The Natural Whey Starter Used in the Production of Grana Padano and Parmigiano Reggiano PDO Cheeses: A Complex Microbial Community †
Abstract
:1. Introduction
2. Cheese as a Complex Microbial Ecosystem
3. Grana Padano and Parmigiano Reggiano Cheeses
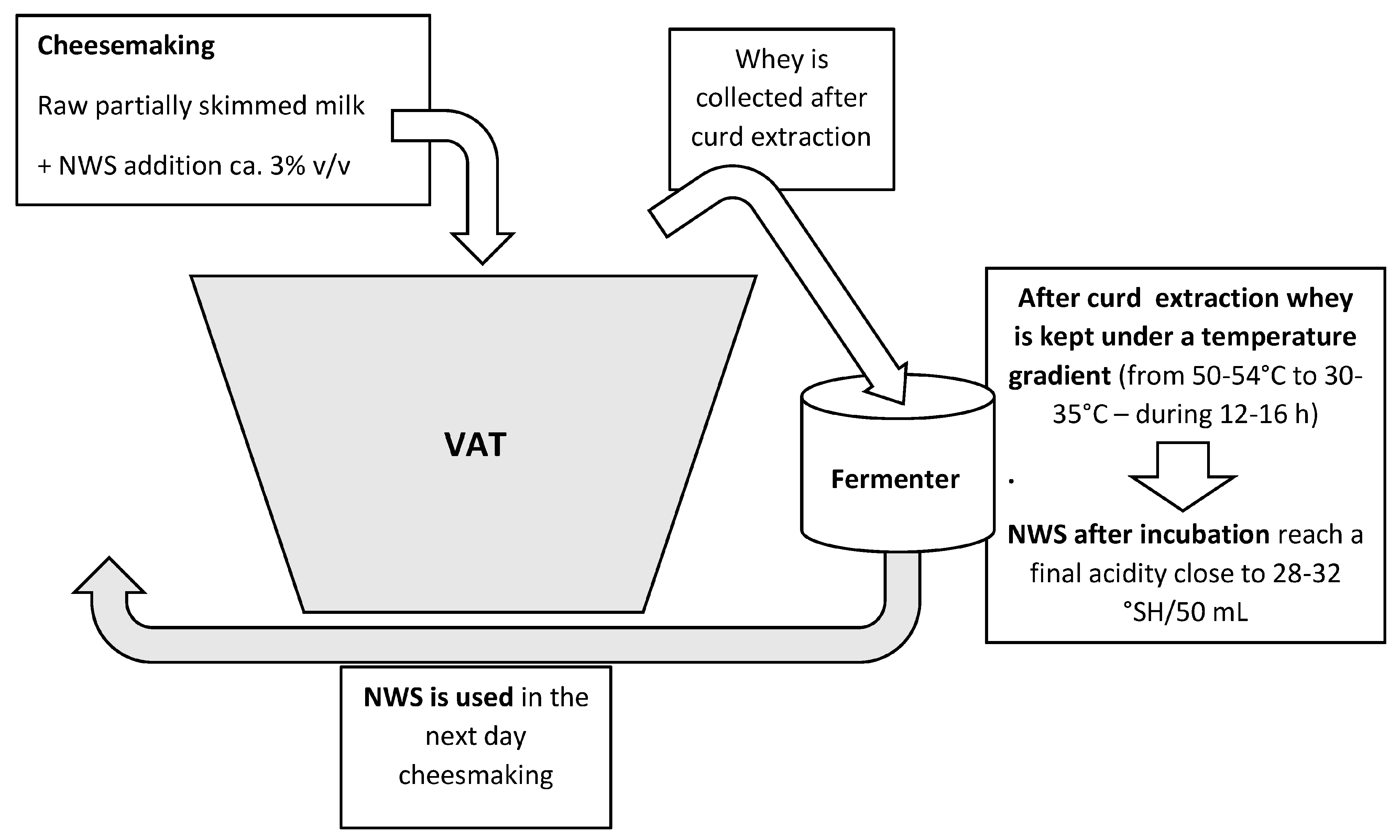
4. Natural Whey Starter—A Peculiar Complex Microbial Ecosystems
5. Microbial Interaction and Complexity in Natural Consortia: NWS as a Model for Functional Microbial Ecosystem
6. Conclusions
Funding
Conflicts of Interest
References
- Avery, S.V. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 2006, 4, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, F.M.; Ameur, H.; Nikoloudaki, O.; Celano, G.; Vacca, M.; JFLemos, W., Jr.; Manzari, C.; Vertè, F.; Di Cagno, R.; Pesole, G.; et al. Metabolic framework of spontaneous and synthetic sourdough metacommunities to reveal microbial players responsible for resilience and performance. Microbiome 2022, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Gobbetti, M.; Neviani, E.; Calasso, M.; De Angelis, M.; Fox, F.P.; Di Cagno, R. Drivers that establish and assembly the lactic acid bacteria biota in cheeses. Trends Food Sci. Technol. 2018, 78, 244–254. [Google Scholar] [CrossRef]
- Gatti, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Mucchetti, G. Invited Review: Microbial evolution in raw milk, long-ripened cheeses produced using undefined natural whey starters. J. Dairy Sci. 2014, 97, 573–591. [Google Scholar] [CrossRef]
- Neviani, E.; Levante, A.; Gatti, M. The Microbial Community of Natural Whey Starter: Why Is It a Driver for the Production of the Most Famous Italian Long-Ripened Cheeses? Fermentation 2024, 10, 186–200. [Google Scholar] [CrossRef]
- De Pasquale, I.; Di Cagno, R.; Buchin, S.; De Angelis, M.; Gobbetti, M. Microbial ecology dynamics reveal a succession in the core microbiota that is involved in the ripening of pasta-filata Caciocavallo Pugliese cheese. Appl. Environ. Microbiol. 2014, 80, 6243–6255. [Google Scholar] [CrossRef]
- Giraffa, G.; Mucchetti, G.; Neviani, E. Interactions among thermophilic lactobacilli during growth in cheese whey. J. Appl. Bacteriol. 1996, 80, 199. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Pot, B.; Tsakalidou, E. How microbes adapt to a diversity of food niches. Curr. Opin. Food Sci. 2015, 2, 29–35. [Google Scholar] [CrossRef]
- Juillard, V.; Spinnler, H.E.; Desmazeaud, M.J.; Boquien, C.Y. Phénomènes de coopération et d’inhibition entre les bactéries lactiques utilisées en industrie laitière. Lait 1987, 67, 149–172. [Google Scholar] [CrossRef]
- Fox, P.F.; Guinee, T.P.; Cogan, T.M.; McSweeney, P.L.H. Fundamentals of Cheese Science, 2nd ed.; Springer International Publishing: New York, NY, USA, 2000. [Google Scholar]
- Alais, C. Sciences du Lait, Principes des Techniques Laitières, 4ème éd.; SEPAIC: Paris, France, 1984. [Google Scholar]
- Gobbetti, M.; Neviani, E.; Fox, P. The Cheeses of Italy: Science and Technology; Springer International Publishing: New York, NY, USA, 2018. [Google Scholar]
- Afsharia, R.; Pillidgea, C.J.; Diasb, D.A.; Osborna, A.M.; Gilla, H. Cheesomics: The future pathway to understanding cheese flavour and quality. Crit. Rev. Food Sci. Nutr. 2020, 60, 33–47. [Google Scholar] [CrossRef]
- Ercolini, D. Secrets of the cheese microbiome. Nat. Food 2020, 1, 466–467. [Google Scholar] [CrossRef] [PubMed]
- Parente, E.; Ricciardi, A.; Zotta, T. The microbiota of dairy milk: A review. Int. Dairy J. 2020, 107, 104714. [Google Scholar] [CrossRef]
- Parente, E.; Guidone, A.; Matera, A.; De Filippis, F.; Mauriello, G.; Ricciardi, A. Microbial community dynamics in thermophilic undefined milk starter cultures. Int. J. Food Microbiol. 2016, 217, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Cocolin, L.; Gobbetti, M.; Neviani, E.; Daffonchio, D. Ensuring safety in artisanal food microbiology. Nat. Microbiol. 2016, 1, 16171. [Google Scholar] [CrossRef]
- Mucchetti, G.; Neviani, E. Microbiologia e Tecnologia Lattiero-Casearia, Qualità e Sicurezza; Tecniche Nuove: Milan, Italy, 2006; ISBN 8848118178. [Google Scholar]
- Mucchetti, G.; Neviani, E. Tecnologia Casearia Dall’empirismo All’industria; CEA Casa Editrice Ambrosiana: Milan, Italy, 2022. [Google Scholar]
- McSweeney, P.L.H.; Fox, P.F.; Cotter, P.D.; Everett, W.D. Cheese: Chemistry, Physics and Microbiology, 4th ed.; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Randazzo, C.L.; Caggia, C.; Neviani, E. Cheese Ripening: Quality, Safety and Health Aspects; Nova Science Publishers: Hauppauge NY, USA, 2013. [Google Scholar]
- Gatti, M.; Fornasari, M.E.; Mucchetti, G.; Addeo, F.; Neviani, E. Presence of peptidase activities in different varietes of cheese. Lett. Appl. Microbiol. 1999, 28, 368. [Google Scholar] [CrossRef]
- Gatti, M.; De Dea Lindner, J.; Gardini, F.; Mucchetti, G.; Bevacqua, D.; Fornasari, M.E.; Neviani, E. A Model to Assess Microbial Enzyme Activities in Parmigiano Reggiano. J. Dairy Sci. 2008, 91, 4129–4137. [Google Scholar] [CrossRef]
- Sgarbi, E.; Bottari, B.; Gatti, M.; Neviani, E. Investigation of the ability of dairy nonstarter lactic acid bacteria to grow using cell lysates of other lactic acid bacteria as the exclusive source of nutrients. Int. J. Dairy Technol. 2014, 67, 342–347. [Google Scholar] [CrossRef]
- Rossetti, L.; Fornasari, M.E.; Monica, G.; Lazzi, C.; Neviani, E.; Giraffa, G. Grana Padano cheese whey starters: Microbial composition and strain distribution. Int. J. Food Microbiol. 2008, 127, 168–171. [Google Scholar] [CrossRef]
- Santarelli, M.; Bottari, B.; Lazzi, C.; Neviani, E.; Gatti, M. Survey on the community and dynamics of lactic acid bacteria in Grana Padano cheese. Syst. Appl. Microbiol. 2013, 36, 593–600. [Google Scholar] [CrossRef]
- Bertani, G.; Levante, A.; Lazzi, C.; Bottari, B.; Gatti, M.; Neviani, E. Dynamics of a natural bacterial community under technological and environmental pressures: The case of natural whey starter for Parmigiano Reggiano cheese. Food Res. Int. 2020, 129, 108860. [Google Scholar] [CrossRef]
- Bottari, B.; Santarelli, M.; Neviani, E.; Gatti, M. Natural whey starter for Parmigiano Reggiano: Culture-independent approach. J. Appl. Microbiol. 2010, 108, 1676–1684. [Google Scholar] [CrossRef]
- Gatti, M.; Bernini, V.; Lazzi, C.; Neviani, E. Fluorescence microscopy for studing the viability of micro-organisms in natural whey starters. Lett. Appl. Microbiol. 2006, 42, 338–343. [Google Scholar] [CrossRef]
- Zago, M.; Fornasari, M.E.; Rossetti, L.; Bonvini, B.; Scano, L.; Carminati, D. Population dynamics of lactobacilli in Grana cheese. Ann. Microbiol. 2007, 57, 349–353. [Google Scholar] [CrossRef]
- Giraffa, G.; Neviani, E. Different Lactobacillus helveticus strain populations dominate during Grana Padano cheesemaking. Food Microbiol. 1999, 16, 205–210. [Google Scholar] [CrossRef]
- Giraffa, G.; Rossetti, L.; Mucchetti, G.; Addeo, F.; Neviani, E. Influence of the temperature gradient on the growth of thermophilic lactobacilli used as natural starter in Grana cheese. J. Dairy Sci. 1998, 81, 31–36. [Google Scholar] [CrossRef]
- Giraffa, G.; Lazzi, C.; Gatti, M.; Rossetti, L.; Mora, D.; Neviani, E. Molecular typing of Lactobacillus delbrueckii of dairy origin by RFLP of protein coding genes. Int.J. Food Microbiol. 2003, 82, 163–172. [Google Scholar] [CrossRef]
- Gatti, M.; Lazzi, C.; Rossetti, L.; Mucchetti, G.; Neviani, E. Biodiversity in Lactobacillus helveticus strains in natural whey starter used for Parmigiano Reggiano Cheese. J. Appl. Microbiol. 2003, 95, 463–480. [Google Scholar] [CrossRef]
- Giraffa, G.; Andrighetto, C.; Antonello, C.; Gatti, M.; Lazzi, C.; Marcazzan, G.; Lombardi, A.; Neviani, E. Genotypic and phenotypic diversity of Lactobacillus delbrueckii subsp. lactis strains of dairy origin. Int. J. Food Microbiol. 2004, 91, 129–139. [Google Scholar] [CrossRef]
- Gatti, M.; Trivisano, C.; Fabrizi, E.; Neviani, E.; Gardini, F. Biodiversity within Lactobacillus helveticus isolated from different natural whey starter cultures as revealed by classification trees. Appl. Environ. Microbiol. 2004, 70, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Lazzi, C.; Rossetti, L.; Zago, M.; Neviani, E.; Giraffa, G. Evaluation of bacterial comunities belonging to natural whey starters by lenghth heterogeneity-PCR. J. Appl. Microbiol. 2004, 96, 481–490. [Google Scholar] [CrossRef]
- De Dea Lindner, J.; Bernini, V.; De Lorentiis, A.; Pecorari, A.; Neviani, E.; Gatti, M. Parmigiano Reggiano cheese: Evolution of cultivable and total lactic microflora and peptidase activities during manufacture and ripening. Dairy Sci. Technol. 2008, 88, 511–523. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román CM, B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s microbes: Tools for distinguishing the living from the dead in microbial ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Carminati, D.; Mazzucotelli, L.; Giraffa, G.; Neviani, E. Incidence of inducible bacteriophage in Lactobacillus helveticus strains isolated from natural whey starter cultures. J. Dairy Sci. 1997, 80, 1505–1511. [Google Scholar] [CrossRef]
- Mancini, A.; Rodriguez, M.C.; Zago, M.; Cologna, N.; Goss, A.; Carafa, I.; Tuohy, K.; Merz, A.; Franciosi, E. Massive Survey on Bacterial-Bacteriophages Biodiversity and Quality of Natural Whey Starter Cultures in Trentingrana Cheese Production. Front. Microbiol. 2021, 12, 678012. [Google Scholar] [CrossRef]
- Spus, M.; Li, M.; Alexeeva, S.; Zwietering, M.H.; Abee, T.; Smid, E.J. Strain diversity and phage resistance in complex dairy starter cultures. J. Dairy Sci. 2015, 98, 5173–5182. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar]
- Bleuven, C.; Landry, C.R. Molecular and cellular bases of adaptation to a changing environment in microorganisms. Proc. Biol. Sci. 2016, 283, 20161458. [Google Scholar] [CrossRef]
- Margulis, L.; Sagan, D. Acquiring Genomes: A Theory of the Origin of Species; Basic Books: New York, NY, USA, 2002. [Google Scholar]
- Margulis, L. The Symbiotic Planet. A New Look at Evolution; Phoenix: London, UK, 1999. [Google Scholar]
- Margulis, L.; Fester, R. Symbiosis as a Source of Evolutionary Innovation: Special Ion and Morphogenesis; Lynn Margulis and MIT Press: Cambridge, MA, USA; London, UK, 1991. [Google Scholar]
- Shapiro, J.A. Bacteria as multicellular organisms. Sci. Am. 1988, 256, 82–89. [Google Scholar] [CrossRef]
- Shapiro, J.A. Thinking about bacterial populations as multicellular organisms. Annu. Rev. Microbiol. 1998, 52, 81–104. [Google Scholar] [CrossRef]
- Bell, G. Experimental macroevolution. Proc. Biol. Sci. 2016, 13, 283. [Google Scholar] [CrossRef]
- Elena, S.F.; Lenski, R.E. Evolution experiments with microorganisms: The dynamics and genetic bases of adaptation. Nat. Rev. Genet. 2003, 4, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Brookfield, J.F.Y. Evolution and evolvability: Celebrating Darwin 200. Biol. Lett. 2009, 5, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Booth, I.R. Stress and the single cell: Intrapopulation diversity is a mechanism to ensure survival upon exposure to stress. Int. J. Food Microbiol. 2002, 78, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Jeanson, S.; Floury, J.; Gagnaire, V.; Lortal, S.; Thierry, A. Bacterial Colonies in Solid Mediaand Foods: A Review on Their Growth and Interactions with the Micro-Environment. Front. Microbiol. 2015, 6, 1284. [Google Scholar] [CrossRef] [PubMed]
- Skandamis, P.N.; Jeanson, S. Colonial vs. planktonic type of growth: Mathematical modeling of microbial dynamics on surfaces and in liquid, semiliquid and solid foods. Front. Microbiol. 2015, 6, 1178. [Google Scholar] [CrossRef]
- Ryall, B.; Eydallin, G.; Ferenci, T. Culture History and Population Heterogeneity as Determinants of Bacterial Adaptation: The Adaptomics of a Single Environmental Transition. Microbiol. Mol. Biol. Rev. 2012, 76, 597–625. [Google Scholar] [CrossRef]
- Jousset, A.; Schmid, B.; Scheu, S.; Eisenhauer, N. Genotypic richness and dissimilarity opposingly affect ecosystem functioning. Ecol. Lett. 2011, 14, 537–545. [Google Scholar] [CrossRef]
- Konopka, A. What is microbial community ecology? ISME J. 2009, 3, 1223–1230. [Google Scholar] [CrossRef]
- O’Manley, M.A. Phylosopy of Microbiology; Cambridge University Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Friedman, J.; Higgins, L.M.; Gore, J. Community structure follows simple assembly rules in microbial microcosms. Nat. Ecol. Evol. 2017, 1, 109. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neviani, E. The Natural Whey Starter Used in the Production of Grana Padano and Parmigiano Reggiano PDO Cheeses: A Complex Microbial Community. Microorganisms 2024, 12, 2443. https://doi.org/10.3390/microorganisms12122443
Neviani E. The Natural Whey Starter Used in the Production of Grana Padano and Parmigiano Reggiano PDO Cheeses: A Complex Microbial Community. Microorganisms. 2024; 12(12):2443. https://doi.org/10.3390/microorganisms12122443
Chicago/Turabian StyleNeviani, Erasmo. 2024. "The Natural Whey Starter Used in the Production of Grana Padano and Parmigiano Reggiano PDO Cheeses: A Complex Microbial Community" Microorganisms 12, no. 12: 2443. https://doi.org/10.3390/microorganisms12122443
APA StyleNeviani, E. (2024). The Natural Whey Starter Used in the Production of Grana Padano and Parmigiano Reggiano PDO Cheeses: A Complex Microbial Community. Microorganisms, 12(12), 2443. https://doi.org/10.3390/microorganisms12122443





