Macrogenomic Analysis Reveals Soil Microbial Diversity in Different Regions of the Antarctic Peninsula
Abstract
1. Introduction
2. Materials and Methods
2.1. Sites and Sampling
2.2. DNA Extraction and Sequencing
2.3. Data Quality Control
2.4. Species and Functional Annotation
2.5. Statistical Analysis and Data Visualisation
3. Results
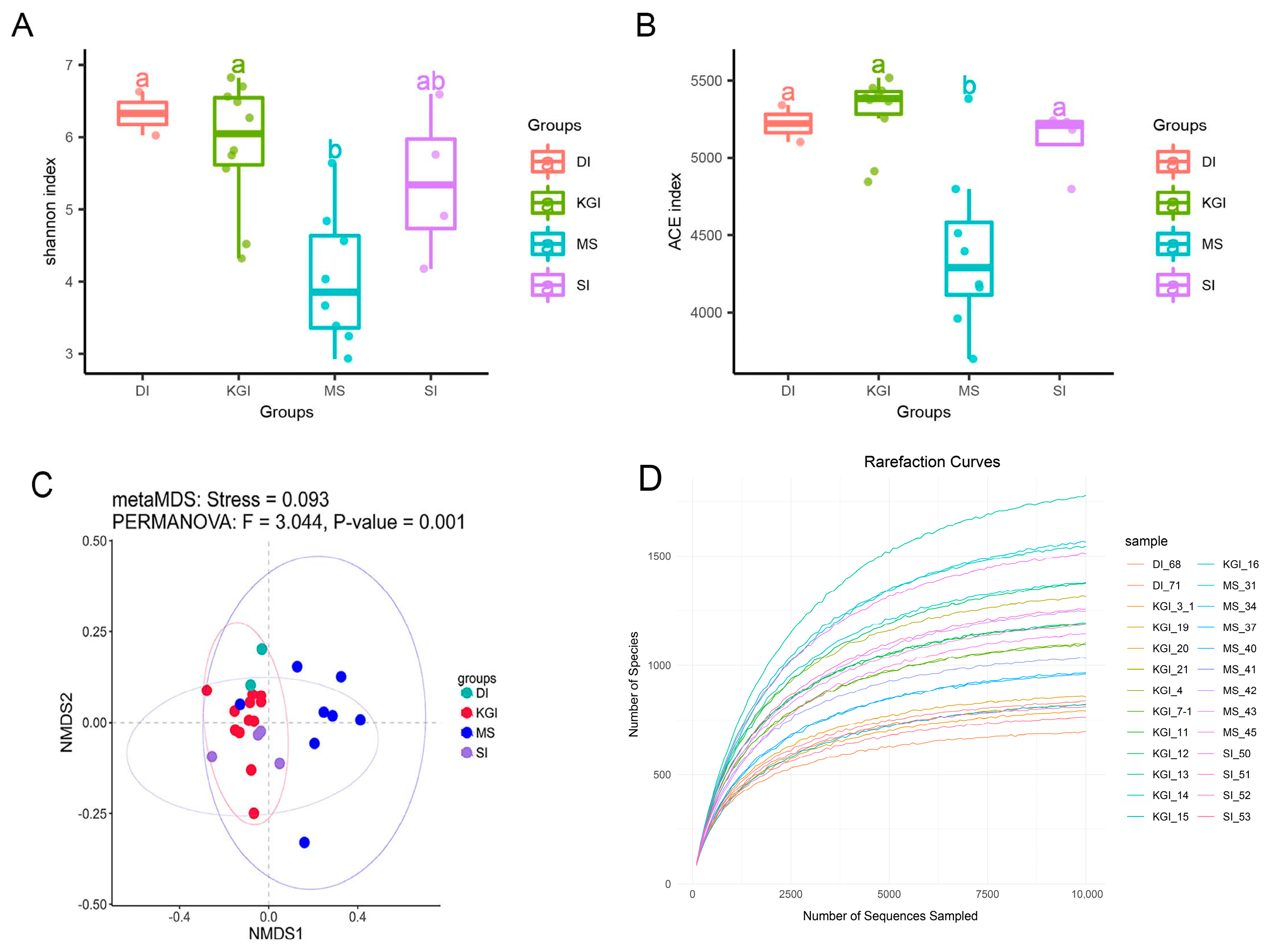
3.1. Species Diversity Analysis
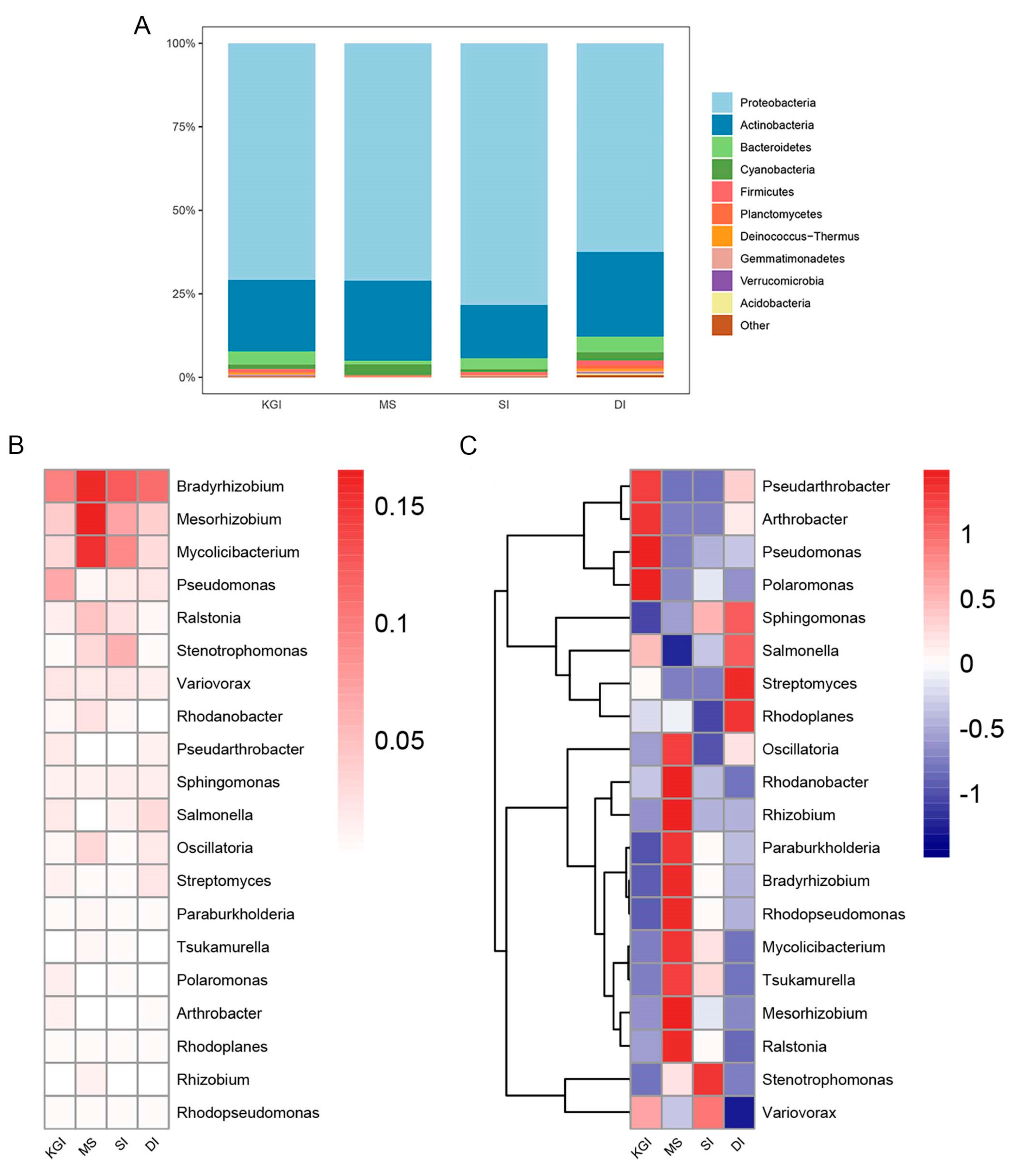
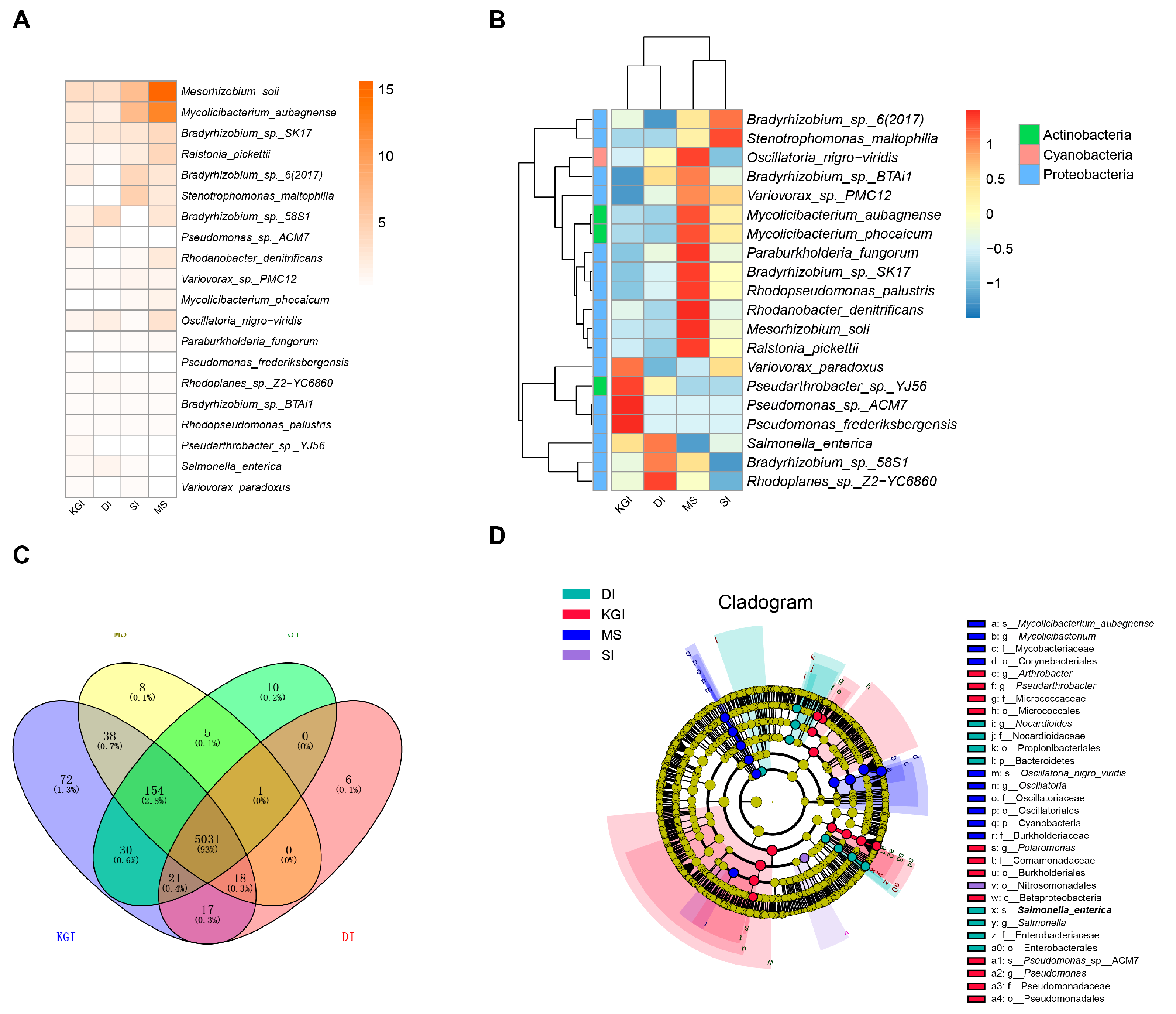
3.2. Taxonomic Profiling of Soil Metagenomes of Antarctic Peninsula
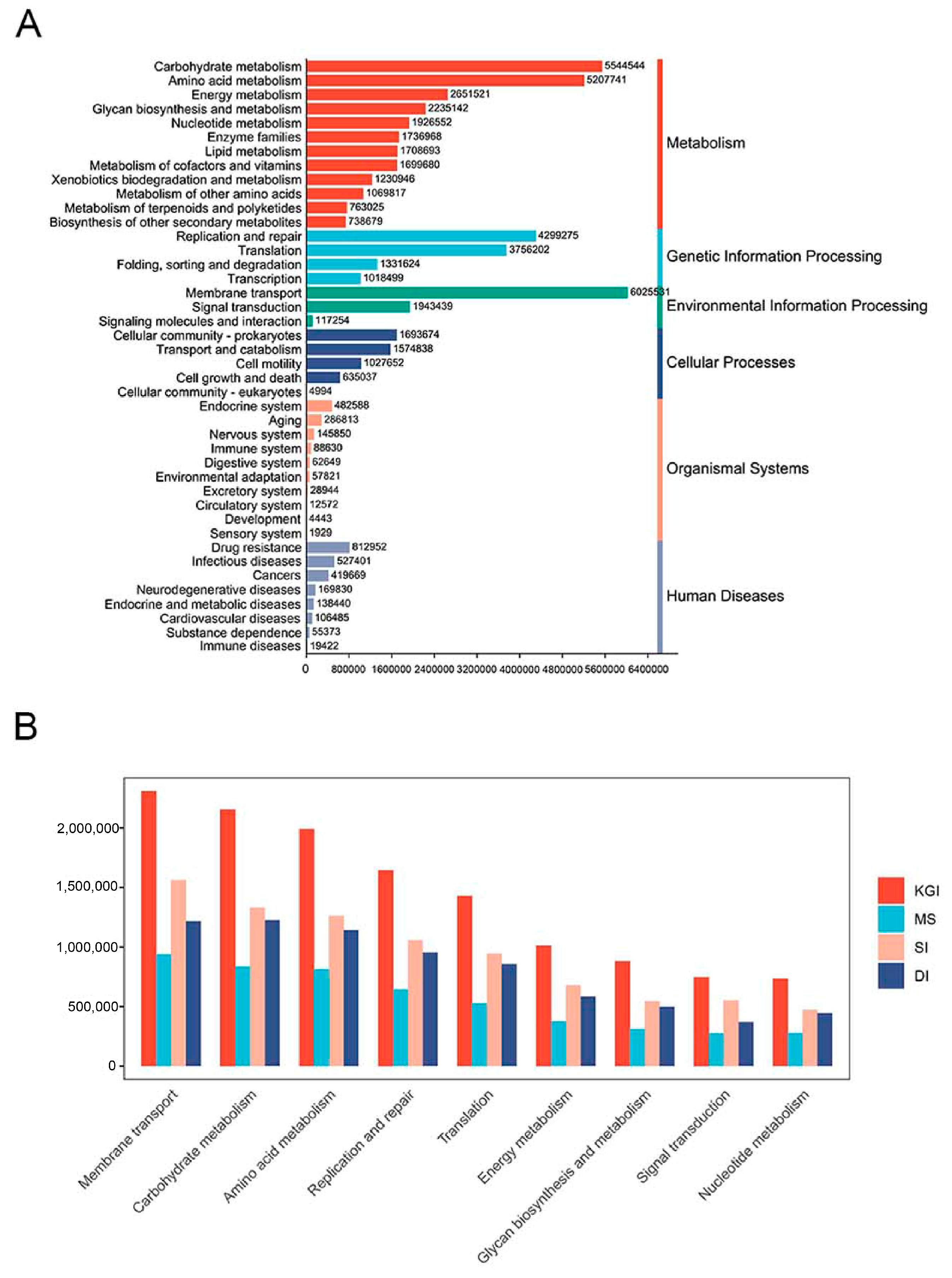
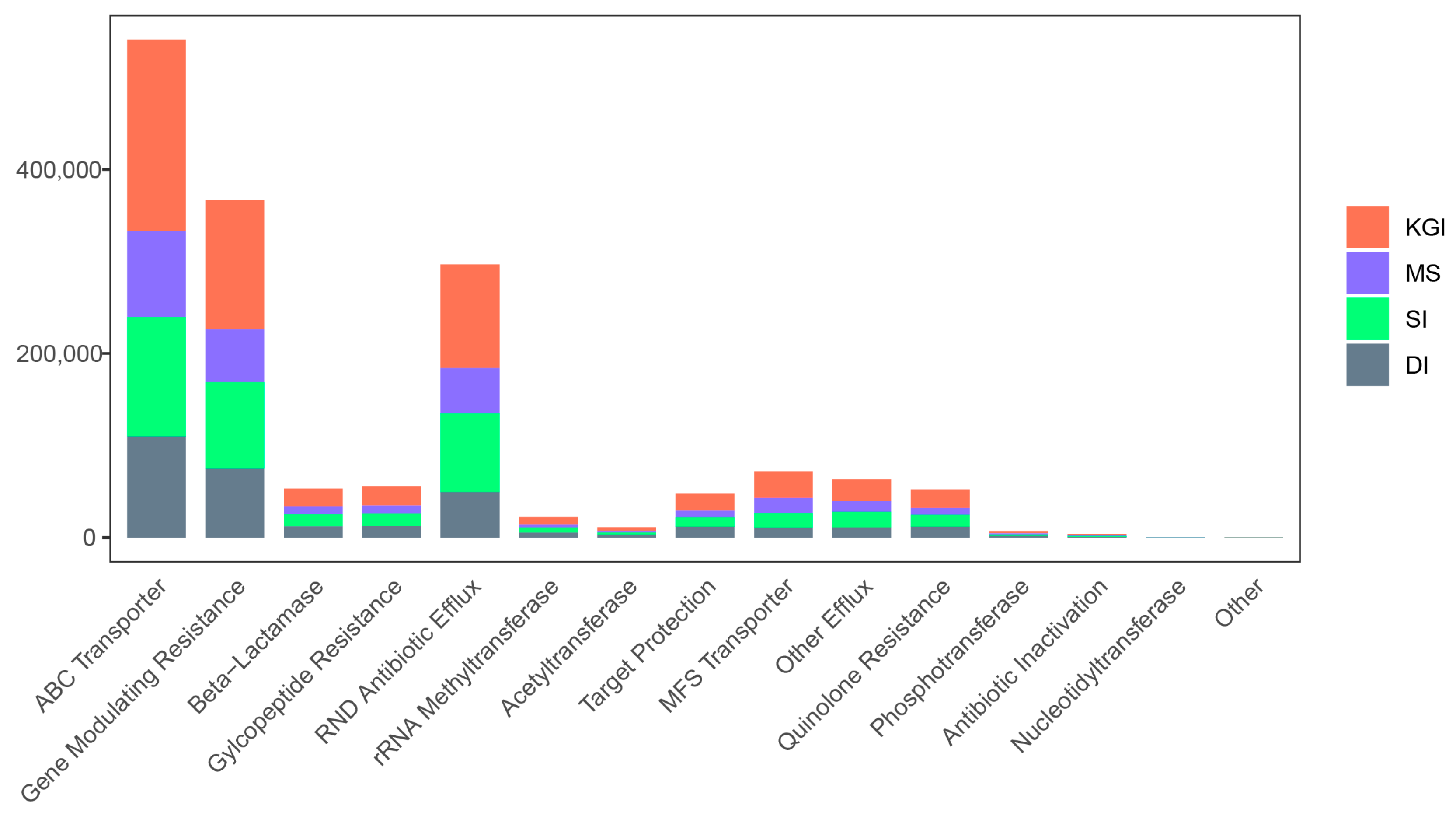
3.3. Functional Composition and Pathways of Soil Microorganisms in the Antarctic Peninsula Region
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruberto, L.; Dias, R.; Lo Balbo, A.; Vazquez, S.C.; Hernandez, E.A.; Mac Cormack, W.P. Influence of nutrients addition and bioaugmentation on the hydrocarbon biodegradation of a chronically contaminated Antarctic soil. J. Appl. Microbiol. 2009, 106, 1101–1110. [Google Scholar] [CrossRef]
- Rampelotto, P.H.; Barboza, A.D.; Pereira, A.B.; Triplett, E.W.; Schaefer, C.E.; de Oliveira Camargo, F.A.; Roesch, L.F. Distribution and interaction patterns of bacterial communities in an ornithogenic soil of Seymour Island, Antarctica. Microb. Ecol. 2015, 69, 684–694. [Google Scholar] [CrossRef]
- Youyu, X. Geomorphologic features and environmental evolution of the Great Wall of China Station area (Fildes Peninsula, King George Island), Antarctica. Chin. Sci. Bull. 1987, 32, 1175–1179. [Google Scholar] [CrossRef]
- Kinasz, C.T.; Kreusch, M.G.; Bendia, A.G.; Pellizari, V.H.; Duarte, R.T.D. Taxonomic and functional diversity from Antarctic ice-tephra microbial community: Ecological insights and potential for bioprospection. An. Acad. Bras. Cienc. 2022, 94, e20210621. [Google Scholar] [CrossRef] [PubMed]
- Kraus, S.; Poblete, F.; Arriagada, C. Dike systems and their volcanic host rocks on King George Island, Antarctica: Implications on the geodynamic history based on a multidisciplinary approach. Tectonophysics 2010, 495, 269–297. [Google Scholar] [CrossRef]
- Global Volcanic Activity Program—Penguin Island. Available online: https://volcano.si.edu/volcano.cfm?vn=390031#:~:text=The%201.4%20x%201.7%20km%20Penguin%20Island%20is,crater%20on%20the%20SW%20side%20of%20the%20island (accessed on 12 November 2024).
- Devil Island—Vega Island. Available online: https://www.ats.aq/devAS/Ats/Guideline/6c6875d4-f0cd-4c67-a5b1-4c8a80cb7322 (accessed on 16 October 2024).
- Almela, P.; Justel, A.; Quesada, A. Heterogeneity of microbial communities in soils from the Antarctic Peninsula region. Front. Microbiol. 2021, 12, 628792. [Google Scholar] [CrossRef] [PubMed]
- Chown, S.L.; Clarke, A.; Fraser, C.I.; Cary, S.C.; Moon, K.L.; McGeoch, M.A. The changing form of Antarctic biodiversity. Nature 2015, 522, 431–438. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, T.; Zhang, F.; Wang, E.T.; He, J.F.; Ding, H.; Zhang, B.T.; Liu, J.; Ran, X.B.; Zang, J.Y. Diversity and structure of soil bacterial communities in the fildes region (maritime Antarctica) as revealed by 454 pyrosequencing. Front. Microbiol. 2015, 6, 1188. [Google Scholar] [CrossRef]
- Tin, T.; Fleming, Z.L.; Hughes, K.A.; Ainley, D.G.; Convey, P.; Moreno, C.A.; Pfeiffer, S.; Scott, J.; Snape, I. Impacts of local human activities on the Antarctic environment. Antarct. Sci. 2009, 21, 3–33. [Google Scholar] [CrossRef]
- Peng, Z.; Liu, G.; Huang, K. Cold Adaptation Mechanisms of a Snow Alga Chlamydomonas nivalis During Temperature Fluctuations. Front. Microbiol. 2020, 11, 611080. [Google Scholar] [CrossRef]
- Hounslow, E.; Evans, C.A.; Pandhal, J.; Sydney, T.; Couto, N.; Pham, T.K.; Gilmour, D.J.; Wright, P.C. Quantitative proteomic comparison of salt stress in Chlamydomonas reinhardtii and the snow alga Chlamydomonas nivalis reveals mechanisms for salt-triggered fatty acid accumulation via reallocation of carbon resources. Biotechnol. Biofuels 2021, 14, 121. [Google Scholar] [CrossRef]
- Grattepanche, J.D.; Jeffrey, W.H.; Gast, R.J.; Sanders, R.W. Diversity of Microbial Eukaryotes Along the West Antarctic Peninsula in Austral Spring. Front. Microbiol. 2022, 13, 844856. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Cheng, Y.; Shi, Q.; Ge, X.; Yang, Y.; Huang, Y. Metagenomic analyses reveal microbial communities and functional differences between Daqu from seven provinces. Food Res. Int. 2023, 172, 113076. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Wen, H.; Shang, Z.; Zou, Y.; Zhao, W.; He, Y.; Yang, S.; Zhang, H.; Qin, J.; Zhu, S.; et al. Macrogenomics reveal the effects of inter-cropping perilla on kiwifruit: Impact on inter-root soil microbiota and gene expression of carbon, nitrogen, and phosphorus cycles in kiwifruit. Front. Microbiol. 2024, 15, 1349305. [Google Scholar] [CrossRef] [PubMed]
- Varliero, G.; Lebre, P.H.; Adams, B.; Chown, S.L.; Convey, P.; Dennis, P.G.; Fan, D.; Ferrari, B.; Frey, B.; Hogg, I.D.; et al. Biogeographic survey of soil bacterial communities across Antarctica. Microbiome 2024, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Schlitzer, R. Data Analysis and Visualization with Ocean Data View. CMOS Bull. SCMO 2015, 43, 9–13. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Menon, R.; Garg, G.; Gasser, R.B.; Ranganathan, S. TranSeqAnnotator: Large-scale analysis of transcriptomic data. BMC Bioinform. 2012, 13 (Suppl. S17), S24. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 623–656. [Google Scholar] [CrossRef]
- Chao, A.; Yang, M.C.K. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 1993, 80, 193–201. [Google Scholar] [CrossRef]
- Wojtowicz, K.; Steliga, T.; Kapusta, P.; Brzeszcz, J.; Skalski, T. Evaluation of the effectiveness of the biopreparation in combination with the polymer γ-PGA for the biodegradation of petroleum contaminants in soil. Materials 2022, 15, 400. [Google Scholar] [CrossRef]
- Njoku, K.L.; Ude, E.O.; Jegede, T.O.; Adeyanju, O.Z.; Iheme, P.O. Characterization of hydrocarbon degrading microorganisms from Glycine max and Zea mays phytoremediated crude oil contaminated soil. Environ. Anal. Health Toxicol. 2022, 37, e2022008. [Google Scholar] [CrossRef]
- Lv, L.; Sun, L.; Yuan, C.; Han, Y.; Huang, Z. The combined enhancement of RL, nZVI and AQDS on the microbial anaerobic-aerobic degradation of PAHs in soil. Chemosphere 2022, 307, 135609. [Google Scholar] [CrossRef]
- Chen, F.; Chen, Y.; Chen, C.; Feng, L.; Dong, Y.; Chen, J.; Lan, J.; Hou, H. High-efficiency degradation of phthalic acid esters (PAEs) by Pseudarthrobacter defluvii E5: Performance, degradative pathway, and key genes. Sci. Total Environ. 2021, 794, 148719. [Google Scholar] [CrossRef] [PubMed]
- Perfumo, A.; Banat, I.M.; Marchant, R. Going green and cold: Biosurfactants from low-temperature environments to biotechnology applications. Trends Biotechnol. 2018, 36, 277–289. [Google Scholar] [CrossRef]
- Tindall, B.J. Prokaryotic diversity in the Antarctic: The tip of the iceberg. Microb. Ecol. 2004, 47, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Li, H.R.; Yuan, M.; Zhang, X.H.; Yu, Y. Deinococcus antarcticus sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 331–335. [Google Scholar] [CrossRef] [PubMed]
- Radziemska, M.; Gusiatin, M.Z.; Cydzik-Kwiatkowska, A.; Blazejczyk, A.; Majewski, G.; Jaskulska, I.; Brtnicky, M. Effect of freeze–thaw manipulation on phytostabilization of industrially contaminated soil with halloysite nanotubes. Sci. Rep. 2023, 13, 22175. [Google Scholar] [CrossRef] [PubMed]
- Gosai, H.B.; Panseriya, H.Z.; Patel, P.G.; Patel, A.C.; Shankar, A.; Varjani, S.; Dave, B.P. Exploring bacterial communities through metagenomics during bioremediation of polycyclic aromatic hydrocarbons from contaminated sediments. Sci. Total Environ. 2022, 842, 156794. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Microbiological Study in Petrol-Spiked Soil. Molecules 2021, 26, 2664. [Google Scholar] [CrossRef]
- Li, J.; Luo, C.; Zhang, D.; Song, M.; Cai, X.; Jiang, L.; Zhang, G. Autochthonous Bioaugmentation-Modified Bacterial Diversity of Phenanthrene Degraders in PAH-Contaminated Wastewater as Revealed by DNA-Stable Isotope Probing. Environ. Sci. Technol. 2018, 52, 2934–2944. [Google Scholar] [CrossRef]
- Campbell, B.J.; Polson, S.W.; Hanson, T.E.; Mack, M.C.; Schuur, E.A. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ. Microbiol. 2010, 12, 1842–1854. [Google Scholar] [CrossRef]
- Zielińska, S.; Matkowski, A.; Dydak, K.; Czerwińska, M.E.; Dziągwa-Becker, M.; Kucharski, M.; Wójciak, M.; Sowa, I.; Plińska, S.; Fijałkowski, K.; et al. Bacterial nanocellulose fortified with antimicrobial and anti-inflammatory natural products from Chelidonium majus plant cell cultures. Materials 2021, 15, 16. [Google Scholar] [CrossRef]
- Allen, E.E.; Facciotti, D.; Bartlett, D.H. Monounsaturated but not polyunsaturated fatty acids are required for growth of the deep-sea bacterium Photobacterium profundum SS9 at high pressure and low temperature. Appl. Environ. Microbiol. 1999, 65, 1710–1720. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002, 357, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Araki, T. An analysis of the effect of changes in growth temperature on proteolysis in vivo in the psychrophilic bacterium Vibrio sp. strain ANT-300. J. Gen. Microbiol. 1992, 138, 2075–2082. [Google Scholar] [CrossRef] [PubMed]
- Johnston, A.W.; Todd, J.D.; Curson, A.R.; Lei, S.; Nikolaidou-Katsaridou, N.; Gelfand, M.S.; Rodionov, D.A. Living without Fur: The subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other α-proteobacteria. BioMetals 2007, 20, 501–511. [Google Scholar] [CrossRef]
- Alam, M.; Fernandes, S.; Mandal, S.; Rameez, M.J.; Bhattacharya, S.; Peketi, A.; Mazumdar, A.; Ghosh, W. 34S enrichment as a signature of thiosulfate oxidation in the “Proteobacteria”. FEMS Microbiol. Lett. 2021, 368, fnab073. [Google Scholar] [CrossRef]
- Alwali, A.Y.; Parkinson, E.I. Small molecule inducers of actinobacteria natural product biosynthesis. J. Ind. Microbiol. Biotechnol. 2023, 50, kuad019. [Google Scholar] [CrossRef]
- Golinska, P.; Wypij, M.; Agarkar, G.; Rathod, D.; Dahm, H.; Rai, M. Endophytic actinobacteria of medicinal plants: Diversity and bioactivity. Antonie van Leeuwenhoek 2015, 108, 267–289. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, S.; Nie, Q.; He, H.; Tan, H.; Geng, F.; Ji, H.; Hu, J.; Nie, S. Gut firmicutes: Relationship with dietary fiber and role in host homeostasis. Crit. Rev. Food Sci. Nutr. 2023, 63, 12073–12088. [Google Scholar] [CrossRef]
- Pereira, B.K.; Rosa, R.M.; Silva, J.d.; Guecheva, T.N.; Oliveira, I.M.d.; Ianistcki, M.; Benvegnú, V.C.; Furtado, G.V.; Ferraz, A.; Richter, M.F.; et al. Protective effects of three extracts from Antarctic plants against ultraviolet radiation in several biological models. J. Photochem. Photobiol. B Biol. 2009, 96, 117–129. [Google Scholar] [CrossRef]
- Cordero, R.R.; Feron, S.; Damiani, A.; Redondas, A.; Carrasco, J.; Sepúlveda, E.; Jorquera, J.; Fernandoy, F.; Llanillo, P.; Rowe, P.M.; et al. Persistent extreme ultraviolet irradiance in Antarctica despite the ozone recovery onset. Sci. Rep. 2022, 12, 1266. [Google Scholar] [CrossRef]
- Okada, U.; Murakami, S. Structural and functional characteristics of the tripartite ABC transporter. Microbiology 2022, 168, 001257. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ecker, G.F. A Structure-Based View on ABC-Transporter Linked to Multidrug Resistance. Molecules 2023, 28, 495. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Picazo, P.; Roscales, G.; Toribio-Avedillo, D.; Gómez-Gómez, C.; Avila, C.; Ballesté, E.; Muniesa, M.; Rodríguez-Rubio, L. Antibiotic Resistance Genes in Phage Particles from Antarctic and Mediterranean Seawater Ecosystems. Microorganisms 2020, 8, 1293. [Google Scholar] [CrossRef]
- Jara, D.; Bello-Toledo, H.; Domínguez, M.; Cigarroa, C.; Fernández, P.; Vergara, L.; Quezada-Aguiluz, M.; Opazo-Capurro, A.; Lima, C.A.; González-Rocha, G. Antibiotic resistance in bacterial isolates from freshwater samples in Fildes Peninsula, King George Island, Antarctica. Sci. Rep. 2020, 10, 3145. [Google Scholar] [CrossRef]
- Akram, F.; Haq, I.U.; Shah, F.I.; Aqeel, A.; Ahmed, Z.; Mir, A.S.; Qureshi, S.S.; Raja, S.I. Genus Thermotoga: A valuable home of multifunctional glycoside hydrolases (GHs) for industrial sustainability. Bioorg. Chem. 2022, 127, 105942. [Google Scholar] [CrossRef]
- McArthur, J.B.; Chen, X. Glycosyltransferase engineering for carbohydrate synthesis. Biochem. Soc. Trans. 2016, 44, 129–142. [Google Scholar] [CrossRef]
- Aakko, J.; Pietilä, S.; Toivonen, R.; Rokka, A.; Mokkala, K.; Laitinen, K.; Elo, L.; Hänninen, A. A carbohydrate-active enzyme (CAZy) profile links successful metabolic specialization of Prevotella to its abundance in gut microbiota. Sci. Rep. 2020, 10, 12411. [Google Scholar] [CrossRef]
- Song, Q.; Wang, B.; Han, Y.; Zhou, Z. Metagenomics Reveals the Diversity and Taxonomy of Carbohydrate-Active Enzymes and Antibiotic Resistance Genes in Suancai Bacterial Communities. Genes 2022, 13, 773. [Google Scholar] [CrossRef]
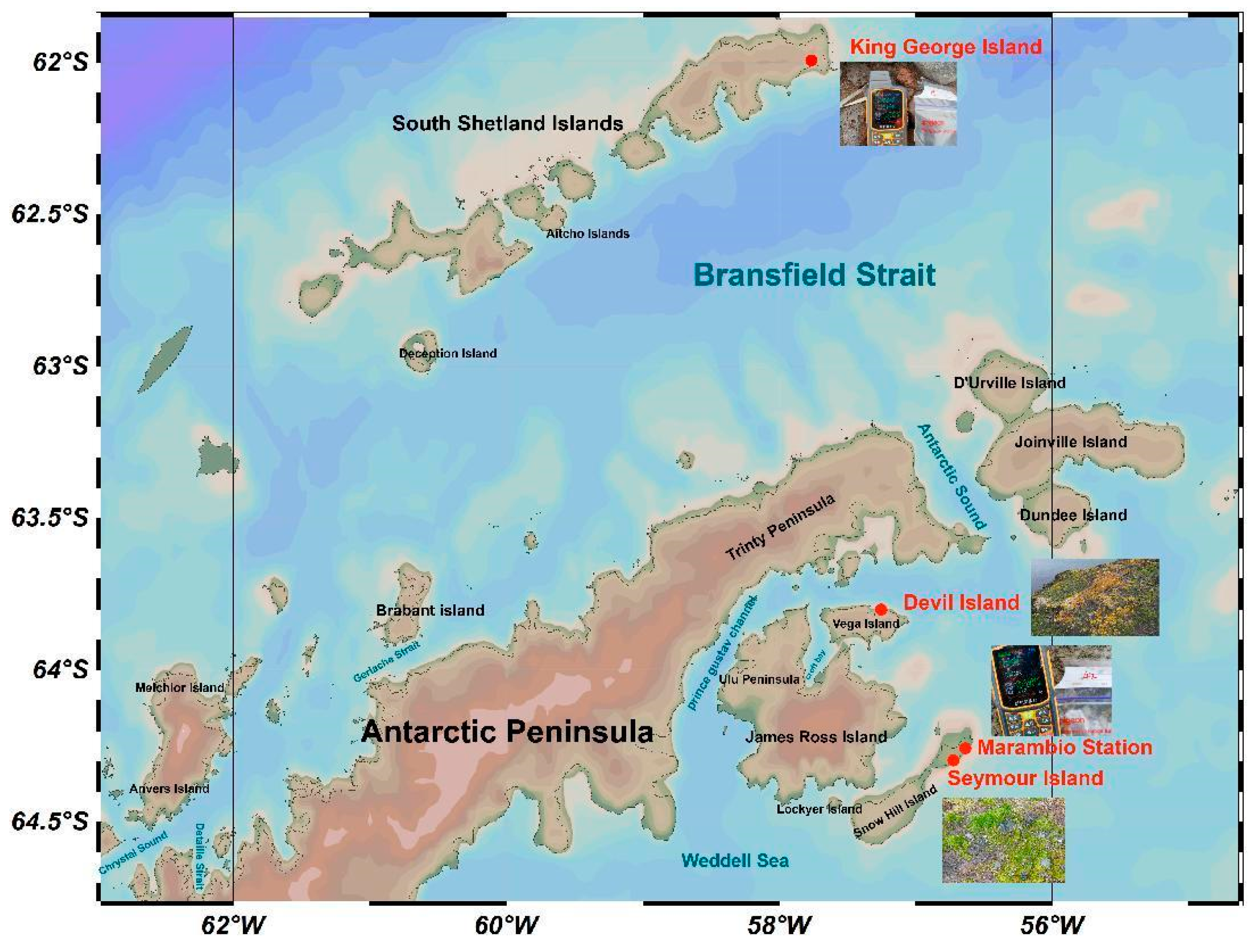



| Sample Number | Sampling Area | Longitude | Latitude | Altitude (m.a.s.l) | Sampling Time | Accession No. |
|---|---|---|---|---|---|---|
| KGI-3-1 | KGI | −57.7076 | −62.0197 | 7 | 31 January 2020 | SRR22742764 |
| KGI-4 | KGI | −57.7076 | −62.0197 | 8 | 31 January 2020 | SRR22742763 |
| KGI-7-1 | KGI | −57.7078 | −62.0199 | 9 | 31 January 2020 | SRR22742752 |
| KGI-11 | KGI | −57.7062 | −62.0213 | 10 | 31 January 2020 | SRR22742745 |
| KGI-12 | KGI | −57.7062 | −62.0213 | 10 | 31 January 2020 | SRR22742744 |
| KGI-13 | KGI | −57.7058 | −62.0214 | 14 | 31 January 2020 | SRR22742743 |
| KGI-14 | KGI | −57.7056 | −62.0215 | 17 | 31 January 2020 | SRR22742742 |
| KGI-15 | KGI | −57.7054 | −62.0217 | 23 | 31 January 2020 | SRR22742741 |
| KGI-16 | KGI | −57.7054 | −62.0219 | 23 | 31 January 2020 | SRR22742740 |
| KGI-19 | KGI | −57.7042 | −62.0225 | 41 | 31 January 2020 | SRR22742739 |
| KGI-20 | KGI | −57.6983 | −62.0213 | 86 | 31 January 2020 | SRR22742762 |
| KGI-21 | KGI | −57.6946 | −62.0212 | 123 | 31 January 2020 | SRR22742761 |
| MS-31 | MS | −56.6122 | −64.2539 | 34 | 2 February 2020 | SRR22742760 |
| MS-34 | MS | −56.6118 | −64.2536 | 26 | 2 February 2020 | SRR22742759 |
| MS-37 | MS | −56.6118 | −64.2534 | 19 | 2 February 2020 | SRR22742758 |
| MS-40 | MS | −56.6108 | −64.2531 | 13 | 2 February 2020 | SRR22742757 |
| MS-41 | MS | −56.6108 | −64.2531 | 15 | 2 February 2020 | SRR22742756 |
| MS-42 | MS | −56.6108 | −64.2531 | 24 | 2 February 2020 | SRR22742755 |
| MS-43 | MS | −56.6108 | −64.2530 | 22 | 2 February 2020 | SRR22742753 |
| MS-45 | MS | −56.6108 | −64.2530 | 25 | 2 February 2020 | SRR22742754 |
| SI-50 | SI | −56.68889 | −64.2899 | 8 | 4 February 2020 | SRR22742751 |
| SI-51 | SI | −56.68951 | −64.2897 | 11 | 4 February 2020 | SRR22742750 |
| SI-52 | SI | −56.6914 | −64.2894 | 9 | 4 February 2020 | SRR22742748 |
| SI-53 | SI | −56.6906 | −64.2896 | 8 | 4 February 2020 | SRR22742749 |
| DI-68 | DI | −57.2944 | −63.7997 | 78 | 5 February 2020 | SRR22742747 |
| DI-71 | DI | −57.2901 | −63.8022 | 23 | 5 February 2020 | SRR22742746 |
| Resistance Gene | Mechanism Classification |
|---|---|
| AAC6-Ia | Acetyltransferase |
| Subclass B2 | β-lactamase |
| VIM | β-lactamase |
| SubclassB1 | β-lactamase |
| ErmA | rRNA Methyltransferase |
| adeA-adeI | RND Antibiotic Efflux |
| mecR1 | Gene modulating resistance |
| ErmC | rRNA methyltransferase |
| TEM | β-lactamase |
| tolC | ABC transporter |
| AAC6-Ib | Acetyltransferase |
| ANT | Nucleotidyltransferase |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, J.; Lu, X.; Liu, T.; Qu, Y.; Xing, Z.; Wang, S.; Jing, S.; Zheng, L.; Wang, L.; Wang, X. Macrogenomic Analysis Reveals Soil Microbial Diversity in Different Regions of the Antarctic Peninsula. Microorganisms 2024, 12, 2444. https://doi.org/10.3390/microorganisms12122444
Qu J, Lu X, Liu T, Qu Y, Xing Z, Wang S, Jing S, Zheng L, Wang L, Wang X. Macrogenomic Analysis Reveals Soil Microbial Diversity in Different Regions of the Antarctic Peninsula. Microorganisms. 2024; 12(12):2444. https://doi.org/10.3390/microorganisms12122444
Chicago/Turabian StyleQu, Jiangyong, Xiaofei Lu, Tianyi Liu, Ying Qu, Zhikai Xing, Shuang Wang, Siluo Jing, Li Zheng, Lijun Wang, and Xumin Wang. 2024. "Macrogenomic Analysis Reveals Soil Microbial Diversity in Different Regions of the Antarctic Peninsula" Microorganisms 12, no. 12: 2444. https://doi.org/10.3390/microorganisms12122444
APA StyleQu, J., Lu, X., Liu, T., Qu, Y., Xing, Z., Wang, S., Jing, S., Zheng, L., Wang, L., & Wang, X. (2024). Macrogenomic Analysis Reveals Soil Microbial Diversity in Different Regions of the Antarctic Peninsula. Microorganisms, 12(12), 2444. https://doi.org/10.3390/microorganisms12122444





