Evolution of SUMO Function and Chain Formation in Insects. Mol. Biol. Evol. 2016, 33, 568–584. [Google Scholar] [CrossRef]
- Sahili, A.E.I.; Lescar, J. Dengue Virus Non-Structural Protein. Viruses 2017, 9, 91. [Google Scholar] [CrossRef]
- Golubeva, V.A.; Nepomuceno, T.C.; Gregoriis, G.D.; Mesquita, R.D.; Li, X.L.; Dash, S.; Garcez, P.P.; Suarez-Kurtz, G.; Izumi, V.; Koomen, J. Network of Interactions between ZIKA Virus Non-Structural Proteins and Human Host Proteins. Cells 2020, 9, 153. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.I.; Xia, H.J.; Aguilera-Aguirre, L.; Hage, A.; Tol, S.V.; Shan, C.; Xie, X.P.; Sturdevant, G.L.; Robertson, S.J.; McNally, K.L.; et al. Envelope Protein Ubiquitination Drives Zika Virus Entry and Pathogenesis. Nature 2020, 585, 414–419. [Google Scholar] [CrossRef]
- Lan, Y.; van Leur, S.W.; Fernando, J.A.; Wong, H.H.; Kampmann, M.; Siu, L.; Zhang, J.S.; Li, M.Y.; Nicholls, J.M.; Sanyal, S. Viral subversion of selective autophagy is critical for biogenesis of virus replication organelles. Nat. Commun. 2023, 14, 2698. [Google Scholar] [CrossRef]
- Feng, T.T.; Deng, L.; Lu, X.C.; Pan, W.; Wu, Q.H.; Dai, J.F. Ubiquitin-conjugating enzyme UBE2J1 negatively modulates interferon pathway and promotes RNA virus infection. Virol. J. 2018, 15, 132. [Google Scholar] [CrossRef]
- Terradas, G.; McGraw, E.A. Using genetic variation in Aedes aegypti to identify candidate anti-dengue virus genes. BMC Infect. Dis. 2019, 19, 580. [Google Scholar] [CrossRef] [PubMed]
- Valerdi, K.M.; Hage, A.; Tol, S.V.; Rajsbaum, R.; Giraldo, M.I. The Role of the Host Ubiquitin System in Promoting Replication of Emergent Viruses. Viruses 2021, 13, 369. [Google Scholar] [CrossRef] [PubMed]
- Aguilar Briseño, J.A.; Ramos Pereira, L.; van der Laan, M.; Pauzuolis, M.; ter Ellen, B.M.; Upasani, V.; Moser, J.; de Souza Ferreira, L.C.; Smit, J.M.; Rodenhuis-Zybert, I.A. TLR2 axis on peripheral blood mononuclear cells regulates inflammatory responses to non-infectious immature dengue virus particles. PLoS Pathog. 2022, 18, e1010499. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Zheng, X.; Wang, X.; Zhong, D.; Zhou, G. E2 Ubiquitin-Conjugating Enzymes Regulates Dengue Virus-2 Replication in Aedes albopictus. Microorganisms 2024, 12, 2508. https://doi.org/10.3390/microorganisms12122508
Wang J, Zheng X, Wang X, Zhong D, Zhou G. E2 Ubiquitin-Conjugating Enzymes Regulates Dengue Virus-2 Replication in Aedes albopictus. Microorganisms. 2024; 12(12):2508. https://doi.org/10.3390/microorganisms12122508
Chicago/Turabian StyleWang, Jiaqi, Xueli Zheng, Xuexue Wang, Daibin Zhong, and Guofa Zhou. 2024. "E2 Ubiquitin-Conjugating Enzymes Regulates Dengue Virus-2 Replication in Aedes albopictus" Microorganisms 12, no. 12: 2508. https://doi.org/10.3390/microorganisms12122508
APA StyleWang, J., Zheng, X., Wang, X., Zhong, D., & Zhou, G. (2024). E2 Ubiquitin-Conjugating Enzymes Regulates Dengue Virus-2 Replication in Aedes albopictus. Microorganisms, 12(12), 2508. https://doi.org/10.3390/microorganisms12122508




