Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Viral Nucleic Acid Extraction
2.3. Sequencing of SARS-CoV-2 Isolates
2.4. Genetic Analysis
2.5. Phylogenetic Analysis
3. Results
3.1. COVID Lineage Analysis
3.2. Evaluation of the RdRp Complex Variability
3.2.1. Catalytic Subunit
3.2.2. Accessory Subunits
3.3. Evaluation of Nsp5 Variability
3.4. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malone, B.; Urakova, N.; Snijder, E.J.; Campbell, E.A. Structures and functions of coronavirus replication-transcription complexes and their relevance for SARS-CoV-2 drug design. Nat. Rev. Mol. Cell Biol. 2022, 23, 21–39. [Google Scholar] [CrossRef] [PubMed]
- Van Cleemput, J.; van Snippenberg, W.; Lambrechts, L.; Dendooven, A.; D’Onofrio, V.; Couck, L.; Trypsteen, W.; Vanrusselt, J.; Theuns, S.; Vereecke, N.; et al. Organ-specific genome diversity of replication-competent SARS-CoV-2. Nat. Commun. 2021, 12, 6612, Erratum in Nat. Commun. 2022, 13, 6247. [Google Scholar] [CrossRef] [PubMed]
- Trypsteen, W.; Van Cleemput, J.; Snippenberg, W.; Gerlo SVandekerckhove, L. On the whereabouts of SARS-CoV-2 in the human body: A systematic review. PLoS Pathog. 2020, 16, e1009037. [Google Scholar] [CrossRef] [PubMed]
- Lou, Z.; Rao, Z. The Life of SARS-CoV-2 Inside Cells: Replication-Transcription Complex Assembly and Function. Annu. Rev. Biochem. 2022, 91, 381–401. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Bonilla, H.; Jagannathan, P.; Shafer, R.W. SARS-CoV-2 Antiviral Therapy. Clin. Microbiol. Rev. 2021, 34, e0010921. [Google Scholar] [CrossRef] [PubMed]
- Malone, B.; Campbell, E.A. Molnupiravir: Coding for catastrophe. Nat. Struct. Mol. Biol. 2021, 28, 706–708, Erratum in Nat. Struct. Mol. Biol. 2021, 28, 955. [Google Scholar] [CrossRef] [PubMed]
- Rubin, D.; Chan-Tack, K.; Farley, J.; Sherwat, A. FDA Approval of Remdesivir—A Step in the Right Direction. N. Engl. J. Med. 2020, 383, 2598–2600. [Google Scholar] [CrossRef]
- Wise, J. COVID-19: Remdesivir is recommended for authorisation by European Medicines Agency. BMJ 2020, 369, m2610. [Google Scholar] [CrossRef]
- Vicenti, I.; Zazzi, M.; Saladini, F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 2021, 31, 325–337. [Google Scholar] [CrossRef]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, L.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef]
- Bravo, J.P.K.; Dangerfield, T.L.; Taylor, D.W.; Johnson, K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell 2021, 81, 1548–1552.e4. [Google Scholar] [CrossRef] [PubMed]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Höbartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Medicines and Healthcare Products Regulatory Agency. Regulatory Approval of Lagevrio (Molnupiravir). Available online: https://www.gov.uk/government/publications/regulatory-approval-of-lagevrio-molnupiravir (accessed on 4 November 2021).
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes Additional Oral Antiviral for Treatment of COVID-19 in Certain Adults. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-additional-oral-antiviral-treatment-covid-19-certain (accessed on 23 December 2021).
- Kabinger, F.; Stiller, C.; Schmitzová, J.; Dienemann, C.; Kokic, G.; Hillen, H.S.; Höbartner, C.; Cramer, P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021, 28, 740–746. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports. BMJ 2021, 375, n2422. [Google Scholar] [CrossRef] [PubMed]
- European Medicines Agency. Withdrawal of Marketing Authorizarion Application for LAGEVRIO, Molnupiravir, 200 mg Hard Capsules, EMEA/H/C/005789. Available online: https://www.ema.europa.eu/en/documents/withdrawal-letter/withdrawal-letter-lagevrio_en.pdf (accessed on 21 January 2023).
- Food and Drug Administration. Coronavirus (COVID-19) Update: FDA Authorizes First Oral Antiviral for Treatment of COVID-19. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-first-oral-antiviral-treatment-covid-19 (accessed on 22 December 2021).
- European Medicines Agency. COVID-19: EMA Recommends Conditional Marketing Authorisation for Paxlovid. Available online: https://www.ema.europa.eu/en/news/covid-19-ema-recommends-conditional-marketing-authorisation-paxlovid#:~:text=COVID%2D19%3A%20EMA%20recommends%20conditional%20marketing%20authorisation%20for%20Paxlovid,-Share&text=Update%3A%20Paxlovid%20is%20now%20authorised,Commission%20on%2028%20January%202022 (accessed on 27 January 2022).
- Kneller, D.W.; Phillips, G.; O’Neill, H.M.; Jedrzejczak, R.; Stols, L.; Langan, P.; Joachimiak, A.; Coates, L.; Kovalevsky, A. Structural plasticity of SARS-CoV-2 3CL Mpro active site cavity revealed by room temperature X-ray crystallography. Nat. Commun. 2020, 11, 3202. [Google Scholar] [CrossRef]
- Lee, J.; Worrall, L.J.; Vuckovic, M.; Rosell, F.I.; Gentile, F.; Ton, A.T.; Caveney, N.A.; Ban, F.; Cherkasov, A.; Paetzel, M.; et al. Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site. Nat. Commun. 2020, 11, 5877. [Google Scholar] [CrossRef]
- Mahase, E. COVID-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness, company reports. BMJ 2021, 375, n2713. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef]
- Vere Hodge, A.; Field, H.J. General Mechanisms of Antiviral Resistance. In Genetics and Evolution of Infectious Disease; Elsevier: Amsterdam, The Netherlands, 2011; pp. 339–362. [Google Scholar] [CrossRef]
- Koyama, T.; Platt, D.; Parida, L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef]
- Mason, S.; Devincenzo, J.P.; Toovey, S.; Wu, J.Z.; Whitley, R.J. Comparison of antiviral resistance across acute and chronic viral infections. Antivir. Res. 2018, 158, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Ono, S.K.; Bassit, L.; Verma, K.; Amblard, F.; Schinazi, R.F. Assessment of a Computational Approach to Predict Drug Resistance Mutations for HIV, HBV and SARS-CoV-2. Molecules 2022, 27, 5413. [Google Scholar] [CrossRef] [PubMed]
- Pachetti, M.; Marini, B.; Benedetti, F.; Giudici, F.; Mauro, E.; Storici, P.; Masciovecchio, C.; Angeletti, S.; Ciccozzi, M.; Gallo, R.C.; et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 2020, 18, 179. [Google Scholar] [CrossRef] [PubMed]
- Eskier, D.; Suner, A.; Karakülah, G.; Oktay, Y. Mutation density changes in SARS-CoV-2 are related to the pandemic stage but to a lesser extent in the dominant strain with mutations in spike and RdRp. PeerJ 2020, 8, e9703. [Google Scholar] [CrossRef] [PubMed]
- Martinot, M.; Jary, A.; Fafi-Kremer, S.; Leducq, V.; Delagreverie, H.; Garnier, M.; Pacanowski, J.; Mékinian, A.; Pirenne, F.; Tiberghien, P.; et al. Emerging RNA-Dependent RNA Polymerase Mutation in a Remdesivir-Treated B-cell Immunodeficient Patient with Protracted Coronavirus Disease 2019. Clin. Infect. Dis. 2021, 73, e1762–e1765. [Google Scholar] [CrossRef] [PubMed]
- Szemiel, A.M.; Merits, A.; Orton, R.J.; MacLean, O.A.; Pinto, R.M.; Wickenhagen, A.; Lieber, G.; Turnbull, M.L.; Wang, S.; Furnon, W.; et al. In vitro selection of Remdesivir resistance suggests evolutionary predictability of SARS-CoV-2. PLoS Pathog. 2021, 17, e1009929. [Google Scholar] [CrossRef] [PubMed]
- Ip, J.D.; Chu, A.W.-H.; Chan, W.-M.; Leung, R.C.-Y.; Abdullah, S.M.U.; Sun, Y.; To, K.K.-W. Global prevalence of SARS-CoV-2 3CL protease mutations associated with nirmatrelvir or ensitrelvir resistance. EBioMedicine 2023, 91, 104559. [Google Scholar] [CrossRef]
- Peloquin, D.; DiMaio, M.; Bierer, B.; Barnes, M. Disruptive and avoidable: GDPR challenges to secondary research uses of data. Eur. J. Hum. Genet. 2020, 28, 697–705. [Google Scholar] [CrossRef]
- Italian Data Protection Authority. Authorization n. 9/2016—General Authorization to Process Personal Data for Scientific Research Purposes. 2016. Available online: https://www.garanteprivacy.it/home/docweb/-/docweb-display/docweb/5805552 (accessed on 15 December 2016).
- De Sabato, L.; Vaccari, G.; Knijn, A.; Ianiro, G.; Di Bartolo, I.; Morabito, S. SARS-CoV-2 RECoVERY: A multi-platform open-source bioinformatic pipeline for the automatic construction and analysis of SARS-CoV-2 genomes from NGS sequencing data. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Dragelj, J.; Mroginski, M.A.; Ebrahimi, K.H. Hidden in Plain Sight: Natural Products of Commensal Microbiota as an Environmental Selection Pressure for the Rise of New Variants of SARS-CoV-2. Chembiochem 2021, 22, 2946–2950, Erratum in Chembiochem 2022, 23, e202200362. [Google Scholar] [CrossRef] [PubMed]
- De, R.; Dutta, S. Role of the Microbiome in the Pathogenesis of COVID-19. Front. Cell. Infect. Microbiol. 2022, 12, 736397. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Wang, X.; Yin, W.; Gao, Z.; Xia, B. Human microbiota is a reservoir of SARS-CoV-2 advantageous mutations. bioRxiv 2023. [Google Scholar] [CrossRef]
- Chand, G.B.; Banerjee, A.; Azad, G.K. Identification of novel mutations in RNA-dependent RNA polymerases of SARS-CoV-2 and their implications on its protein structure. PeerJ 2020, 8, e9492. [Google Scholar] [CrossRef] [PubMed]
- Reshamwala, S.M.S.; Likhite, V.; Degani, M.S.; Deb, S.S.; Noronha, S.B. Mutations in SARS-CoV-2 Nsp7 and Nsp8 proteins and their predicted impact on replication/transcription complex structure. J. Med. Virol. 2021, 93, 4616–4619. [Google Scholar] [CrossRef]
- Ilmjärv, S.; Abdul, F.; Acosta-Gutiérrez, S.; Estarellas, C.; Galdadas, I.; Casimir, M.; Alessandrini, M.; Gervasio, F.L.; Krause, K.H. Concurrent mutations in RNA-dependent RNA polymerase and spike protein emerged as the epidemiologically most successful SARS-CoV-2 variant. Sci. Rep. 2021, 11, 13705. [Google Scholar] [CrossRef]
- Mohammad, A.; Al-Mulla, F.; Wei, D.Q.; Abubaker, J.; Remdesivir, M.D. Simulations Suggest a More Favourable Binding to SARS-CoV-2 RNA Dependent RNA Polymerase Mutant P323L Than Wild-Type. Biomolecules 2021, 11, 919. [Google Scholar] [CrossRef]
- Kim, S.M.; Kim, E.H.; Casel, M.A.B.; Kim, Y.I.; Sun, R.; Kwak, M.J.; Yoo, J.S.; Yu, M.; Yu, K.M.; Jang, S.G.; et al. SARS-CoV-2 variants with NSP12 P323L/G671S mutations display enhanced virus replication in ferret upper airways and higher transmissibility. Cell Rep. 2023, 42, 113077. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Peng, R.; Yuan, B.; Zhao, J.; Wang, M.; Wang, X.; Wang, Q.; Sun, Y.; Fan, Z.; Qi, J.; et al. Structural and Biochemical Characterization of the Nsp12-Nsp7-Nsp8 Core Polymerase Complex from SARS-CoV-2. Cell Rep. 2020, 31, 107774. [Google Scholar] [CrossRef] [PubMed]
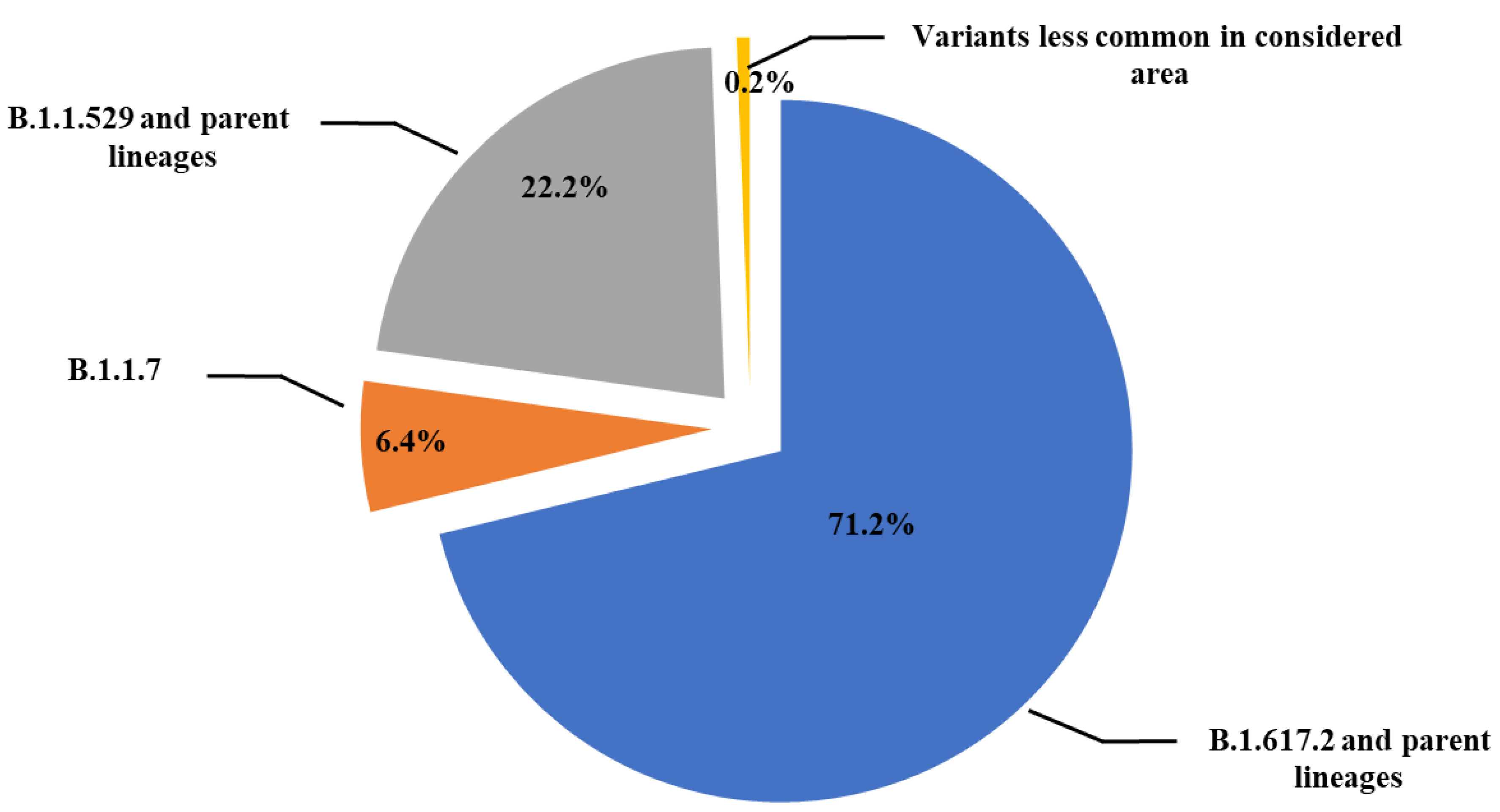
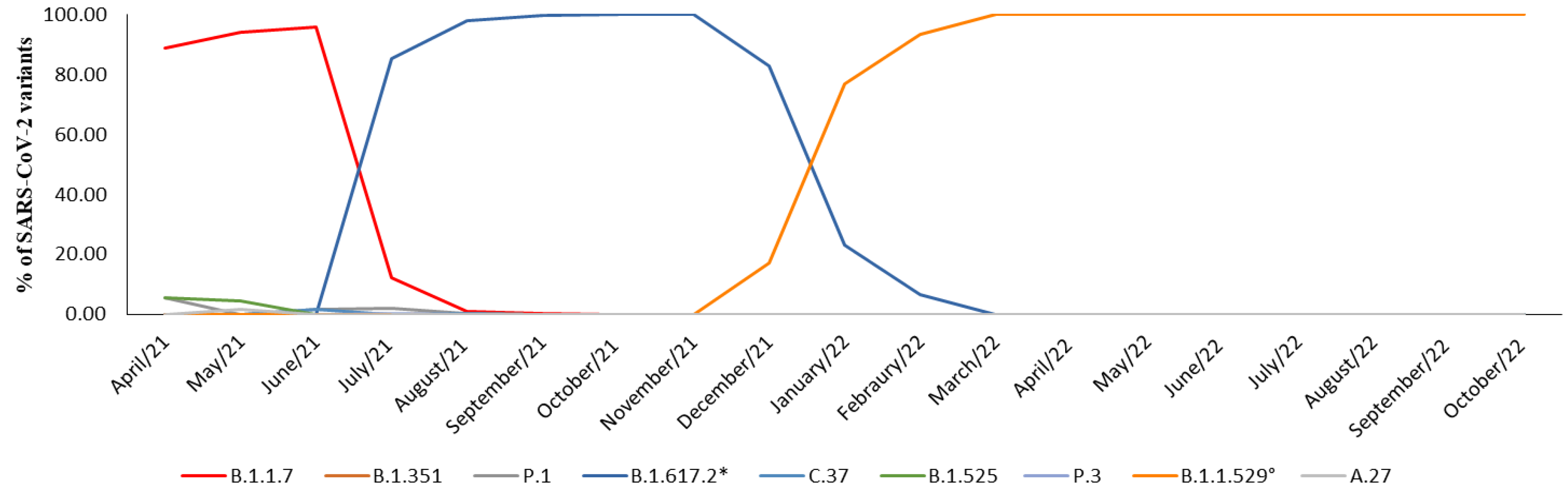

| WHO Label | Alpha | Beta | Gamma | Delta | Lambda | Eta | Theta | Omicron | None |
|---|---|---|---|---|---|---|---|---|---|
| Pango Lineage | B.1.1.7 | B.1.351 | P.1 | B.1.617.2 * | C.37 | B.1.525 | P.3 | B.1.1.529 ° | A.27 |
| April/2021 | 88.8% | - | 5.6% | - | - | 5.6% | - | - | - |
| May/2021 | 94.1% | - | - | - | - | 4.4% | - | - | 1.5% |
| June/2021 | 96% | 0.8% | 1.6% | - | 1.6% | - | - | - | - |
| July/2021 | 12.2% | - | 2% | 85.5% | - | - | 0.3% | - | - |
| August/2021 | 1.20% | 0.1% | 0.3% | 98% | 0.4% | - | - | - | - |
| September/2021 | 0.2% | - | - | 99.8% | - | - | - | - | - |
| October/2021 | - | - | - | 100% | - | - | - | - | - |
| November/2021 | - | - | - | 100% | - | - | - | - | - |
| December/2021 | - | - | - | 83% | - | - | - | 17% | - |
| January/2022 | - | - | - | 23.1% | - | - | - | 76.9% | - |
| February/2022 | - | - | - | 6.5% | - | - | - | 93.5% | - |
| March/2022 | - | - | - | - | - | - | - | 100% | - |
| April/2022 | - | - | - | - | - | - | - | 100% | - |
| May/2022 | - | - | - | - | - | - | - | 100% | - |
| June/2022 | - | - | - | - | - | - | - | 100% | - |
| July/2022 | - | - | - | - | - | - | - | 100% | - |
| August/2022 | - | - | - | - | - | - | - | 100% | - |
| September/2022 | - | - | - | - | - | - | - | 100% | - |
| October/2022 | - | - | - | - | - | - | - | 100% | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lombardo, D.; Musolino, C.; Chines, V.; Caminiti, G.; Palermo, C.; Cacciola, I.; Raffa, G.; Pollicino, T. Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients. Microorganisms 2024, 12, 2. https://doi.org/10.3390/microorganisms12010002
Lombardo D, Musolino C, Chines V, Caminiti G, Palermo C, Cacciola I, Raffa G, Pollicino T. Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients. Microorganisms. 2024; 12(1):2. https://doi.org/10.3390/microorganisms12010002
Chicago/Turabian StyleLombardo, Daniele, Cristina Musolino, Valeria Chines, Giuseppe Caminiti, Claudia Palermo, Irene Cacciola, Giuseppina Raffa, and Teresa Pollicino. 2024. "Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients" Microorganisms 12, no. 1: 2. https://doi.org/10.3390/microorganisms12010002
APA StyleLombardo, D., Musolino, C., Chines, V., Caminiti, G., Palermo, C., Cacciola, I., Raffa, G., & Pollicino, T. (2024). Assessing Genomic Mutations in SARS-CoV-2: Potential Resistance to Antiviral Drugs in Viral Populations from Untreated COVID-19 Patients. Microorganisms, 12(1), 2. https://doi.org/10.3390/microorganisms12010002







