Targeted Variant Assessments of Human Endogenous Retroviral Regions in Whole Genome Sequencing Data Reveal Retroviral Variants Associated with Papillary Thyroid Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Cancer Genome Atlas and 1000 Genomes Project Data
2.2. Identification of HERVs Near or Within Differentially Expressed Cancer Predisposition Genes
2.3. Targeted Variant Discovery in TCGA Data
2.4. Determination of Ancestral Populations
2.5. Statistical Analysis
2.6. Functional in Silico Predictions
2.7. Chemicals and Reagents
2.8. Cell Culture
2.9. Genomic DNA Extraction
2.10. Targeted Genotyping by Sanger Sequencing
2.11. Visualization and Data Processing
3. Results
3.1. Cancer Predisposition Genes (CPGs) in PTC Include a Total of 3725 HERV Sequences Within or in Close Proximity
3.2. Targeted Variant Calling Revealed 612,603 High-Quality Variants Within CPG-Associated HERV Regions
3.3. Multivariate Analyses Revealed Strong Confounding Effects of Gender and Ancestral Profile on HERV Variants
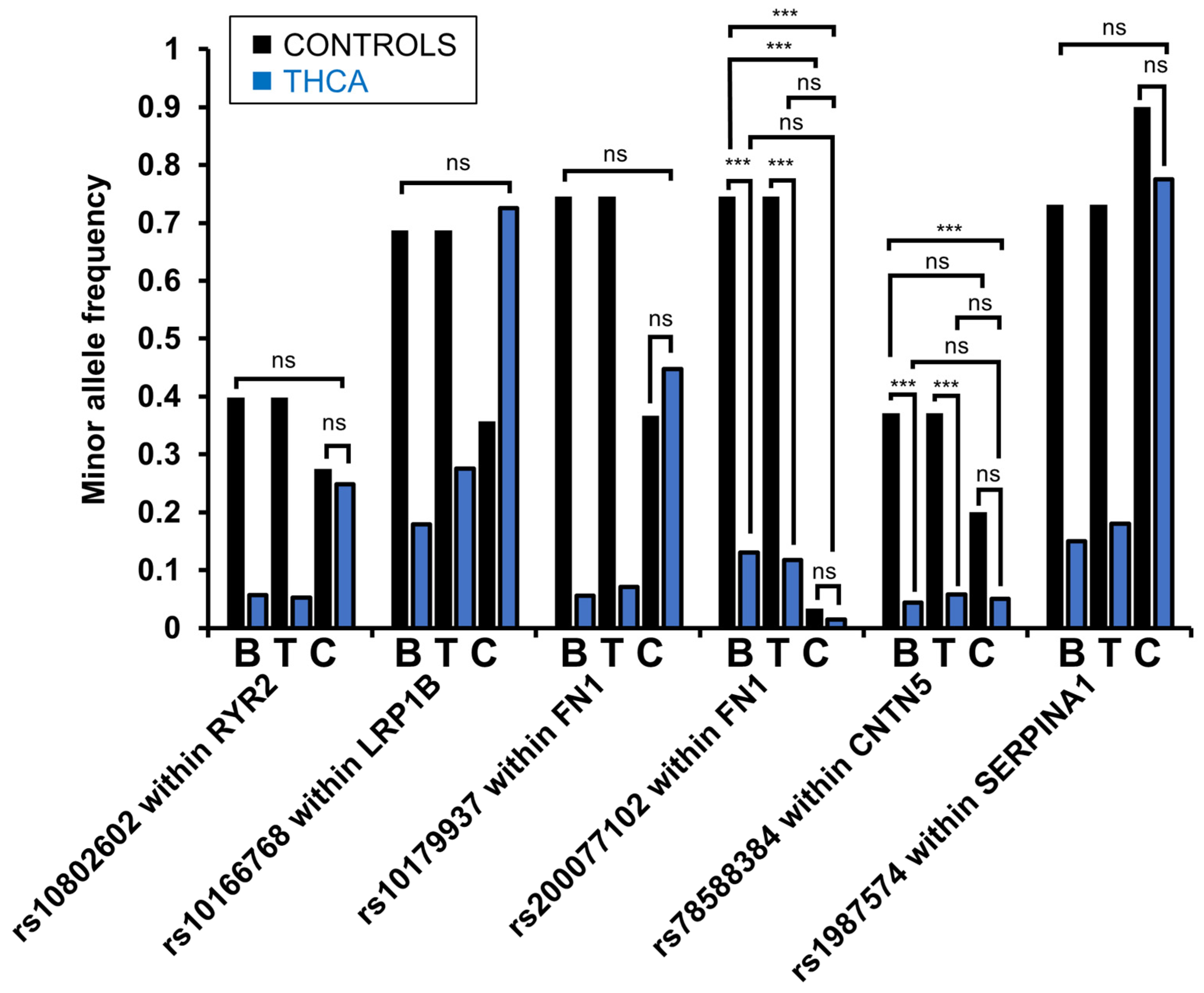
3.4. Evaluation of Common Variants Exposed 15 HERV Variants Significantly Different in Frequency Between PTC and Healthy Controls
3.5. Rare Variants Affect the Poly(A)-Tail Length of Several Alu Elements


| Name | CPG | REF>ALT | Protein Bound a | Motifs b | Chromatin State | Histone Mark c |
|---|---|---|---|---|---|---|
| rs10802602 | RYR2 | C>G | YY1 CEBPA, CEBPB, CEBPG | PAX-8, THAP1, YY1 | - | Enhancer |
| rs2618671 | RYR2 | C>G | - | AHR, KLF9 | Hetero-chromatin | Promoter, Enhancer |
| rs2779420 | RYR2 | C>T | - | EGR1, FOXP1, RREB1 | - | - |
| rs10166768 | LRP1B | C>T,G | - | SOX4, SOX15 | - | Enhancer |
| rs10179937 | FN1 | T>A | POL2 | FOXP1, KLF9, RREB1 | Strong transcription | Promoter, Enhancer |
| rs200077102 | FN1 | T>A | POL2 | FOXP1, RREB1, SOX3, SOX15 | Strong transcription | Promoter, Enhancer |
| rs12543616 | RUNX1T1 | G>A | - | EWSR1, IRF1, STAT1, STAT2 | - | Enhancer |
| rs200093832 | TRPM3 | A>G | - | EP300, EWSR1-FLI1, IRF1, HDAC2, PRDM1, SPI1 | - | Enhancer |
| rs78588384 | CNTN5 | G>C | - | ATF7, FOXP1, IRF1, RREB1, SPI1 | - | Promoter |
| rs1987574 | SERPINA1 | T>A | - | CUX1, EP300, EVI1, FOXP1, HDAC2, HMGA1, HOMEZ, IRF1–4, ZNF35, ZNF384 | monocyte eQTL | Enhancer |
| rs78393784 | SERPINA1 | T>A | - | EP300, EVI1, FOXP1, HDAC2, HOMEZ, IRF1, POU6F1, ZNF35, ZNF384 | - | Enhancer |
| rs112385920 | CD70 | C>T | - | EWSR1-FLI1, HDAC2, SP1, SPZ1, STATTCF12, ZNF143, ZNF263 | Weak Repressed polyComb | Promoter, Enhancer |
| rs2076859 | RUNX1 | T>C | - | SMAD2, SMAD3 | - | Promoter |
| rs2754876 | PCDH11X | G>C | - | BCL6B | - | - |
| rs2750652 | PCDH11X | A>G | - | - | - | Enhancer |
3.6. In Vitro Analyses of Thyroid Cancer Cell Lines Mirrored Low Variant Frequencies for rs200077102 Within FN1 and rs78588384 Within CNTN5 Detected in PTC Samples
3.7. The Genomic Context of rs200077102 and rs78588384 Indicates Transcriptional Dysregulation Caused by the Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, G.; Zhang, H.; Hao, S.; Liu, C.; Xu, J.; Ning, J.; Wu, G.; Jiang, L.; Li, G.; Zheng, H.; et al. Patterns and clinical significance of cervical lymph node metastasis in papillary thyroid cancer patients with Delphian lymph node metastasis. Oncotarget 2017, 8, 57089–57098. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Konturek, A.; Barczyński, M.; Stopa, M.; Nowak, W. Trends in Prevalence of Thyroid Cancer Over Three Decades: A Retrospective Cohort Study of 17,526 Surgical Patients. World J. Surg. 2016, 40, 538–544. [Google Scholar] [CrossRef]
- Surveillance Research Program, National Cancer Institute. SEER*Explorer: An Interactive Website for SEER Cancer Statistics. Available online: https://seer.cancer.gov/statistics-network/explorer/ (accessed on 26 April 2023).
- Abdullah, M.I.; Junit, S.M.; Ng, K.L.; Jayapalan, J.J.; Karikalan, B.; Hashim, O.H. Papillary Thyroid Cancer: Genetic Alterations and Molecular Biomarker Investigations. Int. J. Med. Sci. 2019, 16, 450–460. [Google Scholar] [CrossRef]
- Blackburn, B.E.; Ganz, P.A.; Rowe, K.; Snyder, J.; Wan, Y.; Deshmukh, V.; Newman, M.; Fraser, A.; Smith, K.; Herget, K.; et al. Aging-Related Disease Risks among Young Thyroid Cancer Survivors. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Surveillance, Epidemiology, and End Results (SEER) Program. SEER*Stat Database: Incidence and Mortality—SEER Research Data, 8 Registries, Nov 2021 Sub (1975–2020)—Linked to County Attributes—Time Dependent (1990–2020) Income/Rurality, 1969–2020 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2023, based on the November 2022 Submission. Underlying Mortality Data Provided by NCHS. Available online: www.cdc.gov/nchs (accessed on 1 June 2023).
- Voutilainen, P.E.; Multanen, M.M.; Leppäniemi, A.K.; Haglund, C.H.; Haapiainen, R.K.; Franssila, K.O. Prognosis after lymph node recurrence in papillary thyroid carcinoma depends on age. Thyroid Off. J. Am. Thyroid Assoc. 2001, 11, 953–957. [Google Scholar] [CrossRef]
- Pezzi, T.A.; Sandulache, V.C.; Pezzi, C.M.; Turkeltaub, A.E.; Feng, L.; Cabanillas, M.E.; Williams, M.D.; Lai, S.Y. Treatment and survival of patients with insular thyroid carcinoma: 508 cases from the National Cancer Data Base. Head Neck 2016, 38, 906–912. [Google Scholar] [CrossRef]
- Agrawal, N.; Akbani, R.; Aksoy, B.A.; Ally, A.; Arachchi, H.; Asa, S.L.; Auman, J.T.; Balasundaram, M.; Balu, S.; Baylin, S.B.; et al. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Nissen, K.K.; Laska, M.J.; Hansen, B.; Terkelsen, T.; Villesen, P.; Bahrami, S.; Petersen, T.; Pedersen, F.S.; Nexø, B.A. Endogenous retroviruses and multiple sclerosis-new pieces to the puzzle. BMC Neurol. 2013, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Brütting, C.; Emmer, A.; Kornhuber, M.; Staege, M.S. A survey of endogenous retrovirus (ERV) sequences in the vicinity of multiple sclerosis (MS)-associated single nucleotide polymorphisms (SNPs). Mol. Biol. Rep. 2016, 43, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Otowa, T.; Tochigi, M.; Rogers, M.; Umekage, T.; Kato, N.; Sasaki, T. Insertional polymorphism of endogenous retrovirus HERV-K115 in schizophrenia. Neurosci. Lett. 2006, 408, 226–229. [Google Scholar] [CrossRef]
- Nyegaard, M.; Demontis, D.; Thestrup, B.B.; Hedemand, A.; Sørensen, K.M.; Hansen, T.; Werge, T.; Hougaard, D.M.; Yolken, R.H.; Mortensen, P.B.; et al. No association of polymorphisms in human endogenous retrovirus K18 and CD48 with schizophrenia. Psychiatr. Genet. 2012, 22, 146–148. [Google Scholar] [CrossRef]
- Marguerat, S.; Wang, W.Y.S.; Todd, J.A.; Conrad, B. Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes 2004, 53, 852–854. [Google Scholar] [CrossRef] [PubMed]
- Dickerson, F.; Rubalcaba, E.; Viscidi, R.; Yang, S.; Stallings, C.; Sullens, A.; Origoni, A.; Leister, F.; Yolken, R. Polymorphisms in human endogenous retrovirus K-18 and risk of type 2 diabetes in individuals with schizophrenia. Schizophr. Res. 2008, 104, 121–126. [Google Scholar] [CrossRef]
- Freimanis, G.; Hooley, P.; Ejtehadi, H.D.; Ali, H.A.; Veitch, A.; Rylance, P.B.; Alawi, A.; Axford, J.; Nevill, A.; Murray, P.G.; et al. A role for human endogenous retrovirus-K (HML-2) in rheumatoid arthritis: Investigating mechanisms of pathogenesis. Clin. Exp. Immunol. 2010, 160, 340–347. [Google Scholar] [CrossRef]
- Büscher, K.; Trefzer, U.; Hofmann, M.; Sterry, W.; Kurth, R.; Denner, J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005, 65, 4172–4180. [Google Scholar] [CrossRef]
- Wang-Johanning, F.; Liu, J.; Rycaj, K.; Huang, M.; Tsai, K.; Rosen, D.G.; Chen, D.-T.; Lu, D.W.; Barnhart, K.F.; Johanning, G.L. Expression of multiple human endogenous retrovirus surface envelope proteins in ovarian cancer. Int. J. Cancer 2007, 120, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Rycaj, K.; Geng, S.; Li, M.; Plummer, J.B.; Yin, B.; Liu, H.; Xu, X.; Zhang, Y.; Yan, Y.; et al. Expression of Human Endogenous Retrovirus Type K Envelope Protein is a Novel Candidate Prognostic Marker for Human Breast Cancer. Genes Cancer 2011, 2, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Goering, W.; Ribarska, T.; Schulz, W.A. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 2011, 32, 1484–1492. [Google Scholar] [CrossRef] [PubMed]
- Signorini, L.; Villani, S.; Bregni, M.; Ferrante, P.; Delbue, S. Do the Human Endogenous Retroviruses Play a Role in Colon Cancer? Adv. Tumor Virol. 2016, 6, 11–21. [Google Scholar] [CrossRef]
- Kassiotis, G. Endogenous retroviruses and the development of cancer. J. Immunol. 2014, 192, 1343–1349. [Google Scholar] [CrossRef]
- Garazha, A.; Ivanova, A.; Suntsova, M.; Malakhova, G.; Roumiantsev, S.; Zhavoronkov, A.; Buzdin, A. New bioinformatic tool for quick identification of functionally relevant endogenous retroviral inserts in human genome. Cell Cycle 2015, 14, 1476–1484. [Google Scholar] [CrossRef]
- Buzdin, A.A.; Prassolov, V.; Garazha, A.V. Friends-Enemies: Endogenous Retroviruses Are Major Transcriptional Regulators of Human DNA. Front. Chem. 2017, 5, 35. [Google Scholar] [CrossRef]
- Crosslin, D.R.; Carrell, D.S.; Burt, A.; Kim, D.S.; Underwood, J.G.; Hanna, D.S.; Comstock, B.A.; Baldwin, E.; de Andrade, M.; Kullo, I.J.; et al. Genetic variation in the HLA region is associated with susceptibility to herpes zoster. Genes Immun. 2015, 16, 1–7. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef]
- Ohnuki, M.; Tanabe, K.; Sutou, K.; Teramoto, I.; Sawamura, Y.; Narita, M.; Nakamura, M.; Tokunaga, Y.; Nakamura, M.; Watanabe, A.; et al. Dynamic regulation of human endogenous retroviruses mediates factor-induced reprogramming and differentiation potential. Proc. Natl. Acad. Sci. USA 2014, 111, 12426–12431. [Google Scholar] [CrossRef]
- Durruthy-durruthy, J.; Sebastiano, V.; Wossidlo, M.; Cepeda, D.; Cui, J.; Grow, E.J.; Davila, J.; Mall, M.; Wong, W.H.; Wysocka, J.; et al. The primate-specific noncoding RNA HPAT5 regulates pluripotency during human preimplantation development and nuclear reprogramming. Nat. Genet. 2016, 48, 44–52. [Google Scholar] [CrossRef]
- Frendo, J.-L.; Olivier, D.; Cheynet, V.; Blond, J.-L.; Bouton, O.; Vidaud, M.; Rabreau, M.; Evain-Brion, D.; Mallet, F. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 2003, 23, 3566–3574. [Google Scholar] [CrossRef]
- Soygur, B.; Sati, L. The role of syncytins in human reproduction and reproductive organ cancers. Reproduction 2016, 152, R167–R178. [Google Scholar] [CrossRef]
- Ting, C.N.; Rosenberg, M.P.; Snow, C.M.; Samuelson, L.C.; Meisler, M.H. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992, 6, 1457–1465. [Google Scholar] [CrossRef]
- Gogvadze, E.; Stukacheva, E.; Buzdin, A.; Sverdlov, E. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J. Virol. 2009, 83, 6098–6105. [Google Scholar] [CrossRef]
- Emera, D.; Casola, C.; Lynch, V.J.; Wildman, D.E.; Agnew, D.; Wagner, G.P. Convergent Evolution of Endometrial Prolactin Expression in Primates, Mice, and Elephants Through the Independent Recruitment of Transposable Elements. Mol. Biol. Evol. 2012, 29, 239–247. [Google Scholar] [CrossRef]
- Tuan, D.; Pi, W. In Human Beta-Globin Gene Locus, ERV-9 LTR Retrotransposon Interacts with and Activates Beta- but Not Gamma-Globin Gene. Blood 2014, 124, 2686. [Google Scholar] [CrossRef]
- Seifarth, W.; Frank, O.; Zeilfelder, U.; Spiess, B.; Greenwood, A.D.; Hehlmann, R.; Leib-Mösch, C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005, 79, 341–352. [Google Scholar] [CrossRef]
- Ito, J.; Kimura, I.; Soper, A.; Coudray, A.; Koyanagi, Y.; Nakaoka, H.; Inoue, I.; Turelli, P.; Trono, D.; Sato, K. Endogenous retroviruses drive KRAB zinc-finger family protein expression for tumor suppression. bioRxiv 2020. [Google Scholar] [CrossRef]
- Glinsky, G.V. Transposable Elements and DNA Methylation Create in Embryonic Stem Cells Human-Specific Regulatory Sequences Associated with Distal Enhancers and Noncoding RNAs. Genome Biol. Evol. 2015, 7, 1432–1454. [Google Scholar] [CrossRef]
- Pavlicev, M.; Hiratsuka, K.; Swaggart, K.A.; Dunn, C.; Muglia, L. Detecting endogenous retrovirus-driven tissue-specific gene transcription. Genome Biol. Evol. 2015, 7, 1082–1097. [Google Scholar] [CrossRef]
- Chang, T.-C.; Goud, S.; Torcivia-Rodriguez, J.; Hu, Y.; Pan, Q.; Kahsay, R.; Blomberg, J.; Mazumder, R. Investigation of somatic single nucleotide variations in human endogenous retrovirus elements and their potential association with cancer. PLoS ONE 2019, 14, e0213770. [Google Scholar] [CrossRef]
- Wallace, A.D.; Wendt, G.A.; Barcellos, L.F.; de Smith, A.J.; Walsh, K.M.; Metayer, C.; Costello, J.F.; Wiemels, J.L.; Francis, S.S. To ERV Is Human: A Phenotype-Wide Scan Linking Polymorphic Human Endogenous Retrovirus-K Insertions to Complex Phenotypes. Front. Genet. 2018, 9, 298. [Google Scholar] [CrossRef]
- Burns, K.H.; Boeke, J.D. Human transposon tectonics. Cell 2012, 149, 740–752. [Google Scholar] [CrossRef]
- Goodier, J.L.; Kazazian, H.H., Jr. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef]
- Hancks, D.C.; Kazazian, H.H., Jr. Active human retrotransposons: Variation and disease. Curr. Opin. Genet. Dev. 2012, 22, 191–203. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef]
- Batzer, M.A.; Deininger, P.L. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002, 3, 370–379. [Google Scholar] [CrossRef]
- Kriegs, J.O.; Churakov, G.; Jurka, J.; Brosius, J.; Schmitz, J. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 2007, 23, 158–161. [Google Scholar] [CrossRef]
- Rogers, J.H.; Willison, K.R. A major rearrangement in the H-2 complex of mouse t haplotypes. Nature 1983, 304, 549–552. [Google Scholar] [CrossRef]
- Weiner, A.M.; Deininger, P.L.; Efstratiadis, A. Nonviral retroposons: Genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu. Rev. Biochem. 1986, 55, 631–661. [Google Scholar] [CrossRef]
- Mathias, S.L.; Scott, A.F.; Kazazian, H.H., Jr.; Boeke, J.D.; Gabriel, A. Reverse transcriptase encoded by a human transposable element. Science 1991, 254, 1808–1810. [Google Scholar] [CrossRef]
- Feng, Q.; Moran, J.V.; Kazazian, H.H., Jr.; Boeke, J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 1996, 87, 905–916. [Google Scholar] [CrossRef]
- Roy-Engel, A.M.; Salem, A.H.; Oyeniran, O.O.; Deininger, L.; Hedges, D.J.; Kilroy, G.E.; Batzer, M.A.; Deininger, P.L. Active Alu element “A-tails”: Size does matter. Genome Res. 2002, 12, 1333–1344. [Google Scholar] [CrossRef]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Tomczak, K.; Czerwinska, P.; Wiznerowicz, M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp. Oncol. 2015, 19, A68–A77. [Google Scholar] [CrossRef]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef]
- Zalunin, V.; Leinonen, R.; Duckart, F.; Xue, Z.; Ashton, P. Cramtools; Version 3.0; github: San Francisco, CA, USA, 2018. [Google Scholar]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Rahman, N. Realizing the promise of cancer predisposition genes. Nature 2014, 505, 302–308. [Google Scholar] [CrossRef]
- Zhang, J.; Walsh, M.F.; Wu, G.; Edmonson, M.N.; Gruber, T.A.; Easton, J.; Hedges, D.; Ma, X.; Zhou, X.; Yergeau, D.A.; et al. Germline Mutations in Predisposition Genes in Pediatric Cancer. N. Engl. J. Med. 2015, 373, 2336–2346. [Google Scholar] [CrossRef]
- Repana, D.; Nulsen, J.; Dressler, L.; Bortolomeazzi, M.; Venkata, S.K.; Tourna, A.; Yakovleva, A.; Palmieri, T.; Ciccarelli, F.D. The Network of Cancer Genes (NCG): A comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019, 20, 1. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Wan, Q.; Dingerdissen, H.; Fan, Y.; Gulzar, N.; Pan, Y.; Wu, T.J.; Yan, C.; Zhang, H.; Mazumder, R. BioXpress: An integrated RNA-seq-derived gene expression database for pan-cancer analysis. Database 2015, 2015, bav019. [Google Scholar] [CrossRef]
- Dingerdissen, H.M.; Torcivia-Rodriguez, J.; Hu, Y.; Chang, T.C.; Mazumder, R.; Kahsay, R. BioMuta and BioXpress: Mutation and expression knowledgebases for cancer biomarker discovery. Nucleic Acids Res. 2018, 46, D1128–D1136. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- GATK. GATK Resource Bundle; GATK: Cambridge, MA, USA, 2024. [Google Scholar]
- Glusman, G.; Caballero, J.; Mauldin, D.E.; Hood, L.; Roach, J.C. Kaviar: An accessible system for testing SNV novelty. Bioinformatics 2011, 27, 3216–3217. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Halder, I.; Shriver, M.; Thomas, M.; Fernandez, J.R.; Frudakis, T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: Utility and applications. Hum. Mutat. 2008, 29, 648–658. [Google Scholar] [CrossRef]
- Archer, N.P.; Perez-Andreu, V.; Scheurer, M.E.; Rabin, K.R.; Peckham-Gregory, E.C.; Plon, S.E.; Zabriskie, R.C.; De Alarcon, P.A.; Fernandez, K.S.; Najera, C.R.; et al. Family-based exome-wide assessment of maternal genetic effects on susceptibility to childhood B-cell acute lymphoblastic leukemia in hispanics. Cancer 2016, 122, 3697–3704. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Urbanek, S.; Horner, J. Cairo: R Graphics Device using Cairo Graphics Library for Creating High-Quality Bitmap (PNG, JPEG, TIFF), Vector (PDF, SVG, PostScript) and Display (X11 and Win32) Output; Version 1.6-2; R Core Team: Vienna, Austria, 2022. [Google Scholar]
- Wickham, H.; François, R.; Henry, L.; Müller, K. Dplyr: A Grammar of Data Manipulation; Version 1.0.7; R Core Team: Vienna, Austria, 2021. [Google Scholar]
- Wickham, H. stringr: Simple, Consistent Wrappers for Common String Operations; Version 1.5.1; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Knaus, B.J.; Grunwald, N.J. vcfr: A package to manipulate and visualize variant call format data in R. Mol. Ecol. Resour. 2017, 17, 44–53. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated Tool for Genome-wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Sicko, R.J.; Stevens, C.F.; Hughes, E.E.; Leisner, M.; Ling, H.; Saavedra-Matiz, C.A.; Caggana, M.; Kay, D.M. Validation of a Custom Next-Generation Sequencing Assay for Cystic Fibrosis Newborn Screening. Int. J. Neonatal. Screen 2021, 7, 73. [Google Scholar] [CrossRef]
- Tongyoo, P.; Avihingsanon, Y.; Prom-On, S.; Mutirangura, A.; Mhuantong, W.; Hirankarn, N. EnHERV: Enrichment analysis of specific human endogenous retrovirus patterns and their neighboring genes. PLoS ONE 2017, 12, e0177119. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S.; Cox, N.J.; Goldstein, D.B.; Hindorff, L.A.; Hunter, D.J.; McCarthy, M.I.; Ramos, E.M.; Cardon, L.R.; Chakravarti, A.; et al. Finding the missing heritability of complex diseases. Nature 2009, 461, 747–753. [Google Scholar] [CrossRef]
- Hamann, M.V.; Adiba, M.; Lange, U.C. Confounding factors in profiling of locus-specific human endogenous retrovirus (HERV) transcript signatures in primary T cells using multi-study-derived datasets. BMC Med. Genom. 2023, 16, 68. [Google Scholar] [CrossRef] [PubMed]
- Wildschutte, J.H.; Williams, Z.H.; Montesion, M.; Subramanian, R.P.; Kidd, J.M.; Coffin, J.M. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc. Natl. Acad. Sci. USA 2016, 113, E2326–E2334. [Google Scholar] [CrossRef]
- Chen, S.; Francioli, L.C.; Goodrich, J.K.; Collins, R.L.; Kanai, M.; Wang, Q.; Alföldi, J.; Watts, N.A.; Vittal, C.; Gauthier, L.D.; et al. A genome-wide mutational constraint map quantified from variation in 76,156 human genomes. bioRxiv 2022. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg v4: Systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016, 44, D877–D881. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Consortium, E.P. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 9, e1001046. [Google Scholar] [CrossRef]
- Berger, M.F.; Philippakis, A.A.; Qureshi, A.M.; He, F.S.; Estep, P.W., 3rd; Bulyk, M.L. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 2006, 24, 1429–1435. [Google Scholar] [CrossRef]
- Berger, M.F.; Badis, G.; Gehrke, A.R.; Talukder, S.; Philippakis, A.A.; Pena-Castillo, L.; Alleyne, T.M.; Mnaimneh, S.; Botvinnik, O.B.; Chan, E.T.; et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 2008, 133, 1266–1276. [Google Scholar] [CrossRef]
- Badis, G.; Berger, M.F.; Philippakis, A.A.; Talukder, S.; Gehrke, A.R.; Jaeger, S.A.; Chan, E.T.; Metzler, G.; Vedenko, A.; Chen, X.; et al. Diversity and complexity in DNA recognition by transcription factors. Science 2009, 324, 1720–1723. [Google Scholar] [CrossRef] [PubMed]
- Kheradpour, P.; Kellis, M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014, 42, 2976–2987. [Google Scholar] [CrossRef] [PubMed]
- Aladal, M.; You, W.; Huang, R.; Huang, J.; Deng, Z.; Duan, L.; Wang, D.; Li, W.; Sun, W. Insights into the implementation of Fibronectin 1 in the cartilage tissue engineering. Biomed. Pharmacother. 2022, 148, 112782. [Google Scholar] [CrossRef]
- Zollinger, A.J.; Smith, M.L. Fibronectin, the extracellular glue. Matrix Biol. 2017, 60–61, 27–37. [Google Scholar] [CrossRef]
- Oguro-Ando, A.; Zuko, A.; Kleijer, K.T.E.; Burbach, J.P.H. A current view on contactin-4, -5, and -6: Implications in neurodevelopmental disorders. Mol. Cell Neurosci. 2017, 81, 72–83. [Google Scholar] [CrossRef]
- Zavadil, J.; Bitzer, M.; Liang, D.; Yang, Y.C.; Massimi, A.; Kneitz, S.; Piek, E.; Bottinger, E.P. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc. Natl. Acad. Sci. USA 2001, 98, 6686–6691. [Google Scholar] [CrossRef]
- Sponziello, M.; Rosignolo, F.; Celano, M.; Maggisano, V.; Pecce, V.; De Rose, R.F.; Lombardo, G.E.; Durante, C.; Filetti, S.; Damante, G.; et al. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol. Cell Endocrinol. 2016, 431, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Lock, F.E.; Rebollo, R.; Miceli-Royer, K.; Gagnier, L.; Kuah, S.; Babaian, A.; Sistiaga-Poveda, M.; Lai, C.B.; Nemirovsky, O.; Serrano, I.; et al. Distinct isoform of FABP7 revealed by screening for retroelement-activated genes in diffuse large B-cell lymphoma. Proc. Natl. Acad. Sci. USA 2014, 111, E3534–E3543. [Google Scholar] [CrossRef]
- Partridge, E.C.; Chhetri, S.B.; Prokop, J.W.; Ramaker, R.C.; Jansen, C.S.; Goh, S.T.; Mackiewicz, M.; Newberry, K.M.; Brandsmeier, L.A.; Meadows, S.K.; et al. Occupancy maps of 208 chromatin-associated proteins in one human cell type. Nature 2020, 583, 720–728. [Google Scholar] [CrossRef]
- Babaian, A.; Mager, D.L. Endogenous retroviral promoter exaptation in human cancer. Mob. DNA 2016, 7, 24. [Google Scholar] [CrossRef]
- Stricker, E.; Peckham-Gregory, E.C.; Scheurer, M.E. HERVs and Cancer-A Comprehensive Review of the Relationship of Human Endogenous Retroviruses and Human Cancers. Biomedicines 2023, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Xia, F.; Du, X.; Jiang, B.; Chen, Y.; Li, X. Identification of key candidate genes and pathways in follicular variant papillary thyroid carcinoma by integrated bioinformatical analysis. Transl. Cancer Res. 2020, 9, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Hong, C.; Zhang, B.; Lowdon, R.F.; Xing, X.; Li, D.; Zhou, X.; Lee, H.J.; Maire, C.L.; Ligon, K.L.; et al. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat. Genet. 2013, 45, 836–841. [Google Scholar] [CrossRef]
- Camacho Londono, J.; Philipp, S.E. A reliable method for quantification of splice variants using RT-qPCR. BMC Mol. Biol. 2016, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Yin, G.; Liu, C.; Zhang, W.; Shen, Y.; Wang, D.; Lin, Z.; Yang, J.; Mao, J.; Guo, R.; et al. Critically short telomeres derepress retrotransposons to promote genome instability in embryonic stem cells. Cell Discov. 2023, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Kulski, J.K. Long Noncoding RNA HCP5, a Hybrid HLA Class I Endogenous Retroviral Gene: Structure, Expression, and Disease Associations. Cells 2019, 8, 840. [Google Scholar] [CrossRef] [PubMed]
- The All of Us Research Program Genomics Investigators. Genomic data in the All of Us Research Program. Nature 2024, 627, 340–346. [Google Scholar] [CrossRef]
- Halder, I.; Shriver, M.D. Measuring and using admixture to study the genetics of complex diseases. Hum. Genom. 2003, 1, 52–62. [Google Scholar] [CrossRef]
- Bonilla, C.; Parra, E.J.; Pfaff, C.L.; Dios, S.; Marshall, J.A.; Hamman, R.F.; Ferrell, R.E.; Hoggart, C.L.; McKeigue, P.M.; Shriver, M.D. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann. Hum. Genet. 2004, 68, 139–153. [Google Scholar] [CrossRef]
- Bonilla, C.; Shriver, M.D.; Parra, E.J.; Jones, A.; Fernandez, J.R. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York city. Hum. Genet. 2004, 115, 57–68. [Google Scholar] [CrossRef]
- Atkinson, E.G.; Maihofer, A.X.; Kanai, M.; Martin, A.R.; Karczewski, K.J.; Santoro, M.L.; Ulirsch, J.C.; Kamatani, Y.; Okada, Y.; Finucane, H.K.; et al. Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nat. Genet. 2021, 53, 195–204. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C. Best practices for variant calling in clinical sequencing. Genome Med. 2020, 12, 91. [Google Scholar] [CrossRef] [PubMed]






| Training Set (n = 83; 60%) 1 | Validation Set (n = 55; 40%) 1 | |
|---|---|---|
| Blood samples | ||
| Low coverage only | 50 (64.1%) | 32 (68.1%) |
| High coverage only | 20 (25.6%) | 11 (23.4%) |
| High and low coverage | 8 (10.3%) | 4 (8.5%) |
| Tumor samples | ||
| Low coverage only | 52 (62.7%) | 36 (65.5%) |
| High coverage only | 23 (27.7%) | 15 (27.3%) |
| High and low coverage | 8 (9.6%) | 4 (7.3%) |
| Gender | ||
| Female | 64 (77.1%) | 42 (76.4%) |
| Male | 19 (22.9%) | 13 (23.6%) |
| Age at diagnosis | ||
| Average age | 48.68 | 48.06 |
| Race/Ethnicity (self-reported) | ||
| Non-Hispanic White | 51 (61.4%) | 32 (58.2%) |
| Hispanic White | 2 (2.4%) | 3 (5.5%) |
| Black or African American | 3 (3.6%) | 2 (3.6%) |
| Asian | 5 (6%) | 4 (7.3%) |
| Not reported | 22 (26.5%) | 14 (25.5%) |
| Ancestral population (calculated) | ||
| EUR | 44 (53%) | 31 (56.4%) |
| HIS | 24 (28.9%) | 14 (25.5%) |
| AFR | 2 (2.4%) | 1 (1.8%) |
| EAS | 3 (3.6%) | 3 (5.5%) |
| AMR | 10 (12%) | 6 (10.9%) |
| Vital status | ||
| alive | 81 (97.6%) | 54 (98.2%) |
| dead | 2 (2.4%) | 1 (1.8%) |
| Tumor stage | ||
| I | 43 (51.8%) | 31 (56.4%) |
| II | 9 (10.8%) | 11 (20%) |
| III | 19 (22.9%) | 7 (12.7%) |
| IV | 11 (13.3%) | 6 (10.9%) |
| Not reported | 1 (1.2%) | 0 (0%) |
| Training Set (n = 1224; 60%) 1 | Validation Set (n = 791; 40%) 1 | |
|---|---|---|
| Gender | ||
| Female | 765 (50.8%) | 506 (50.9%) |
| Male | 740 (49.2%) | 489 (49.1%) |
| Superpopulation | ||
| EUR | 300 (19.9%) | 203 (20.4%) |
| AMR | 216 (14.4%) | 131 (13.2%) |
| AFR | 408 (27.1%) | 253 (25.4%) |
| EAS | 300 (19.9%) | 204 (20.5%) |
| Ancestral population (calculated) | ||
| EUR | 206 (13.7%) | 147 (14.8%) |
| AMR | 126 (8.4%) | 75 (7.5%) |
| AFR | 397 (26.4%) | 242 (24.3%) |
| EAS | 301 (20%) | 204 (20.5%) |
| HIS | 194 (12.9%) | 123 (12.4%) |
| Name | CPG | MAF PTC Blood | MAF PTC Tumor | MAF 1KGP | MAF GnomAD |
|---|---|---|---|---|---|
| rs10802602 | RYR2 | 3.8% | 3.3% | 39.6% | 22.3% |
| rs2618671 | RYR2 | 32.5% | 30.9% | 57.1% | 56.7% |
| rs2779420 | RYR2 | 30.1% | 29.3% | 53.6% | 48.3% |
| rs13030271 ◊ | LRP1B | 8.2% | 11.5% | 37.8% | 0.0% |
| rs10166768 (C>T) | LRP1B | 17.9% | 23.0% | 67.7% | 17.9% |
| rs10166768 (C>G) | LRP1B | 50.0% | 32.1% | 0% | 49.3% |
| rs10179937 | FN1 | 8.8% | 7.8% | 74.9% | NA |
| rs200077102 | FN1 | 7.6% | 6.5% | 74.9% | 41.5% |
| rs7682763 † | EPHA5 | 25.8% | 25.0% | 71.6% | NA |
| rs13311049 ◊ | SDK1 | 9.9% | 10.9% | 38.9% | 16.4% |
| rs13311637 ◊ | SDK1 | 6.7% | 6.3% | 50.8% | 2.4% |
| rs611655 ◊ | MMD2 (RADIL) | 0.5% | 2.5% | 43.8% | 0.0% |
| rs12543616 | RUNX1T1 | 18.9% | 22.4% | 76.0% | 48.7% |
| rs10956571 ‡ | ADCY8 | 25.3% | 24.0% | 62.9% | 55.3% |
| rs200093832 | TRPM3 | 26.7% | 18.9% | 53.5% | 71.8% |
| rs61909780 ◊ | CNTN5 | 2.7% | 2.5% | 38.8% | 14.1% |
| rs78588384 | CNTN5 | 4.5% | 5.5% | 36.9% | 29.8% |
| rs1987574 | SERPINA1 | 20.0% | 26.8% | 73.1% | NA |
| rs78393784 | SERPINA1 | 38.6% | 31.2% | 29.4% | NA |
| rs370565365 †,◊ | ACSM2A | 5.3% | NA | 25.2% | 2.0% |
| rs112385920 | CD70 | 50.7% | 51.3% | 79.2% | 81.6% |
| rs2076859 | RUNX1 | 8.1% | 11.7% | 82.7% | NA |
| rs3989120 ◊ | RUNX1 | 22.0% | 21.4% | 82.7% | 0.0% |
| rs13046555 † | RUNX1 | 15.0% | 22.2% | 32.1% | 46.2% |
| rs778825437 | PCDH11X | 3.8% | 3.3% | 39.6% | NA |
| rs2754876 | PCDH11X | 18.7% | 20.7% | 65.0% | 36.9% |
| rs2750652 ◊ | PCDH11X | 39.2% | 40.0% | 73.0% | 35.7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stricker, E.; Peckham-Gregory, E.C.; Lai, S.Y.; Sandulache, V.C.; Scheurer, M.E. Targeted Variant Assessments of Human Endogenous Retroviral Regions in Whole Genome Sequencing Data Reveal Retroviral Variants Associated with Papillary Thyroid Cancer. Microorganisms 2024, 12, 2435. https://doi.org/10.3390/microorganisms12122435
Stricker E, Peckham-Gregory EC, Lai SY, Sandulache VC, Scheurer ME. Targeted Variant Assessments of Human Endogenous Retroviral Regions in Whole Genome Sequencing Data Reveal Retroviral Variants Associated with Papillary Thyroid Cancer. Microorganisms. 2024; 12(12):2435. https://doi.org/10.3390/microorganisms12122435
Chicago/Turabian StyleStricker, Erik, Erin C. Peckham-Gregory, Stephen Y. Lai, Vlad C. Sandulache, and Michael E. Scheurer. 2024. "Targeted Variant Assessments of Human Endogenous Retroviral Regions in Whole Genome Sequencing Data Reveal Retroviral Variants Associated with Papillary Thyroid Cancer" Microorganisms 12, no. 12: 2435. https://doi.org/10.3390/microorganisms12122435
APA StyleStricker, E., Peckham-Gregory, E. C., Lai, S. Y., Sandulache, V. C., & Scheurer, M. E. (2024). Targeted Variant Assessments of Human Endogenous Retroviral Regions in Whole Genome Sequencing Data Reveal Retroviral Variants Associated with Papillary Thyroid Cancer. Microorganisms, 12(12), 2435. https://doi.org/10.3390/microorganisms12122435





