The Association of Blood Eosinophils and Neutrophils Expressing Eosinophilic Surface Markers with the Severity and Outcome of COVID-19
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Clinical Data Collection
2.3. Blood Sample Collection, Processing, and Isolation of White Blood Cells
2.4. Flow Cytometry
2.5. Cytokine Profiling by LEGENDplexTM
2.6. Statistical Analysis
3. Results
3.1. Demographic, Clinical and Prognosis Characteristics
3.2. Increased Blood Neutrophils but Decreased Eosinophils and Lymphocytes in Patients with Severe-to-Very Severe Disease
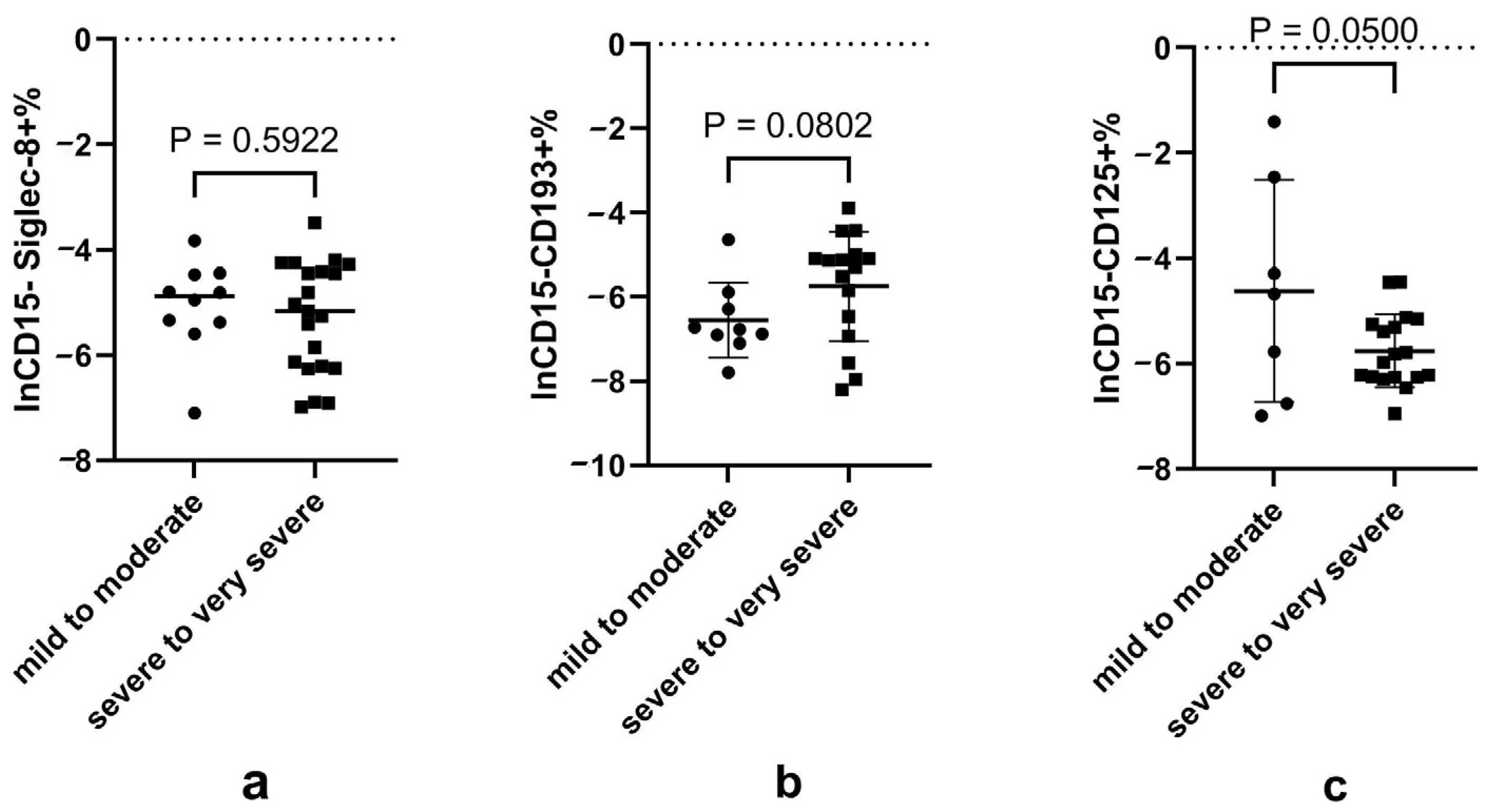
3.3. Decreased CD125+ Eosinophils in Patients with Severe-to-Very Severe Disease
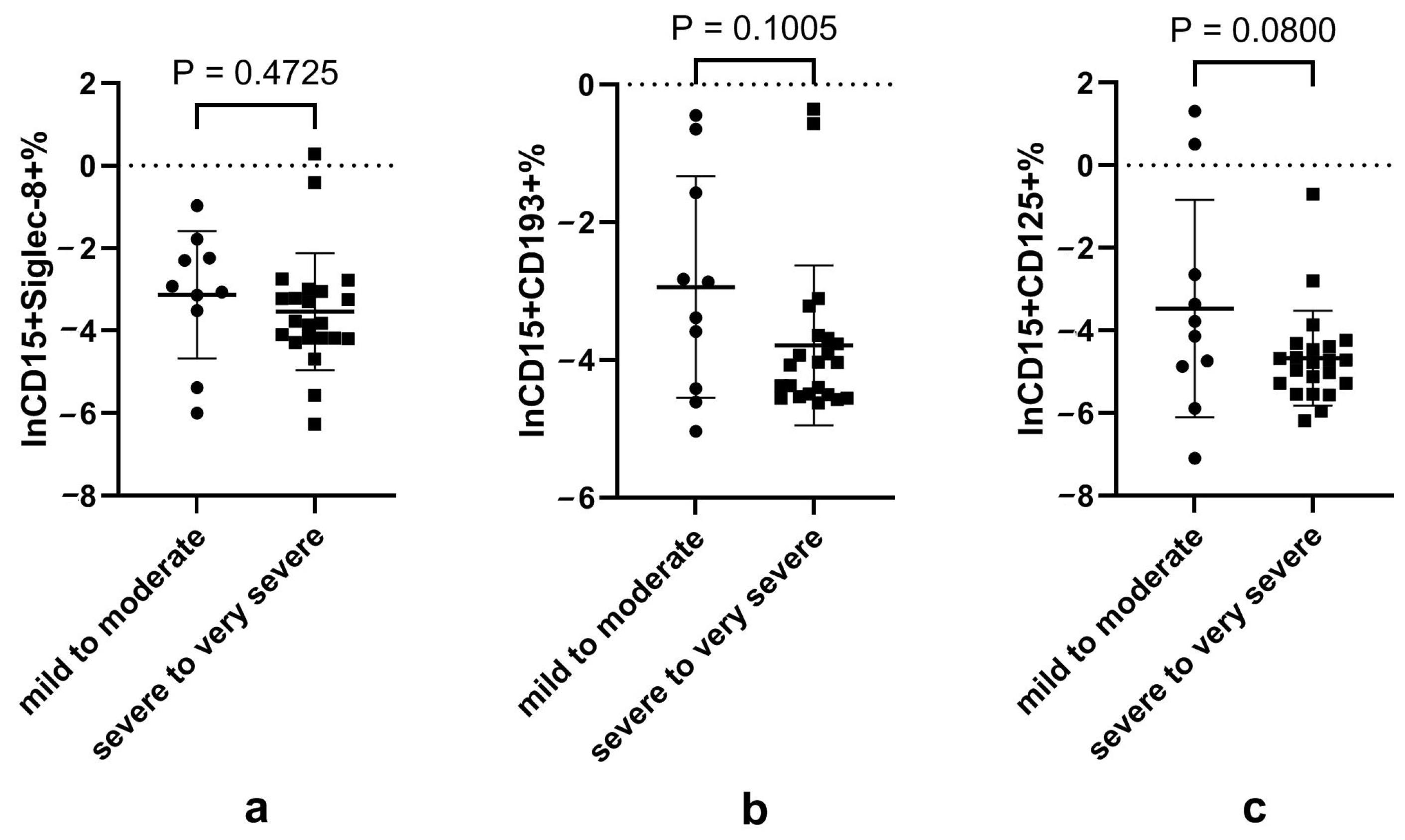
3.4. Disturbed Neutrophil Subsets Expressing Eosinophilic Surface Markers in Patients with Severe-to-Very Severe Disease
3.5. Blood Cytokines in COVID-19 Patients with Different Disease Severities
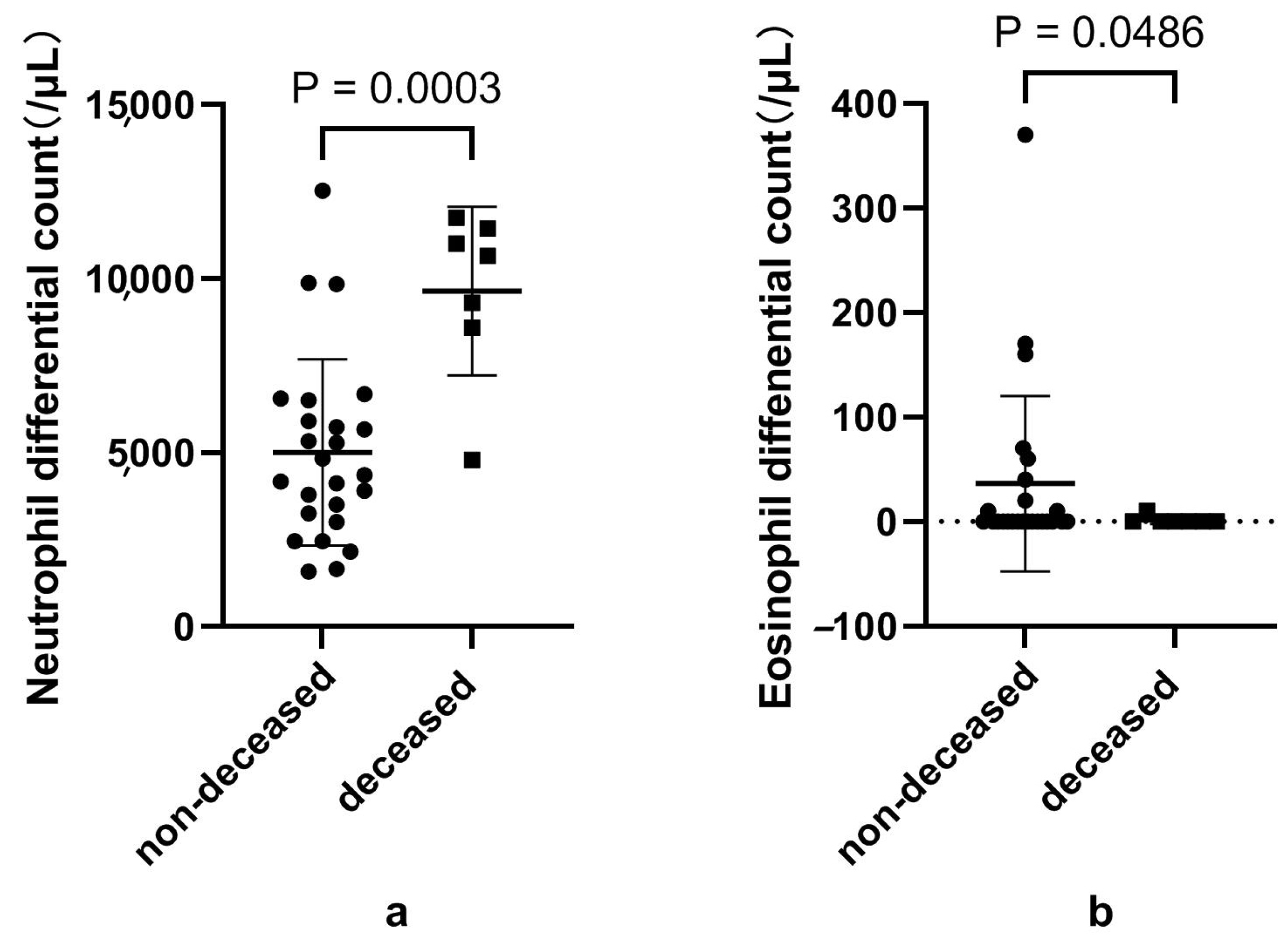
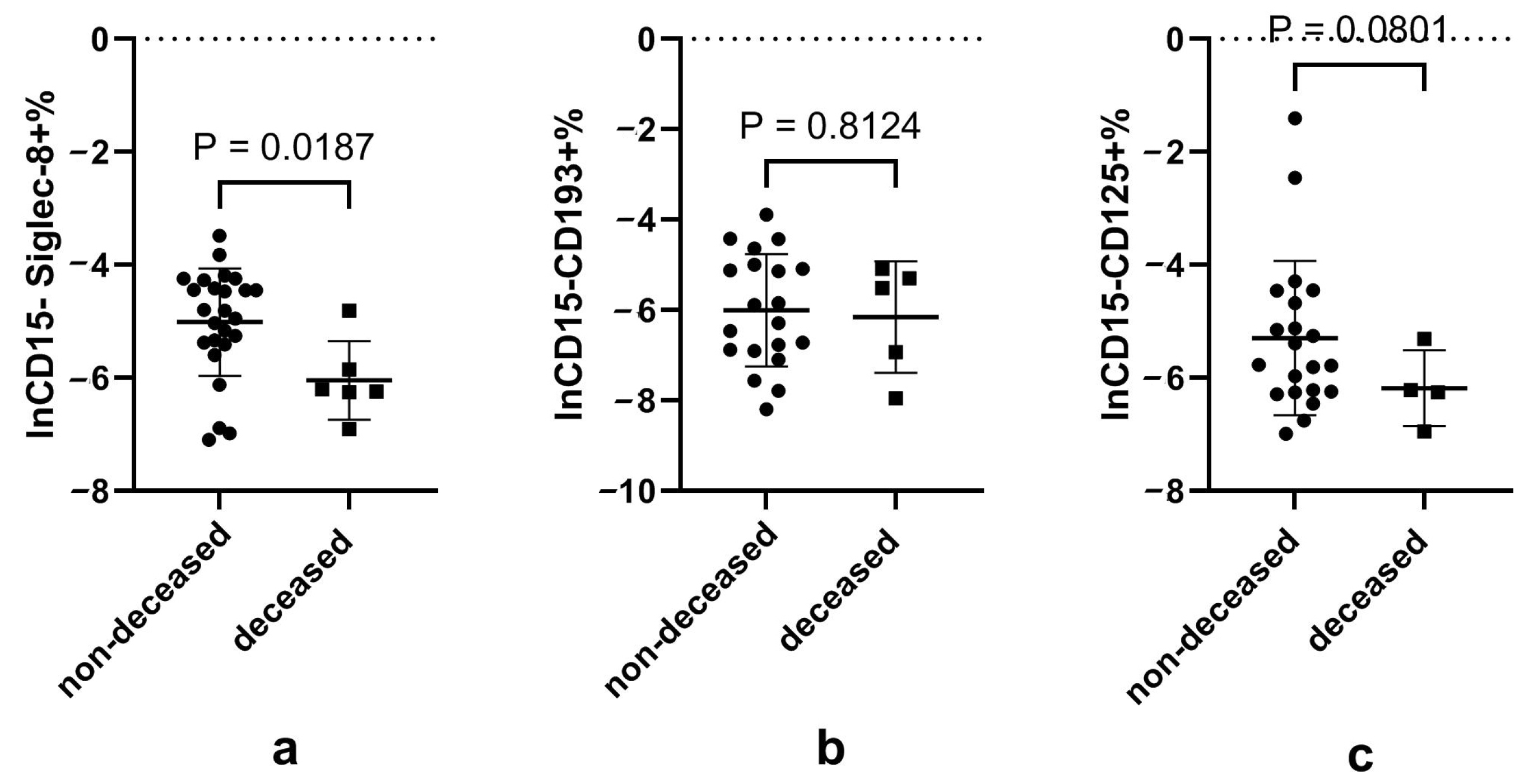
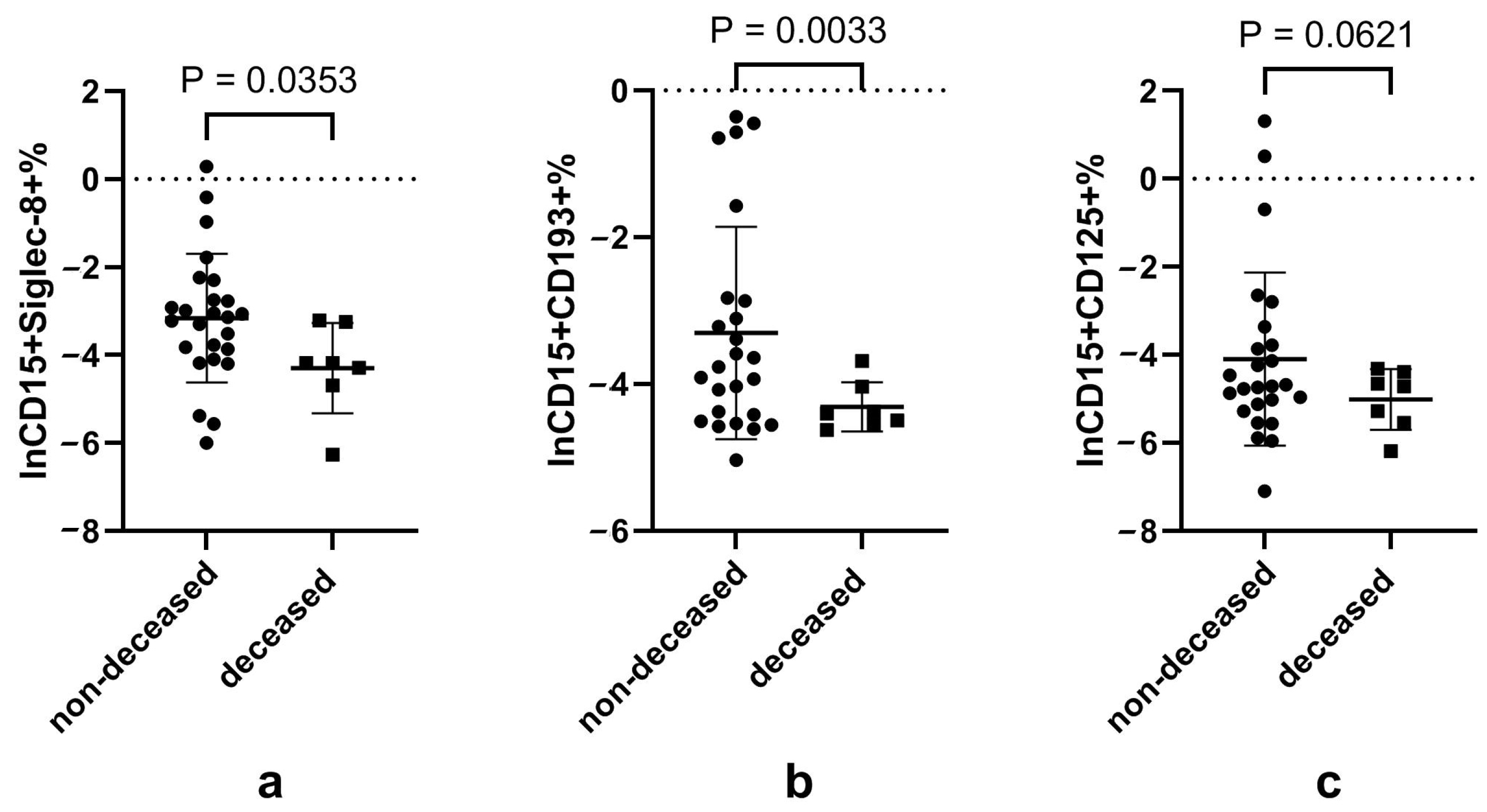
3.6. Disturbed Eosinophils and Neutrophils, and Their Subsets in Patients That Died of COVID-19
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.J.; Dong, X.; Cao, Y.Y. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Allergy 2020, 75, 1730–1741. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Qin, R.; He, L.; Yang, Z.; Jia, N.; Chen, R.; Xie, J.; Fu, W.; Chen, H.; Lin, X.; Huang, R.; et al. Identification of Parameters Representative of Immune Dysfunction in Patients with Severe and Fatal COVID-19 Infection: A Systematic Review and Meta-Analysis. Clin. Rev. Allergy Immunol. 2022, 64, 33–65. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Cai, J.; Tang, Q.; Mo, Y.Y.; Deng, T.; Zhang, X.; Xu, K.; Wu, B.; Tang, H.; Zhang, Z. Circulating eosinophils associated with responsiveness to COVID-19 vaccine and the disease severity in patients with SARS-CoV-2 omicron variant infection. BMC Pulm. Med. 2023, 23, 177. [Google Scholar] [CrossRef] [PubMed]
- Rutkowska, E.; Kwiecień, I.; Pietruszka-Wałęka, E.; Więsik-Szewczyk, E.; Rzepecki, P.; Jahnz-Różyk, K. Analysis of Leukocyte Subpopulations by Flow Cytometry during Hospitalization Depending on the Severity of COVID-19 Course. Biomedicines 2023, 11, 2728. [Google Scholar] [CrossRef] [PubMed]
- Macchia, I.; La Sorsa, V.; Urbani, F.; Moretti, S.; Antonucci, C.; Afferni, C.; Schiavoni, G. Eosinophils as potential biomarkers in respiratory viral infections. Front. Immunol. 2023, 14, 1170035. [Google Scholar] [CrossRef]
- Du, Y.; Tu, L.; Zhu, P.; Mu, M.; Wang, R.; Yang, P.; Wang, X.; Hu, C.; Ping, R.; Hu, P.; et al. Clinical Features of 85 Fatal Cases of COVID-19 from Wuhan. A Retrospective Observational Study. Am. J. Respir. Crit. Care Med. 2020, 201, 1372–1379. [Google Scholar] [CrossRef]
- Wang, X.; Dian, Y.; Zhou, Q.; Deng, G.; Wei, R.; Zeng, F. Association of eosinopenia with worsening prognosis in hospitalized Azvudine-treated COVID-19 patients: A retrospective cohort study. Front. Immunol. 2023, 14, 1320973. [Google Scholar] [CrossRef]
- Yan, B.; Yang, J.; Xie, Y.; Tang, X. Relationship between blood eosinophil levels and COVID-19 mortality. World Allergy Organ. J. 2021, 14, 100521. [Google Scholar] [CrossRef]
- Gambichler, T.; Schuleit, N.; Susok, L.; Becker, J.C.; Scheel, C.H.; Torres-Reyes, C.; Overheu, O.; Reinacher-Schick, A.; Schmidt, W. Prognostic Performance of Inflammatory Biomarkers Based on Complete Blood Counts in COVID-19 Patients. Viruses 2023, 15, 1920. [Google Scholar] [CrossRef]
- Yan, P.D.; Markovic, D.; Hixson, R.Y.; Shover, C.M.; Buhr, R.G.; Salehi-Rad, R.; LeMaster, B.; Tashkin, D.P.; Fulcher, J.A.; Barjaktarevic, I.Z. Eosinophil recovery in hospitalized COVID-19 patients is associated with lower rates of ICU admission and in-hospital mortality: An observational cohort analysis. Respir. Med. Res. 2023, 84, 101031. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Ishida, T.; Nakanishi, Y.; Kobe, H.; Tokioka, F. Eosinopenia is associated with adverse outcomes after COVID-19 infection: A perspective from Japan. Respirology 2023, 28, 677–680. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, A.W.; Schwartz, J.T.; Rothenberg, M.E. Eosinophil responses during COVID-19 infections and coronavirus vaccination. J. Allergy Clin. Immunol. 2020, 146, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rajeevan, H.; Simonov, M.; Lee, S.; Wilson, F.P.; Desir, G.V.; Vinetz, J.M.; Yan, X.; Wang, Z.; Clark, B.J.; et al. Differences in Mortality Among Patients with Asthma and COPD Hospitalized with COVID-19. J. Allergy Clin. Immunol. Pract. 2023, 11, 3383–3390.e3. [Google Scholar] [CrossRef]
- Yang, J.M.; Koh, H.Y.; Moon, S.Y.; Yoo, I.K.; Ha, E.K.; You, S.; Kim, S.Y.; Yon, D.K.; Lee, S.W. Allergic disorders and susceptibility to and severity of COVID-19: A nationwide cohort study. J. Allergy Clin. Immunol. 2020, 146, 790–798. [Google Scholar] [CrossRef]
- Zhu, Z.; Hasegawa, K.; Ma, B.; Fujiogi, M.; Camargo, C.A., Jr.; Liang, L. Association of asthma and its genetic predisposition with the risk of severe COVID-19. J. Allergy Clin. Immunol. 2020, 146, 327–329.e4. [Google Scholar] [CrossRef]
- Camiolo, M.; Gauthier, M.; Kaminski, N.; Ray, A.; Wenzel, S.E. Expression of SARS-CoV-2 receptor ACE2 and coincident host response signature varies by asthma inflammatory phenotype. J. Allergy Clin. Immunol. 2020, 146, 315–324.e7. [Google Scholar] [CrossRef]
- Carli, G.; Cecchi, L.; Stebbing, J.; Parronchi, P.; Farsi, A. Is asthma protective against COVID-19? Allergy 2021, 76, 866–868. [Google Scholar] [CrossRef]
- Adir, Y.; Saliba, W.; Beurnier, A.; Humbert, M. Asthma and COVID-19: An update. Eur. Respir. Rev. 2021, 30, 210152. [Google Scholar] [CrossRef]
- Chen, C.; Song, X.; Murdock, D.J.; Marcus, A.; Hussein, M.; Jalbert, J.J.; Geba, G.P. Association between allergic conditions and COVID-19 susceptibility and outcomes. Ann. Allergy Asthma Immunol. 2024, 132, 637–645.e7. [Google Scholar] [CrossRef]
- Liu, S.; Zhi, Y.; Ying, S. COVID-19 and Asthma: Reflection During the Pandemic. Clin. Rev. Allergy Immunol. 2020, 59, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Bao, W.; Tian, X.; Zhang, Y.; Pan, Y.; Xie, G.; Bao, A.; Yin, D.; Zhang, M.; Zhou, Y. Vitamin D status in hospitalized COVID-19 patients is associated with disease severity and IL-5 production. Virol. J. 2023, 20, 212. [Google Scholar] [CrossRef] [PubMed]
- Ju, X.; Son, K.; Jamil, R.; Culgin, S.; Salter, B.; Miyasaki, K.; Fard, N.E.; Xiao, M.; Patel, Z.; Zhang, K.; et al. Eosinophil-independent IL-5 levels are increased in critically ill COVID-19 patients who survive. Allergy Asthma Clin. Immunol. 2023, 19, 58. [Google Scholar] [CrossRef] [PubMed]
- Aung, H.; McAuley, H.; Porter, K.; Richardson, M.; Wright, A.; Brightling, C.E.; Greening, N.J. Differences in hospital admissions for acute exacerbations of COPD during the COVID-19 pandemic stratified by stable-state blood eosinophil count. Eur. Respir. J. 2023, 62, 2301125. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Berdnikovs, S. The twilight zone: Plasticity and mixed ontogeny of neutrophil and eosinophil granulocyte subsets. Semin. Immunopathol. 2021, 43, 337–346. [Google Scholar] [CrossRef]
- Gorski, S.A.; Lawrence, M.G.; Hinkelman, A.; Spano, M.M.; Steinke, J.W.; Borish, L.; Teague, W.G.; Braciale, T.J. Expression of IL-5 receptor alpha by murine and human lung neutrophils. PLoS ONE 2019, 14, e0221113. [Google Scholar] [CrossRef]
- Lawrence, M.G.; Teague, W.G.; Feng, X.; Welch, C.; Etter, E.; Negri, J.; Spano, M.; Wavell, K.; Braciale, T.; Steinke, J.W.; et al. Interleukin-5 receptor alpha (CD125) expression on human blood and lung neutrophils. Ann. Allergy Asthma Immunol. 2021, 128, 53–60.e3. [Google Scholar] [CrossRef]
- Mesnil, C.; Raulier, S.; Paulissen, G.; Xiao, X.; Birrell, M.A.; Pirottin, D.; Janss, T.; Starkl, P.; Ramery, E.; Henket, M.; et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J. Clin. Investig. 2016, 126, 3279–3295. [Google Scholar] [CrossRef]
- Jeong, B.M.; Walker, M.T.; Rodriguez, R.; Coden, M.E.; Nagasaka, R.; Doan, T.C.; Politanska, Y.; Abdala-Valencia, H.; Berdnikovs, S. More than neutrophils: Lin(+)Ly6G(+)IL-5Rα(+) multipotent myeloid cells (MMCs) are dominant in normal murine bone marrow and retain capacity to differentiate into eosinophils and monocytes. J. Leukoc. Biol. 2021, 111, 113–122. [Google Scholar] [CrossRef]
- Pfirschke, C.; Engblom, C.; Gungabeesoon, J.; Lin, Y.; Rickelt, S.; Zilionis, R.; Messemaker, M.; Siwicki, M.; Gerhard, G.M.; Kohl, A.; et al. Tumor-Promoting Ly-6G(+) SiglecF(high) Cells Are Mature and Long-Lived Neutrophils. Cell Rep. 2020, 32, 108164. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Nagakubo, D.; Satooka, H.; Hirata, T. A novel Siglec-F+ neutrophil subset in the mouse nasal mucosa exhibits an activated phenotype and is increased in an allergic rhinitis model. Biochem. Biophys. Res. Commun. 2020, 526, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Rudd, J.M.; Pulavendran, S.; Ashar, H.K.; Ritchey, J.W.; Snider, T.A.; Malayer, J.R.; Marie, M.; Chow, V.T.K.; Narasaraju, T. Neutrophils Induce a Novel Chemokine Receptors Repertoire During Influenza Pneumonia. Front. Cell Infect. Microbiol. 2019, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.; Krauss-Etschmann, S.; Koller, B.; Hordijk, P.L.; Kuijpers, T.W.; Hoffmann, F.; Hector, A.; Eber, E.; Marcos, V.; Bittmann, I.; et al. Infiltrated Neutrophils Acquire Novel Chemokine Receptor Expression and Chemokine Responsiveness in Chronic Inflammatory Lung Diseases. J. Immunol. 2008, 181, 8053–8067. [Google Scholar] [CrossRef]
- Bekaert, S.; Rocks, N.; Vanwinge, C.; Noel, A.; Cataldo, D. Asthma-related inflammation promotes lung metastasis of breast cancer cells through CCL11–CCR3 pathway. Respir. Res. 2021, 22, 61. [Google Scholar] [CrossRef]
- Huaux, F.; Gharaee-Kermani, M.; Liu, T.; Morel, V.; McGarry, B.; Ullenbruch, M.; Kunkel, S.L.; Wang, J.; Xing, Z.; Phan, S.H. Role of Eotaxin-1 (CCL11) and CC Chemokine Receptor 3 (CCR3) in Bleomycin-Induced Lung Injury and Fibrosis. Am. J. Pathol. 2005, 167, 1485–1496. [Google Scholar] [CrossRef]
- Haruna, N.F.; Politanska, Y.; Connelly, A.R.; O’Connor, K.; Bhattacharya, S.; Miklaszewski, G.E.; Pérez-Leonor, X.G.; Rerko, G.; Hentenaar, I.T.; Nguyen, D.C.; et al. scRNA-seq profiling of human granulocytes reveals expansion of developmentally flexible neutrophil precursors with mixed neutrophil and eosinophil properties in asthma. J. Leukoc. Biol. 2024, 116, 1184–1197. [Google Scholar] [CrossRef]
- Hassani, M.; Leijte, G.; Bruse, N.; Kox, M.; Pickkers, P.; Vrisekoop, N.; Koenderman, L. Differentiation and activation of eosinophils in the human bone marrow during experimental human endotoxemia. J. Leukoc. Biol. 2020, 108, 1665–1671. [Google Scholar] [CrossRef]
- Hassani, M.; van Staveren, S.; van Grinsven, E. Characterization of the phenotype of human eosinophils and their progenitors in the bone marrow of healthy individuals. Haematologica 2020, 105, e52–e56. [Google Scholar] [CrossRef]
- Höchstetter, R.; Dobos, G.; Kimmig, D.; Dulkys, Y.; Kapp, A.; Elsner, J. The CC chemokine receptor 3 CCR3 is functionally expressed on eosinophils but not on neutrophils. Eur. J. Immunol. 2000, 30, 2759–2764. [Google Scholar] [CrossRef]
- Yu, J.; Yan, B.; Shen, S.; Wang, Y.; Li, Y.; Cao, F.; Xiong, W.; Piao, Y.; Hu, C.; Sun, Y.; et al. IgE directly affects eosinophil migration in chronic rhinosinusitis with nasal polyps through CCR3 and predicts the efficacy of omalizumab. J. Allergy Clin. Immunol. 2023, 153, 447–460.e9. [Google Scholar] [CrossRef] [PubMed]
- Floyd, H.; Ni, J.; Cornish, A.L. Siglec-8. A novel eosinophil-specific member of the immunoglobulin superfamily. J. Biol. Chem. 2000, 275, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Kikly, K.K.; Bochner, B.S.; Freeman, S.D.; Tan, K.; Gallagher, K.T.; D’alessio, K.J.; Holmes, S.D.; Abrahamson, J.A.; Erickson-Miller, C.L.; Murdock, P.R.; et al. Identification of SAF-2, a novel siglec expressed on eosinophils, mast cells, and basophils. J. Allergy Clin. Immunol. 2000, 105, 1093–1100. [Google Scholar] [CrossRef]
- Nutku, E.; Aizawa, H.; Hudson, S.A.; Bochner, B.S. Ligation of Siglec-8: A selective mechanism for induction of human eosinophil apoptosis. Blood 2003, 101, 5014–5020. [Google Scholar] [CrossRef]
- Nutku-Bilir, E.; Hudson, S.A.; Bochner, B.S. Interleukin-5 Priming of Human Eosinophils Alters Siglec-8–Mediated Apoptosis Pathways. Am. J. Respir. Cell Mol. Biol. 2008, 38, 121–124. [Google Scholar] [CrossRef]
- Matucci, A.; Nencini, F.; Maggiore, G. High proportion of inflammatory CD62L(low) eosinophils in blood and nasal polyps of severe asthma patients. Clin. Exp. Allergy 2023, 53, 78–87. [Google Scholar] [CrossRef]
- Liao, D.; Zhou, F.; Luo, L.; Xu, M.; Wang, H.; Xia, J.; Gao, Y.; Cai, L.; Wang, Z.; Yin, P.; et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: A retrospective cohort study. Lancet Haematol. 2020, 7, e671–e678. [Google Scholar] [CrossRef]
- Runnstrom, M.C.; Lamothe, P.A.; Faliti, C.E. Patients taking benralizumab, dupilumab, or mepolizumab have lower postvaccination SARS-CoV-2 immunity. J. Allergy Clin. Immunol. 2024, 154, 435–446. [Google Scholar] [CrossRef]
- Wang, H.; Tumes, D.J.; Hercus, T.R.; Yip, K.H.; Aloe, C.; Vlahos, R.; Lopez, A.F.; Wilson, N.; Owczarek, C.M.; Bozinovski, S. Blocking the human common beta subunit of the GM-CSF, IL-5 and IL-3 receptors markedly reduces hyperinflammation in ARDS models. Cell Death Dis. 2022, 13, 137. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, Y.; Zhang, F.; Wang, Q.; Li, T.; Liu, Z.; Wang, J.; Qin, Y.; Zhang, X.; Yan, X.; et al. The use of anti-inflammatory drugs in the treatment of people with severe coronavirus disease 2019 (COVID-19): The Perspectives of clinical immunologists from China. Clin. Immunol. 2020, 214, 108393. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Zivancevic-Simonovic, S.; Jovanovic, D.; Cupurdija, V.; Milosevic-Djordjevic, O.; Stanojevic, M.; Marinkovic, M.; Igrutinovic, N.; Stanojevic, I.; Vojvodic, D.; Mihaljevic, O. Cytokine producing ability of peripheral blood cells from COVID-19 patients after unspecific in vitro stimulation. Inflamm. Res. 2022, 71, 331–341. [Google Scholar] [CrossRef]
| Mild-to-Moderate n = 10 | Severe-to-Very Severe n = 22 | p-Value | |
|---|---|---|---|
| Age (mean ± SD, years) | 67.3 ± 11.0 | 74.6 ± 13.1 | 0.139 |
| Sex (male, %) | 6 (60) | 15 (68.2) | 0.703 |
| BMI (mean ± SD, kg/m2) | 24.2 ± 3.2 | 23.7 ± 3.4 | 0.714 |
| Comorbidity(n., %) | |||
| Hypertension | 8 (80) | 14 (63.6) | 0.440 |
| Diabetes | 4 (40.0) | 5 (22.7) | 0.407 |
| Coronary heart disease | 2 (20) | 2 (9.1) | 0.572 |
| COPD | 1 (10) | 3 (13.6) | 1 |
| Asthma | 1 (10) | 2 (9.1) | 1 |
| Interstitial lung diseases | 0 (0) | 3 (13.6) | 0.534 |
| Onset days (mean ± SD, days) | 8.4 ± 2.5 | 10.7 ± 2.5 | 0.27 |
| Oxygenation index (mean ± SD, mmHg) | 431 ± 129 | 199 ± 109 | <0.001 |
| Invasive ventilation (n., %) | 0 (0) | 3 (13.6) | 0.534 |
| Non-invasive ventilation (n., %) | 0 (0) | 11 (50) | 0.006 |
| ICU admission (n., %) | 0 (0) | 4 (18.2) | 0.071 |
| Length of stay (mean ± SD, days) | 8.2 ± 1.9 | 16.4 ± 15.3 | 0.022 |
| System corticosteroid therapy (n., %) | 3 (30.0) | 17 (77.3) | 0.018 |
| Mortality(n., %) | 0 (0) | 7 (31.8) | 0.07 |
| Mild-to-Moderate n = 10 | Severe-to-Very Severe n = 22 | p-Value | |
|---|---|---|---|
| Neutrophils | |||
| Differential count (mean ± SD, /μL) | 4100 ± 1700 | 6900 ± 3400 | 0.018 |
| Proportion (%) | 70.3 ± 15.0 | 85.6 ± 6.0 | <0.001 |
| Eosinophils | |||
| Differential count (mean ± SD, /μL) | 90 ± 120 | 2 ± 5 | 0.001 |
| Proportion (%) | 1.50 ± 1.90 | 0.03 ± 0.11 | 0.001 |
| Lymphocytes | |||
| Differential count (mean ± SD, /μL) | 1200 ± 600 | 600 ± 300 | 0.002 |
| Proportion (%) | 21.8 ± 12.5 | 9.49 ± 5.05 | <0.001 |
| PCT (mean ± SD, ng/mL) | 0.56 ± 1.22 | 0.24 ± 0.24 | 0.234 |
| CRP (mean ± SD, mg/dL) | 7.0 ± 5.8 | 9.5 ± 9.4 | 0.473 |
| D-dimer (mean ± SD, μg/mL) | 0.4 ± 0.3 | 9.9 ± 35.7 | 0.411 |
| Mild-to-Moderate n = 10 | Severe-and-Very Severe n = 22 | p-Value | |
|---|---|---|---|
| IL5 (median, IQR, pg/mL) | 0.7 (10.6) | 1.1 (2.6) | 0.826 |
| IL13 (median, IQR, pg/mL) | 6.9 (10.6) | 2.1 (18.0) | 0.366 |
| IL2 (median, IQR, pg/mL) | 1.0 (3.6) | 0 (1.9) | 0.345 |
| IL6 (median, IQR, pg/mL) | 10.1 (30.0) | 5.7 (30.0) | 0.952 |
| IL9 (median, IQR, pg/mL) | 1.3 (10.2) | 5.6 (10.4) | 0.483 |
| IL10 (median, IQR, pg/mL) | 6.3 (7.1) | 4.6 (8.3) | 0.675 |
| INFγ (median, IQR, pg/mL) | 26.8 (34.4) | 12.6 (42.5) | 0.305 |
| TNFα (median, IQR, pg/mL) | 0.5 (81.8) | 0.1 (20.2) | 0.704 |
| IL17A (median, IQR, pg/mL) | 0.9 (3.6) | 0 (2.1) | 0.434 |
| IL17F (median, IQR, pg/mL) | 1.1 (8.4) | 0 (1.4) | 0.163 |
| IL4 (median, IQR, pg/mL) | 2.6 (5.3) | 0.8 (3.0) | 0.219 |
| IL22 (median, IQR, pg/mL) | 3.1 (10.9) | 2.2 (7.6) | 0.795 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Li, X.; Ren, J.; Rao, Y.; Qiao, Y.; Sun, L.; Liang, Y.; Chang, C.; Zhou, Q.; Sun, Y. The Association of Blood Eosinophils and Neutrophils Expressing Eosinophilic Surface Markers with the Severity and Outcome of COVID-19. Microorganisms 2024, 12, 2503. https://doi.org/10.3390/microorganisms12122503
Wang J, Li X, Ren J, Rao Y, Qiao Y, Sun L, Liang Y, Chang C, Zhou Q, Sun Y. The Association of Blood Eosinophils and Neutrophils Expressing Eosinophilic Surface Markers with the Severity and Outcome of COVID-19. Microorganisms. 2024; 12(12):2503. https://doi.org/10.3390/microorganisms12122503
Chicago/Turabian StyleWang, Jun, Xin Li, Jiaqi Ren, Yafei Rao, Yixian Qiao, Lina Sun, Ying Liang, Chun Chang, Qingtao Zhou, and Yongchang Sun. 2024. "The Association of Blood Eosinophils and Neutrophils Expressing Eosinophilic Surface Markers with the Severity and Outcome of COVID-19" Microorganisms 12, no. 12: 2503. https://doi.org/10.3390/microorganisms12122503
APA StyleWang, J., Li, X., Ren, J., Rao, Y., Qiao, Y., Sun, L., Liang, Y., Chang, C., Zhou, Q., & Sun, Y. (2024). The Association of Blood Eosinophils and Neutrophils Expressing Eosinophilic Surface Markers with the Severity and Outcome of COVID-19. Microorganisms, 12(12), 2503. https://doi.org/10.3390/microorganisms12122503





