BiP Proteins from Symbiodiniaceae: A “Shocking” Story
Abstract
1. Introduction
1.1. Discovery of BiP Proteins
1.2. Functional Insights of BiP Proteins
2. Stress Responses, Light-Regulated Phosphorylation, and Possible Roles of BiP Proteins in Symbiodiniaceae
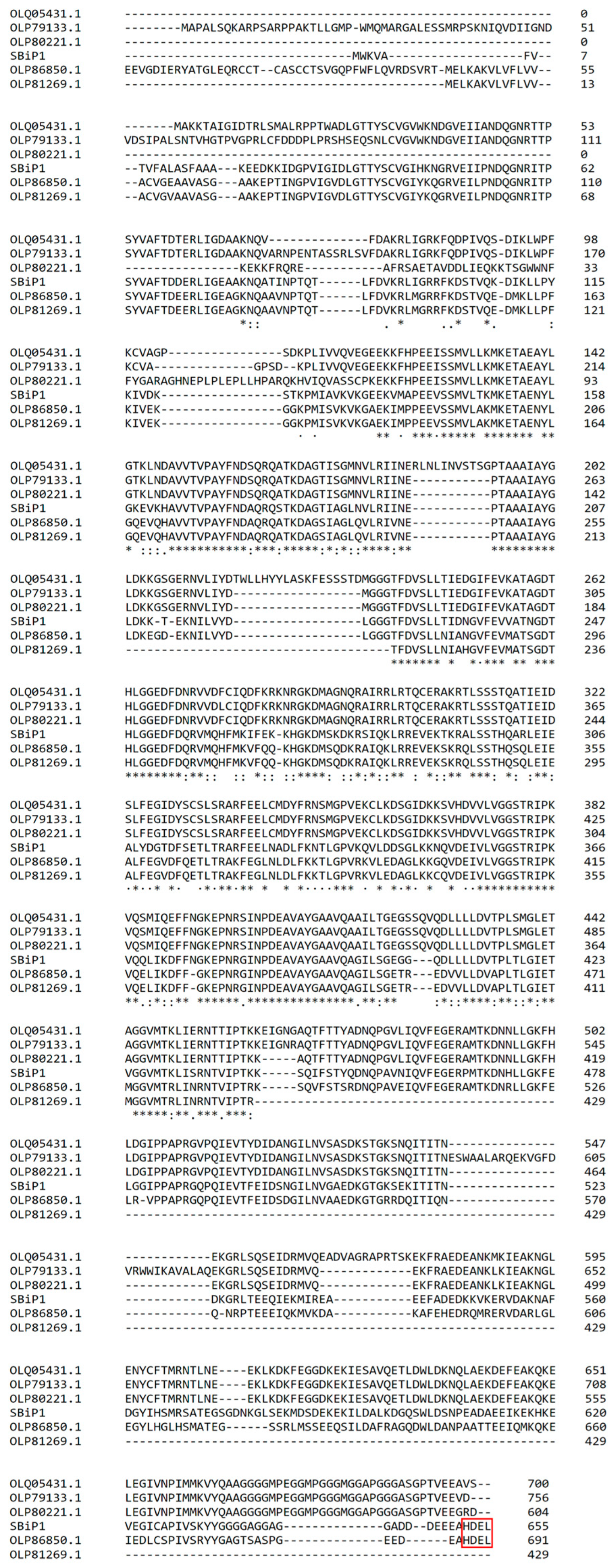
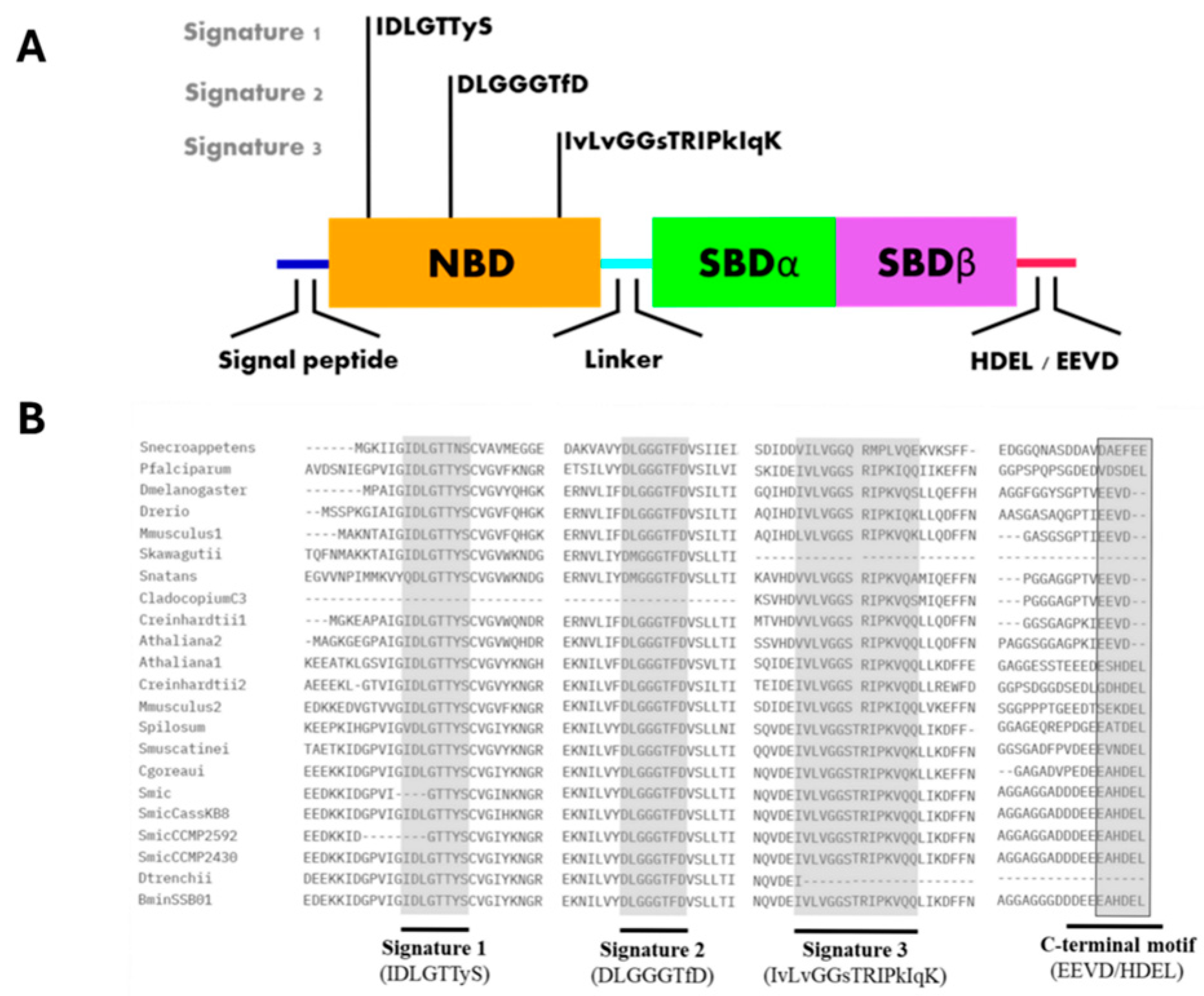
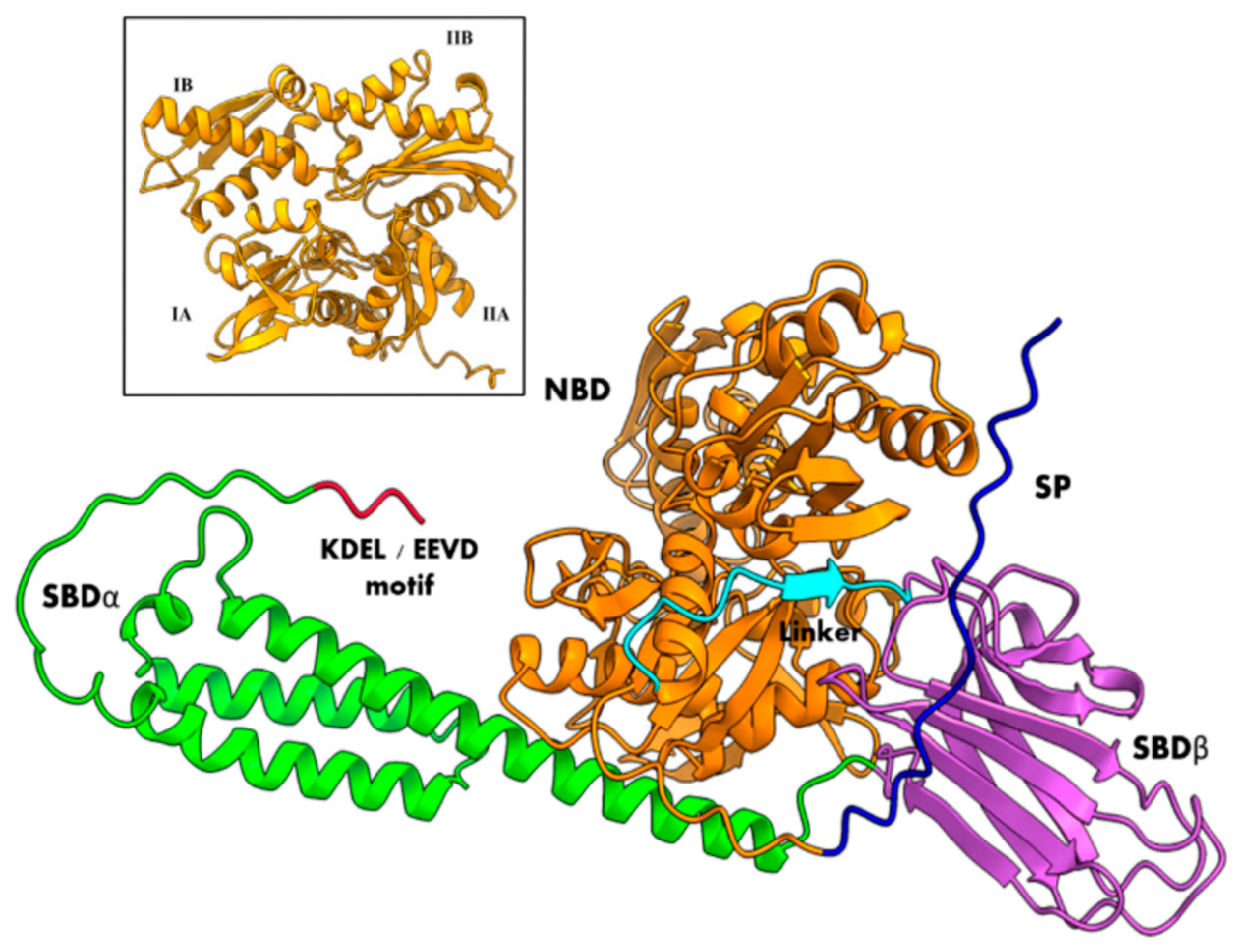
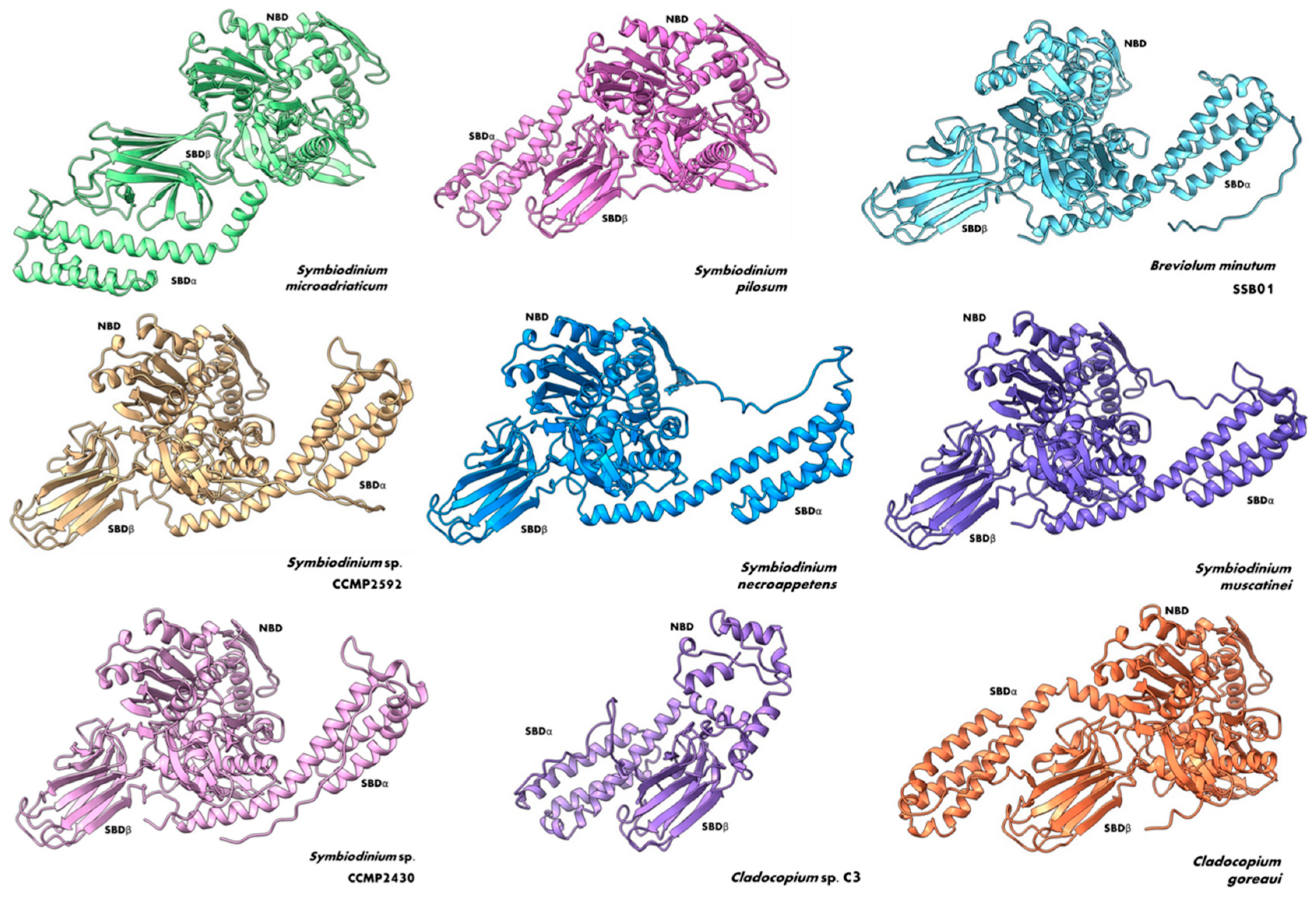
3. Structural Insights of BiP Proteins
4. Regulation of BiP Gene Expression
5. BiP Protein Regulation and Expression Patterns in Dinoflagellates
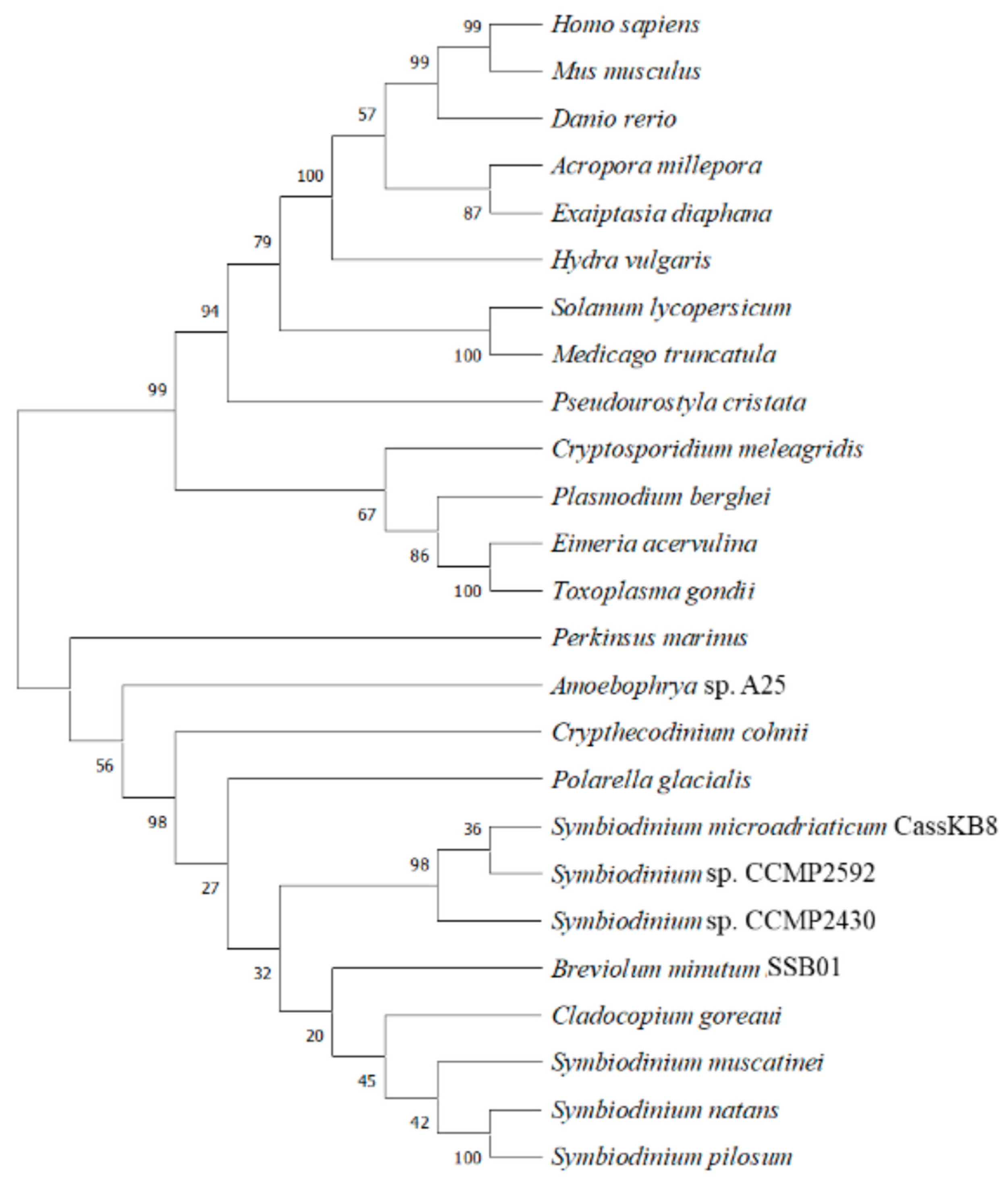
6. Phylogenetic Analysis Shows Conservation and Clustering of Dinoflagellate BiP Proteins
7. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Morrison, S.L.; Scharf, M.D. Heavy chain-producing variants of a mouse myeloma cell line. J. Immunol. 1975, 114, 655–659. [Google Scholar] [CrossRef] [PubMed]
- Haas, I.G.; Wabl, M. Immunoglobulin heavy chain binding protein. Nature 1983, 306, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Haas, I.G.; Wabl, M. Immunoglobulin heavy chain toxicity in plasma cells is neutralized by fusion to pre-B cells. Proc. Natl. Acad. Sci. USA 1984, 81, 7185–7188. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; Bell, J.; Ting, J. Biochemical characterization of the 94- and 78-kilodalton glucose-regulated proteins in hamster fibroblasts. J. Biol. Chem. 1984, 259, 4616–4621. [Google Scholar] [CrossRef] [PubMed]
- Munro, S.; Pelham, H.R. An Hsp70-like protein in the ER: Identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell 1986, 46, 291–300. [Google Scholar] [CrossRef] [PubMed]
- Bole, D.G.; Hendershot, L.M.; Kearney, J.F. Posttranslational association of immunoglobulin heavy chain binding protein with nascent heavy chains in nonsecreting and secreting hybridomas. J. Cell Biol. 1986, 102, 1558–1566. [Google Scholar] [CrossRef]
- Haas, L.G.; Meo, T. cDNA cloning of the immunoglobulin heavy chain binding protein. Proc. Nat. Acad. Sci. USA 1988, 85, 2250–2254. [Google Scholar] [CrossRef]
- Morán Luengo, T.; Mayer, M.P.; Rüdiger, S.G.D. The Hsp70–Hsp90 chaperone cascade in protein folding. Trends Cell Biol. 2019, 29, 164–177. [Google Scholar] [CrossRef]
- Kopp, M.C.; Larburu, N.; Durairaj, V.; Adams, C.J.; Ali, M.M.U. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat. Struct. Mol. Biol. 2019, 26, 1053–1062. [Google Scholar] [CrossRef]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef]
- Mayer, M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Medina, R.E.; Islas-Flores, T.; Villanueva, M.A. Phosphorylation/dephosphorylation response to light stimuli of Symbiodinium proteins: Specific light-induced dephosphorylation of an HSP-like 75 kDa protein from S. microadriaticum. PeerJ 2019, 7, e7406. [Google Scholar] [CrossRef]
- Castillo-Medina, R.E.; Islas-Flores, T.; Villanueva, M.A. Light-stimulated dephosphorylation of the BiP-like protein, SmicHSP75 (SBiP1) from Symbiodinium microadriaticum is inhibited by elevated but not low temperature and suggests regulation of the chaperone function. Acta Biochim. Pol. 2022, 69, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Medina, R.E.; Islas-Flores, T.; Morales-Ruiz, E.; Villanueva, M.A. Biochemical and molecular characterization of the SBiP1 chaperone from Symbiodinium microadriaticum CassKB8 and light parameters that modulate its phosphorylation. PLoS ONE 2023, 18, e0293299. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Medina, R.E.; Islas-Flores, T.; Morales-Ruiz, E.; Villanueva, M.A. Inhibition of protein or glutamine biosynthesis affect the activation of the light stimulated SBiP1 chaperone by dephosphorylation in Symbiodiniaceae. bioRxiv 2024. [Google Scholar] [CrossRef]
- Díaz-Troya, S.; Pérez-Pérez, M.E.; Pérez-Martín, M.; Moes, S.; Jeno, P.; Florencio, F.J.; Crespo, J.L. Inhibition of protein synthesis by TOR inactivation revealed a conserved regulatory mechanism of the BiP chaperone in Chlamydomonas. Plant Physiol. 2011, 157, 730–741. [Google Scholar] [CrossRef]
- Nitika; Truman, A.W. Cracking the chaperone code: Cellular roles for Hsp70 phosphorylation. Trends Biochem. Sci. 2017, 42, 932–935. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase. University of Galway (Republished from AlgaeBase with Permission of M.D. Guiry). 2024. Available online: https://www.marinespecies.org (accessed on 29 May 2024).
- Hoppenrath, M.; Leander, B.S. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS ONE 2010, 5, e13220. [Google Scholar] [CrossRef]
- Stat, M.; Carter, D.; Hoegh-Guldberg, O. The evolutionary history of Symbiodinium and scleractinian hosts—Symbiosis, diversity, and the effect of climate change. Perspect. Plant Ecol. Evol. Syst. 2006, 8, 23–43. [Google Scholar] [CrossRef]
- Bogumil, D.; Alvarez-Ponce, D.; Landan, G.; McInerney, J.O.; Dagan, T. Integration of two ancestral chaperone systems into one: The evolution of eukaryotic molecular chaperones in light of eukaryogenesis. Mol. Biol. Evol. 2014, 31, 410–418. [Google Scholar] [CrossRef][Green Version]
- Rosic, N.N.; Pernice, M.; Dove, S.; Dunn, S.; Hoegh-Guldberg, O. Gene expression profiles of cytosolic heat shock proteins Hsp70 and Hsp90 from symbiotic dinoflagellates in response to thermal stress: Possible implications for coral bleaching. Cell Stress Chaperones 2011, 16, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Leggat, W.; Seneca, F.; Wasmund, K.; Ukani, L.; Yellowlees, D.; Ainsworth, T.D. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS ONE 2011, 6, e26687. [Google Scholar] [CrossRef] [PubMed]
- Levin, R.A.; Beltran, V.H.; Hill, R.; Kjelleberg, S.; McDougald, D.; Steinberg, P.D.; Van Oppen, M.J.H. Sex, scavengers, and chaperones: Transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 2016, 33, 2201–2215. [Google Scholar] [CrossRef] [PubMed]
- Petrou, K.; Nunn, B.L.; Padula, M.P.; Miller, D.J.; Nielsen, D.A. Broad scale proteomic analysis of heat-destabilised symbiosis in the hard coral Acropora millepora. Sci. Rep. 2021, 11, 19061. [Google Scholar] [CrossRef]
- Adams, C.J.; Kopp, M.C.; Larburu, N.; Nowak, P.R.; Ali, M.M.U. Structure and molecular mechanism of ER stress signaling by the Unfolded Protein Response signal activator IRE1. Front. Mol. Biosci. 2019, 6, 11. [Google Scholar] [CrossRef]
- Pobre, K.F.R.; Poet, G.J.; Hendershot, L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J. Biol. Chem. 2019, 294, 2098–2108. [Google Scholar] [CrossRef]
- Singh, A.; Grover, A. Genetic engineering for heat tolerance in plants. Physiol. Mol. Biol. Plants 2008, 14, 155–166. [Google Scholar] [CrossRef]
- Warner, M.E.; Fitt, W.K.; Schmidt, G.W. Damage to photosystem II in symbiotic dinoflagellates: A determinant of coral bleaching. Proc. Nat. Acad. Sci. USA 1999, 96, 8007–8012. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Nakamura, T.; Sakamizu, M.; van Woesik, R.; Yamasaki, H. Repair machinery of symbiotic photosynthesis as the primary target of heat stress for reef-building corals. Plant Cell Physiol. 2004, 45, 251–255. [Google Scholar] [CrossRef]
- Takahashi, S.; Whitney, S.; Itoh, S.; Badger, M. Heat stress causes inhibition of the de novo synthesis of antenna proteins and photobleaching in cultured Symbiodinium. Proc. Nat. Acad. Sci. USA 2008, 105, 4203–4208. [Google Scholar] [CrossRef]
- McGinley, M.P.; Aschaffenburg, M.D.; Pettay, D.T.; Smith, R.T.; LaJeunesse, T.C.; Warner, M.E. Transcriptional response of two core photosystem genes in Symbiodinium spp. exposed to thermal stress. PLoS ONE 2012, 7, e50439. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gierz, S.L.; Gordon, B.R.; Leggat, W. Integral light-harvesting complex expression in Symbiodinium within the coral Acropora aspera under thermal stress. Sci. Rep. 2016, 6, 25081. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.; Li, Y.; Liew, Y.J.; Baumgarten, S.; Simakov, O.; Wilson, M.C.; Piel, J.; Ashoor, H.; Bougouffa, S.; Bajic, V.B.; et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 2016, 6, 39734. [Google Scholar] [CrossRef] [PubMed]
- Castillo Medina, R.E. Caracterización del Estado de Fosforilación de una Proteína tipo HSP70 de Symbiodinium sp. KB8 a Diferentes Condiciones de luz y Temperatura. Ph.D. Thesis, Universidad Nacional Autónoma de México, Mexico City, Mexico, 2020. Available online: http://132.248.9.195/ptd2020/octubre/0804538/Index.html (accessed on 9 December 2020).
- Rädecker, N.; Escrig, S.; Spangenberg, J.E.; Voolstra, C.R.; Meibom, A. Coupled carbon and nitrogen cycling regulates the cnidarian–algal symbiosis. Nat. Commun. 2023, 14, 6948. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Gupta, R.S.; Golding, G.B. Evolution of HSP70 gene and its implications regarding relationships between archaebacteria, eubacteria, and eukaryotes. J. Mol. Evol. 1993, 37, 573–582. [Google Scholar] [CrossRef]
- Liu, J.; Yang, W.J.; Zhu, X.J.; Karouna-Renier, N.K.; Rao, R.K. Molecular cloning and expression of two HSP70 genes in the prawn, Macrobrachium rosenbergii. Cell Stress Chaperones 2004, 9, 313. [Google Scholar] [CrossRef]
- Song, B.; Morse, D.; Song, Y.; Fu, Y.; Lin, X.; Wang, W.; Cheng, S.; Chen, W.; Liu, X.; Lin, S. Comparative genomics reveals two major bouts of gene retroposition coinciding with crucial periods of Symbiodinium evolution. Genome Biol. Evol. 2017, 9, 2037–2047. [Google Scholar] [CrossRef]
- LaJeunesse, T.C.; Parkinson, J.E.; Gabrielson, P.W.; Jeong, H.J.; Reimer, J.D.; Voolstra, C.R.; Santos, S.R. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 2018, 28, 2570–2580. [Google Scholar] [CrossRef]
- Parkinson, J.E.; Baumgarten, S.; Michell, C.T.; Baums, I.B.; LaJeunesse, T.C.; Voolstra, C.R. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol. Evol. 2016, 8, 665–680. [Google Scholar] [CrossRef]
- Yu, L.; Li, T.; Li, L.; Lin, X.; Li, H.; Liu, C.; Guo, C.; Lin, S. SAGER: A database of Symbiodiniaceae and Algal Genomic Resource. Database 2020, 2020, baaa051. [Google Scholar] [CrossRef] [PubMed]
- Rocker, M.M.; Willis, B.L.; Bay, L.K. Thermal stress-related gene expression in corals with different Symbiodinium types. In Proceedings of the 12th International Coral Reef Symposium, Cairns, Australia, 9–13 July 2012; Volume 9, pp. 1–5. [Google Scholar]
- Barshis, D.J.; Ladner, J.T.; Oliver, T.A.; Palumbi, S.R. Lineage-specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol. Biol. Evol. 2014, 31, 1343–1352. [Google Scholar] [CrossRef]
- Bellantuono, A.J.; Dougan, K.E.; Granados-Cifuentes, C.; Rodriguez-Lanetty, M. Free-living and symbiotic lifestyles of a thermotolerant coral endosymbiont display profoundly distinct transcriptomes under both stable and heat stress conditions. Mol. Ecol. 2019, 28, 5265–5281. [Google Scholar] [CrossRef] [PubMed]
- Herath, V.; Gayral, M.; Adhikari, N.; Miller, R.; Verchot, J. Genome-wide identification and characterization of Solanum tuberosum BiP genes reveal the role of the promoter architecture in BiP gene diversity. Sci. Rep. 2020, 10, 11327. [Google Scholar] [CrossRef]
- Imbriano, C.; Bolognese, F.; Gurtner, A.; Piaggio, G.; Mantovani, R. HSP-CBF is an NF-Y-dependent coactivator of the heat shock promoters CCAAT boxes. J. Biol. Chem. 2001, 276, 26332–26339. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, R.; Baranwal, V.K.; Kumar, R.; Sircar, D.; Chauhan, H. Genome-wide identification and expression analysis of Hsp70, Hsp90, and Hsp100 heat shock protein genes in barley under stress conditions and reproductive development. Funct. Integr. Genom. 2019, 19, 1007–1022. [Google Scholar] [CrossRef]
- Hu, Z.; Ban, Q.; Hao, J.; Zhu, X.; Cheng, Y.; Mao, J.; Lin, M.; Xia, E.; Li, Y. Genome-wide characterization of the C-repeat binding factor (CBF) gene family involved in the response to abiotic stresses in tea plant (Camellia sinensis). Front. Plant Sci. 2020, 11, 921. [Google Scholar] [CrossRef]
- Levin, R.A.; Voolstra, C.R.; Agrawal, S.; Steinberg, P.D.; Suggett, D.J.; Van Oppen, M.J.H. Engineering strategies to decode and enhance the genomes of coral symbionts. Front. Microbiol. 2017, 8, 1220. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Haralampidis, K.; Milioni, D.; Rigas, S.; Hatzopoulos, P. Combinatorial interaction of cis elements specifies the expression of the Arabidopsis AtHsp90-1 gene. Plant Physiol. 2002, 129, 1138–1149. [Google Scholar] [CrossRef]
- Von Gromoff, E.D.; Schroda, M.; Oster, U.; Beck, C.F. Identification of a plastid response element that acts as an enhancer within the Chlamydomonas HSP70A promoter. Nucleic Acids Res. 2006, 34, 4767–4779. [Google Scholar] [CrossRef] [PubMed]
- Martínez, I.M.; Chrispeels, M.J. Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 2003, 15, 561–576. [Google Scholar] [CrossRef] [PubMed]
- Fast, N.M.; Xue, L.; Bingham, S.; Keeling, P.J. Re-examining alveolate evolution using multiple protein molecular phylogenies. J. Eukaryot. Microbiol. 2005, 49, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, S.-F.; Lin, L.; Wang, D.-Z. Whole transcriptomic analysis provides insights into molecular mechanisms for toxin biosynthesis in a toxic dinoflagellate Alexandrium catenella (ACHKT). Toxins 2017, 9, 213. [Google Scholar] [CrossRef]
- Supasri, K.M. An Investigation of High-Throughput Proteomic Analysis in Marine Dinoflagellate (Symbiodiniaceae). Ph.D. Thesis, School of Life Sciences, University of Technology Sydney, Ultimo, Australia, 2021. Available online: https://opus.lib.uts.edu.au/bitstream/10453/155987/2/02whole.pdf (accessed on 15 April 2021).
- Sorek, M.; Díaz-Almeyda, E.M.; Medina, M.; Levy, O. Circadian clocks in symbiotic corals: The duet between Symbiodinium algae and their coral host. Mar. Genom. 2014, 14, 47–57. [Google Scholar] [CrossRef]
- Hastings, J.W.; Astrachan, L.; Sweeney, B.M. A persistent daily rhythm in photosynthesis. J. Gen. Physiol. 1961, 45, 69–76. [Google Scholar] [CrossRef]
- Seo, K.S.; Fritz, L. Cell ultrastructural changes correlate with circadian rhythms in Pyrocystis lunula (Pyrrophyta). J. Phycol. 2000, 36, 351–358. [Google Scholar] [CrossRef]
- Gong, S.; Li, G.; Liang, J.; Xu, L.; Tan, Y.; Jin, X.; Xia, X.; Yu, K. Day-night cycle as a key environmental factor affecting coral-Symbiodiniaceae symbiosis. Ecol. Indic. 2023, 146, 109890. [Google Scholar] [CrossRef]
- Fraga, J.; Fernández-Calienes, A.; Montalvo, A.M.; Maes, I.; Deborggraeve, S.; Büscher, P.; Dujardin, J.-C.; Van Der Auwera, G. Phylogenetic analysis of the Trypanosoma genus based on the heat-shock protein 70 gene. Infect. Genet. Evol. 2016, 43, 165–172. [Google Scholar] [CrossRef]
- Noh, S.-J.; Kwon, C.S.; Oh, D.-H.; Moon, J.S.; Chung, W.-I. Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 2003, 311, 81–91. [Google Scholar] [CrossRef]
- Zhu, J.; Hao, P.; Chen, G.; Han, C.; Li, X.; Zeller, F.J.; Hsam, S.L.; Hu, Y.; Yan, Y. Molecular cloning, phylogenetic analysis, and expression profiling of endoplasmic reticulum molecular chaperone BiP genes from bread wheat (Triticum aestivum L.). BMC Plant Biol. 2014, 14, 260. [Google Scholar] [CrossRef]
- Guo, R.; Youn, S.H.; Ki, J.-S. Heat shock protein 70 and 90 genes in the harmful dinoflagellate Cochlodinium polykrikoides: Genomic structures and transcriptional responses to environmental stresses. Int. J. Genom. 2015, 484626. [Google Scholar] [CrossRef]
| Organism | Name | Sequence Type | Subcellular Localization | Cis-Regulatory Elements | Accession No. |
|---|---|---|---|---|---|
| Symbiodinium microadriaticum Cass KB8 | SBiP1 | Protein | ER | CCAAT-box, PRE, CBF/DRE | OP429595 |
| Symbiodinium sp. CCMP2592 | BiP5 | Protein | ER | CCAAT-box, PRE, CBF/DRE | CAE7227403 |
| Symbiodinium microadriaticum | BiP5 | Protein | ER | CCAAT-box, PRE, CBF/DRE | OLP91134 |
| Symbiodinium pilosum | BiP5 | Protein | ER | CCAAT-box, PRE, CBF/DRE, UPR-I | CAE7221339 |
| Symbiodinium natans | - | Protein | ER | CCAAT-box, CBF/DRE | CAE7360486 |
| Symbiodinium necroappetens | carB | Protein | Cytosol | CCAAT-box, CBF/DRE | CAE7932356 |
| Cladocopium sp. C3 | Hsp70 | Protein | Cytosol | NP | ABA28988 |
| Symbiodinium sp. CCMP2430 | Hsp70 | mRNA | ER | NP | HBTH01040005 |
| Breviolum minutum SSB01 | - | mRNA | ER | NP | GICE01029930 |
| Cladocopium goreaui | - | mRNA | ER | NP | ICPI01002597 |
| Durusdinium trenchii | - | mRNA | NP | NP | ICPJ01013860 |
| Symbiodinium muscatinei | - | mRNA | ER | NP | GFDR03033717 |
| Symbiodinium kawagutii CCMP2468 | - | mRNA | NP | NP | KC950716 |
| Organism | Sequence Name | Subcellular Localization | Accession No. |
|---|---|---|---|
| Crypthecodinium cohnii | BiP | ER | AF421538 |
| Prorocentrum minimum | Hsp70 | Cytosol | JN401970 |
| Akashiwo sanguinea | Hsp70 | cytosol | KJ755185 |
| Cochlodinium polykrikoides | Hsp70 | Cytosol | KP010830 |
| Heterocapsa triquetra | Hsp70 | Unknown | AY729868 |
| Polarella glacialis | Unnamed | ER | CAE8584766 |
| Amoebophyra sp. A25 | Unnamed | ER | CAD7976323 |
| Uncultured dinoflagellate | Unnamed | Cytosol | GU555406 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales-Ruiz, E.; Islas-Flores, T.; Villanueva, M.A. BiP Proteins from Symbiodiniaceae: A “Shocking” Story. Microorganisms 2024, 12, 2126. https://doi.org/10.3390/microorganisms12112126
Morales-Ruiz E, Islas-Flores T, Villanueva MA. BiP Proteins from Symbiodiniaceae: A “Shocking” Story. Microorganisms. 2024; 12(11):2126. https://doi.org/10.3390/microorganisms12112126
Chicago/Turabian StyleMorales-Ruiz, Estefanía, Tania Islas-Flores, and Marco A. Villanueva. 2024. "BiP Proteins from Symbiodiniaceae: A “Shocking” Story" Microorganisms 12, no. 11: 2126. https://doi.org/10.3390/microorganisms12112126
APA StyleMorales-Ruiz, E., Islas-Flores, T., & Villanueva, M. A. (2024). BiP Proteins from Symbiodiniaceae: A “Shocking” Story. Microorganisms, 12(11), 2126. https://doi.org/10.3390/microorganisms12112126






