Nanopore Sequencing Allows Recovery of High-Quality Completely Closed Genomes of All Cronobacter Species from Powdered Infant Formula Overnight Enrichments
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Artificial Contamination and Sample Processing
2.3. DNA Extraction and Cronobacter qPCR Detection Following the BAM Procedure
2.4. DNA Extraction and Whole Genome Sequencing and Assembly
2.5. In Silico MLST and Serotyping
2.6. Phylogenetic Relationship of the Strains by Whole Genome Multilocus Typing (Wgmlst) Analysis
3. Results
3.1. Cronobacter-Inoculated Sample-Enrichment Preparation for Nanopore Sequencing
3.2. Nanopore Long-Read Sequencing Results
3.3. Nanopore Long-Read Genome Assembly of Cronobacter PIF-Enriched Samples
3.4. Multilocus Sequence Typing (MLST) and Serotyping Analysis
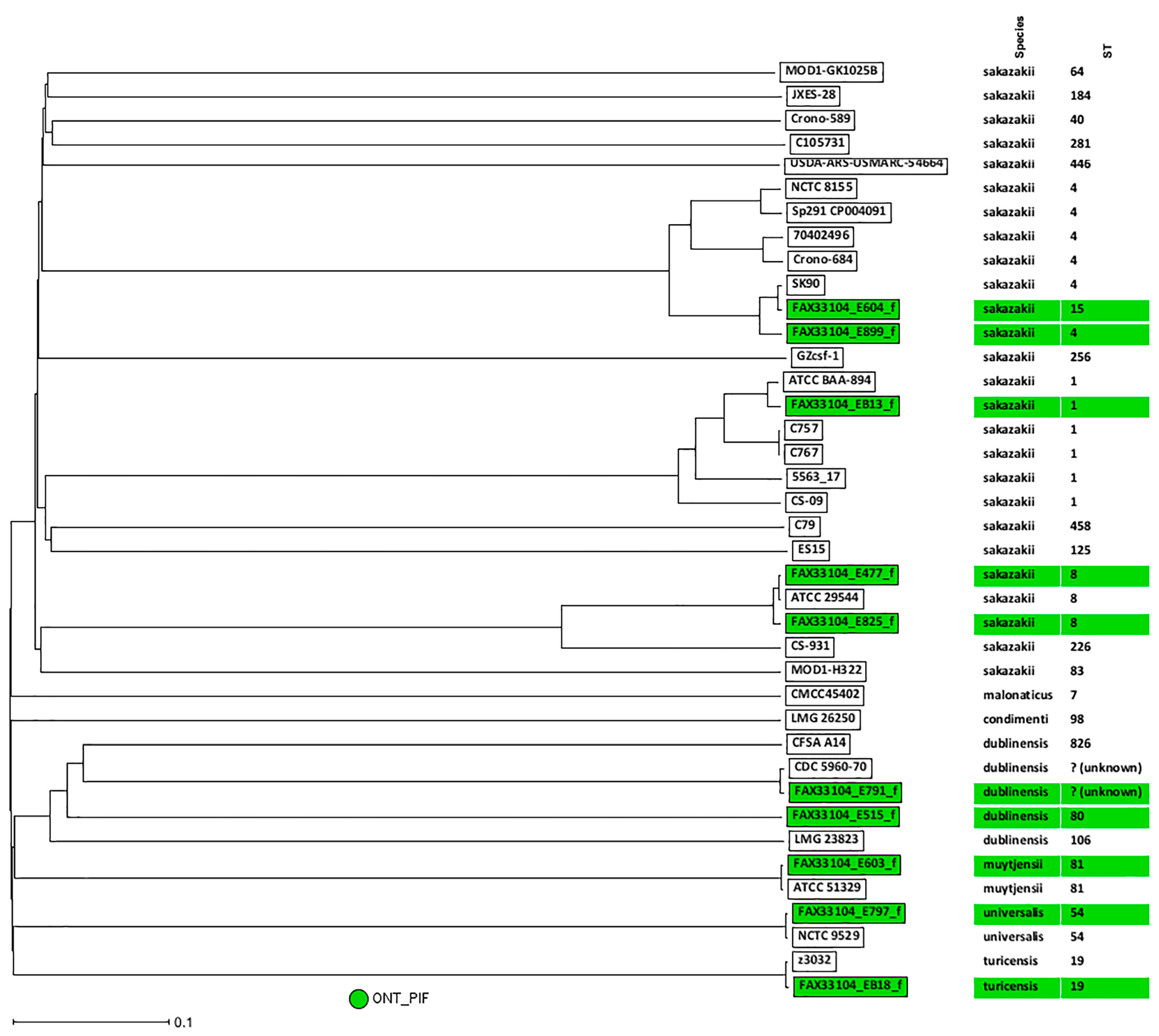
3.5. wgMLST Analysis of Cronobacter PIF Overnight Enriched Samples and Taxa Classification Using a Phylogenetic Tree
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Maguire, M.; Kase, J.A.; Roberson, D.; Muruvanda, T.; Brown, E.W.; Allard, M.; Musser, S.M.; Gonzalez-Escalona, N. Precision long-read metagenomics sequencing for food safety by detection and assembly of shiga toxin-producing Escherichia coli in irrigation water. PLoS ONE 2021, 16, e0245172. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.; Dessai, U.; McGarry, S.; Gerner-Smidt, P. Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog. Dis. 2019, 16, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.D.; Luo, C.; Pena-Gonzalez, A.; Weigand, M.R.; Tarr, C.L.; Konstantinidis, K.T. Metagenomics of two severe foodborne outbreaks provides diagnostic signatures and signs of coinfection not attainable by traditional methods. Appl. Environ. Microbiol. 2017, 83, e02577-16. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Constantinidou, C.; Christner, M.; Rohde, H.; Chan, J.Z.; Quick, J.; Weir, J.C.; Quince, C.; Smith, G.P.; Betley, J.R.; et al. A culture-independent sequence-based metagenomics approach to the investigation of an outbreak of shiga-toxigenic Escherichia coli O104:H4. JAMA 2013, 309, 1502–1510. [Google Scholar] [CrossRef]
- Maguire, M.; Ramachandran, P.; Tallent, S.; Mammel, M.K.; Brown, E.W.; Allard, M.W.; Musser, S.M.; Gonzalez-Escalona, N. Precision metagenomics sequencing for food safety: Hybrid assembly of shiga toxin-producing Escherichia coli in enriched agricultural water. Front. Microbiol. 2023, 14, 1221668. [Google Scholar] [CrossRef]
- Cechin, C.D.F.; Carvalho, G.G.; Bastos, C.P.; Kabuki, D.Y. Cronobacter spp. in foods of plant origin: Occurrence, contamination routes, and pathogenic potential. Crit. Rev. Food Sci. Nutr. 2023, 63, 12398–12412. [Google Scholar] [CrossRef]
- Haston, J.C.; Miko, S.; Cope, J.R.; McKeel, H.; Walters, C.; Joseph, L.A.; Griswold, T.; Katz, L.S.; Andujar, A.A.; Tourdot, L.; et al. Cronobacter sakazakii infections in two infants linked to powdered infant formula and breast pump equipment—United States, 2021 and 2022. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 223–226. [Google Scholar] [CrossRef]
- Li, Y.; Lin, G.; Zhang, L.; Hu, Y.; Hong, C.; Xie, A.; Fang, L. Genomic insights into Cronobacter spp. recovered from food and human clinical cases in Zhejiang province, China (2008–2021). J. Appl. Microbiol. 2023, 134, lxad033. [Google Scholar] [CrossRef]
- Drudy, D.; Mullane, N.R.; Quinn, T.; Wall, P.G.; Fanning, S. Enterobacter sakazakii: An emerging pathogen in powdered infant formula. Clin. Infect. Dis. 2006, 42, 996–1002. [Google Scholar] [CrossRef]
- Strysko, J.; Cope, J.R.; Martin, H.; Tarr, C.; Hise, K.; Collier, S.; Bowen, A. Food safety and invasive Cronobacter infections during early infancy, 1961–2018. Emerg. Infect. Dis. 2020, 26, 857–865. [Google Scholar] [CrossRef]
- Joseph, S.; Sonbol, H.; Hariri, S.; Desai, P.; McClelland, M.; Forsythe, S.J. Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J. Clin. Microbiol. 2012, 50, 3031–3039. [Google Scholar] [CrossRef] [PubMed]
- Iversen, C.; Mullane, N.; McCardell, B.; Tall, B.D.; Lehner, A.; Fanning, S.; Stephan, R.; Joosten, H. Cronobacter gen. Nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. Nov., comb. Nov., Cronobacter malonaticus sp. Nov., Cronobacter turicensis sp. Nov., Cronobacter muytjensii sp. Nov., Cronobacter dublinensis sp. Nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. Nov., Cronobacter dublinensis subsp. lausannensis subsp. Nov. and Cronobacter dublinensis subsp. lactaridi subsp. Nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Hariri, S.; Joseph, S.; Forsythe, S.J. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg. Infect. Dis. 2013, 19, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Jiang, X.T.; Forsythe, S.; Zhang, D.F.; Shen, Y.Z.; Ding, Y.; Wang, J.; Zhang, J.M.; Wu, Q.P.; Ye, Y.W. Food safety risks and contributing factors of Cronobacter spp. Engineering 2022, 12, 128–138. [Google Scholar] [CrossRef]
- Chen, Y.; Miranda, N.E.; Liu, K.C.; Mullins, J.S.; Lampel, K.; Hammack, T. BAM Chapter 29: Cronobacter. Available online: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-29-cronobacter (accessed on 15 November 2024).
- Bertrand, D.; Shaw, J.; Kalathiyappan, M.; Ng, A.H.Q.; Kumar, M.S.; Li, C.; Dvornicic, M.; Soldo, J.P.; Koh, J.Y.; Tong, C.; et al. Hybrid metagenomic assembly enables high-resolution analysis of resistance determinants and mobile elements in human microbiomes. Nat. Biotechnol. 2019, 37, 937–944. [Google Scholar] [CrossRef]
- Maguire, M.; Khan, A.S.; Adesiyun, A.A.; Georges, K.; Gonzalez-Escalona, N. Closed genome sequence of a Salmonella enterica serotype Senftenberg strain carrying the mcr-9 gene isolated from broken chicken eggshells in Trinidad and Tobago. Microbiol. Resour. Announc. 2021, 10, e0146520. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Fan, J.; Huang, S.; Chorlton, S.D. BugSeq: A highly accurate cloud platform for long-read metagenomic analyses. BMC Bioinform. 2021, 22, 160. [Google Scholar] [CrossRef]
- Van Damme, R.; Holzer, M.; Viehweger, A.; Muller, B.; Bongcam-Rudloff, E.; Brandt, C. Metagenomics workflow for hybrid assembly, differential coverage binning, metatranscriptomics and pathway analysis (MUFFIN). PLoS Comput. Biol. 2021, 17, e1008716. [Google Scholar] [CrossRef]
- Gangiredla, J.; Rand, H.; Benisatto, D.; Payne, J.; Strittmatter, C.; Sanders, J.; Wolfgang, W.J.; Libuit, K.; Herrick, J.B.; Prarat, M.; et al. Galaxytrakr: A distributed analysis tool for public health whole genome sequence data accessible to non-bioinformaticians. BMC Genom. 2021, 22, 114. [Google Scholar] [CrossRef]
- Maguire, M.; Khan, A.S.; Adesiyun, A.A.; Georges, K.; Gonzalez-Escalona, N. Genomic comparison of eight closed genomes of multidrug-resistant Salmonella enterica strains isolated from broiler farms and processing plants in Trinidad and Tobago. Front. Microbiol. 2022, 13, 863104. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, N.D.; Hopkins, K.M.V.; Colpus, M.; Parker, M.; Lipworth, S.; Crook, D.; Stoesser, N. Evaluation of the accuracy of bacterial genome reconstruction with Oxford nanopore R10.4.1 long-read-only sequencing. Microb. Genom. 2024, 10, 001246. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.; Fakharuddin, K.; Mulvey, M.R.; Mataseje, L. Do we still need illumina sequencing data? Evaluating Oxford nanopore technologies R10.4.1 flow cells and the rapid v14 library prep kit for Gram negative bacteria whole genome assemblies. Can. J. Microbiol. 2024, 70, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, B.; Van den Bossche, A.; Verhaegen, B.; Delbrassinne, L.; Mattheus, W.; Nouws, S.; Godfroid, M.; Hoffman, S.; Roosens, N.H.C.; De Keersmaecker, S.C.J.; et al. Closing the gap: Oxford nanopore technologies R10 sequencing allows comparable results to illumina sequencing for SNP-based outbreak investigation of bacterial pathogens. J. Clin. Microbiol. 2024, 62, e01576-23. [Google Scholar] [CrossRef]
- Buytaers, F.E.; Verhaegen, B.; Van Nieuwenhuysen, T.; Roosens, N.H.C.; Vanneste, K.; Marchal, K.; De Keersmaecker, S.C.J. Strain-level characterization of foodborne pathogens without culture enrichment for outbreak investigation using shotgun metagenomics facilitated with nanopore adaptive sampling. Front. Microbiol. 2024, 15, 1330814. [Google Scholar] [CrossRef]
- Seo, K.H.; Brackett, R.E. Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J. Food Prot. 2005, 68, 59–63. [Google Scholar] [CrossRef]
- Deer, D.M.; Lampel, K.A.; Gonzalez-Escalona, N. A versatile internal control for use as DNA in real-time PCR and as RNA in real-time reverse transcription PCR assays. Lett. Appl. Microbiol. 2010, 50, 366–372. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Wang, L.; Zhu, W.; Lu, G.; Wu, P.; Wei, Y.; Su, Y.; Jia, T.; Li, L.; Guo, X.; Huang, M.; et al. In silico species identification and serotyping for Cronobacter isolates by use of whole-genome sequencing data. Int. J. Food Microbiol. 2021, 358, 109405. [Google Scholar] [CrossRef]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef] [PubMed]
- Duarte, F.; Cordero, E.; Calderon, M.; Godinez, A.; Ross, B.; Allard, M.; Gonzalez-Escalona, N. Closed genomes of four multidrug resistance Salmonella enterica serotype Infantis Isolated in Costa Rica. Microbiol. Resour. Announc. 2024, 13, e0025723. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Kuang, D.; Xu, X.; Gonzalez-Escalona, N.; Erickson, D.L.; Brown, E.; Meng, J. Genomic analyses of multidrug-resistant Salmonella Indiana, Typhimurium, and Enteritidis isolates using minion and miseq sequencing technologies. PLoS ONE 2020, 15, e0235641. [Google Scholar] [CrossRef]
- Hoffmann, M.; Luo, Y.; Monday, S.R.; Gonzalez-Escalona, N.; Ottesen, A.R.; Muruvanda, T.; Wang, C.; Kastanis, G.; Keys, C.; Janies, D.; et al. Tracing origins of the Salmonella Bareilly strain causing a food-borne outbreak in the United States. J. Infect. Dis. 2016, 213, 502–508. [Google Scholar] [CrossRef]
- Gobin, M.; Hawker, J.; Cleary, P.; Inns, T.; Gardiner, D.; Mikhail, A.; McCormick, J.; Elson, R.; Ready, D.; Dallman, T.; et al. National outbreak of shiga toxin-producing Escherichia coli O157:H7 linked to mixed salad leaves, United Kingdom, 2016. Eurosurveillance 2018, 23, 17-00197. [Google Scholar] [CrossRef]
- Bottichio, L.; Keaton, A.; Thomas, D.; Fulton, T.; Tiffany, A.; Frick, A.; Mattioli, M.; Kahler, A.; Murphy, J.; Otto, M.; et al. Shiga toxin-producing Escherichia coli infections associated with romaine lettuce-United States, 2018. Clin. Infect. Dis. 2020, 71, e323–e330. [Google Scholar] [CrossRef]
- Carleton, H.A.; Besser, J.; Williams-Newkirk, A.J.; Huang, A.; Trees, E.; Gerner-Smidt, P. Metagenomic approaches for public health surveillance of foodborne infections: Opportunities and challenges. Foodborne Pathog. Dis. 2019, 16, 474–479. [Google Scholar] [CrossRef]
- Pena-Gonzalez, A.; Soto-Giron, M.J.; Smith, S.; Sistrunk, J.; Montero, L.; Paez, M.; Ortega, E.; Hatt, J.K.; Cevallos, W.; Trueba, G.; et al. Metagenomic signatures of gut infections caused by different Escherichia coli pathotypes. Appl. Environ. Microbiol. 2019, 85, e01820-19. [Google Scholar] [CrossRef]

| Samples | Cronobacter Species | Source | Country of Origin | Available at NCBI |
|---|---|---|---|---|
| E477 | sakazakii | Human (throat) | Unknown | ATCC 29544 |
| E515 | dublinensis | Water | Switzerland | NA |
| E603 | muytjensii | Unknown | unknown | ATCC 51329 |
| E604 | sakazakii | Human | Canada | SK90 |
| E791 | dublinensis | Human (blood) | USA | CDC 5960-70 |
| E797 | universalis | Water | UK | NCTC 9529 |
| E825 | sakazakii | Human (breast abscess) | USA | NA |
| EB13 | sakazakii | Neonate (meningitis) | Switzerland | NA |
| EB18 | turicensis | Neonate (meningitis) | Switzerland | NA |
| E899 | sakazakii | Clinical | USA | NA |
| Samples | Cronobacter Taxa Expected | Cronobacter Taxa Observed WIMP a | Cronobacter Taxa Observed by Phylogenetic Tree | qPCR CT Value | Estimated Cronobacter CFU/mL b | Cronobacter ONT % DNA Sample |
|---|---|---|---|---|---|---|
| E477 | sakazakii | sakazakii | sakazakii | 13.6 | 1.9 × 108 | 97.0 |
| E515 | dublinensis | dublinensis | dublinensis | 17.0 | 1.5 × 107 | 90.0 |
| E603 | muytjensii | muytjensii | muytjensii | 13.1 | 2.8 × 108 | 99.0 |
| E604 | sakazakii | sakazakii | sakazakii | 12.1 | 5.8 × 108 | 90.0 |
| E791 | dublinensis | dublinensis | dublinensis | 13.4 | 2.2 × 108 | 92.0 |
| E797 | universalis | universalis | universalis | 12.4 | 4.3 × 108 | 99.6 |
| E825 | sakazakii | sakazakii | sakazakii | 12.7 | 3.7 × 108 | 97.1 |
| EB13 | sakazakii | sakazakii | sakazakii | 12.2 | 5.3 × 108 | 98.2 |
| EB18 | turicensis | universalis | turicensis | 12.4 | 4.4 × 108 | 90.4 |
| E899 | sakazakii | sakazakii | sakazakii | 15.3 | 5.3 × 107 | 95.4 |
| Samples | Total Reads | Total Mb | Estimated Coverage Cronobacter Genome All Reads (X) | Reads above 4000 bp | Total Mb above 4000 bp | Estimated Coverage Cronobacter Genome > 4 kb Reads (X) |
|---|---|---|---|---|---|---|
| E477 | 228,000 | 1069 | 238 | 82,998 | 732 | 163 |
| E515 | 70,096 | 354 | 79 | 26,066 | 248 | 55 |
| E603 | 523,730 | 2665 | 592 | 203,038 | 1889 | 420 |
| E604 | 127,684 | 643 | 143 | 47,322 | 455 | 101 |
| E791 | 257,927 | 1148 | 255 | 85,301 | 752 | 167 |
| E797 | 278,012 | 1382 | 307 | 105,162 | 969 | 215 |
| E825 | 210,799 | 1010 | 224 | 77,642 | 697 | 155 |
| EB13 | 239,315 | 1228 | 273 | 91,575 | 870 | 193 |
| EB18 | 319,323 | 1648 | 366 | 125,660 | 1168 | 260 |
| E899 | 191,946 | 941 | 209 | 70,517 | 647 | 144 |
| Samples | Contig No. | %GC Content | Genome Size (bp) | Genome Coverage (X) |
|---|---|---|---|---|
| E477 | 3 | 56.6 | 4,507,829; 93,905; 53,449 | 136; 156; 287 |
| E515 | 1 | 57.9 | 4,487,108 | 65 |
| E603 | 1 | 57.7 | 4,305,928 | 516 |
| E604 | 3 | 56.6 | 4,412,859; 117,865; 52,143 | 115; 123; 179 |
| E791 | 2 | 58.1 | 4,349,860; 166,041 | 208; 239 |
| E797 | 2 | 57.9 | 4,075,540; 129,777 | 273; 306 |
| E825 | 3 | 56.8 | 4,257,543; 97,419; 53,456 | 185; 163; 260 |
| EB13 | 3 | 56.7 | 4,347,023; 131,190; 31,203 | 214; 265; 1269 |
| EB18 | 4 | 57.2 | 4,384,296; 144,804; 53,716; 44,722 | 283; 357; 552; 375 |
| E899 | 2 | 56.7 | 4,340,415; 53,472 | 176; 284 |
| Samples | ST a | Cronobacter Taxa by ST | Serotype b |
|---|---|---|---|
| E477 | 8 | sakazakii | SO1 |
| E515 | 80 | dublinensis | DO2 |
| E603 | 81 | muytjensii | MuO2 |
| E604 | 15 | sakazakii | SO2 |
| E791 | novel | dublinensis | DO1a |
| E797 | 54 | universalis | UO1 |
| E825 | 8 | sakazakii | SO1 |
| EB13 | 1 | sakazakii | SO1 |
| EB18 | 19 | turicensis | TO1 |
| E899 | 4 | sakazakii | SO2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonzalez-Escalona, N.; Kwon, H.J.; Chen, Y. Nanopore Sequencing Allows Recovery of High-Quality Completely Closed Genomes of All Cronobacter Species from Powdered Infant Formula Overnight Enrichments. Microorganisms 2024, 12, 2389. https://doi.org/10.3390/microorganisms12122389
Gonzalez-Escalona N, Kwon HJ, Chen Y. Nanopore Sequencing Allows Recovery of High-Quality Completely Closed Genomes of All Cronobacter Species from Powdered Infant Formula Overnight Enrichments. Microorganisms. 2024; 12(12):2389. https://doi.org/10.3390/microorganisms12122389
Chicago/Turabian StyleGonzalez-Escalona, Narjol, Hee Jin Kwon, and Yi Chen. 2024. "Nanopore Sequencing Allows Recovery of High-Quality Completely Closed Genomes of All Cronobacter Species from Powdered Infant Formula Overnight Enrichments" Microorganisms 12, no. 12: 2389. https://doi.org/10.3390/microorganisms12122389
APA StyleGonzalez-Escalona, N., Kwon, H. J., & Chen, Y. (2024). Nanopore Sequencing Allows Recovery of High-Quality Completely Closed Genomes of All Cronobacter Species from Powdered Infant Formula Overnight Enrichments. Microorganisms, 12(12), 2389. https://doi.org/10.3390/microorganisms12122389








