An Analysis of the Gut Microbiota and Related Metabolites following PCSK9 Inhibition in Statin-Treated Patients with Elevated Levels of Lipoprotein(a)
Abstract
1. Introduction
2. Materials and Methods
2.1. Cohort and Sample Collection
2.2. Isolation of Microbial DNA from Stool Samples, 16S Metabarcoding, Sequencing of the Amplicon Pools, and Processing of the Sequencing Data
2.3. Quantitative Profiling of Bile Acids and Short Chain Fatty Acids
2.4. Statistical Analyses
3. Results
3.1. Study Cohort
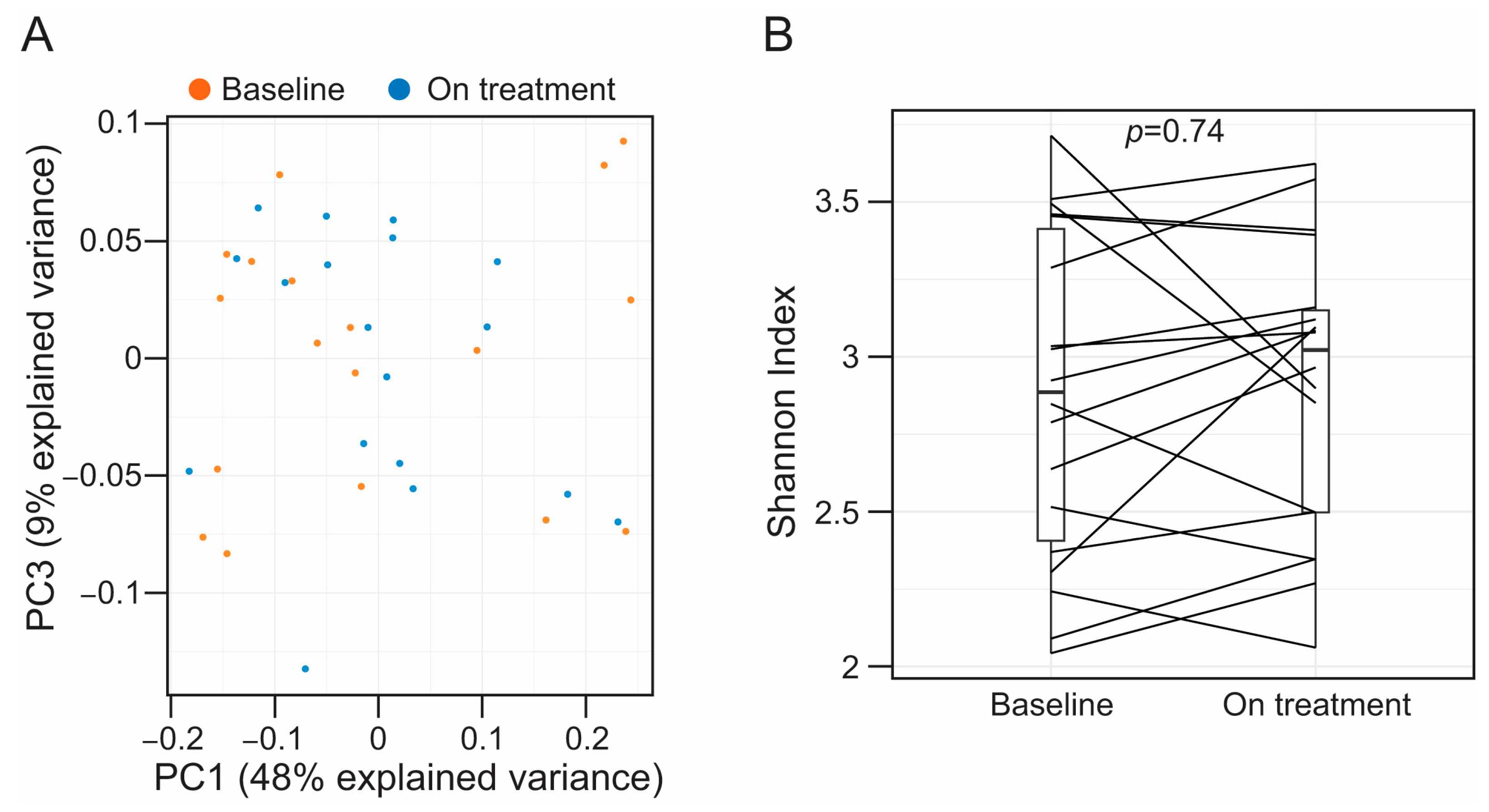
3.2. Alirocumab Is Not Associated with Alterations in the Taxonomic Profiles of the Fecal-Associated Bacterial Biota
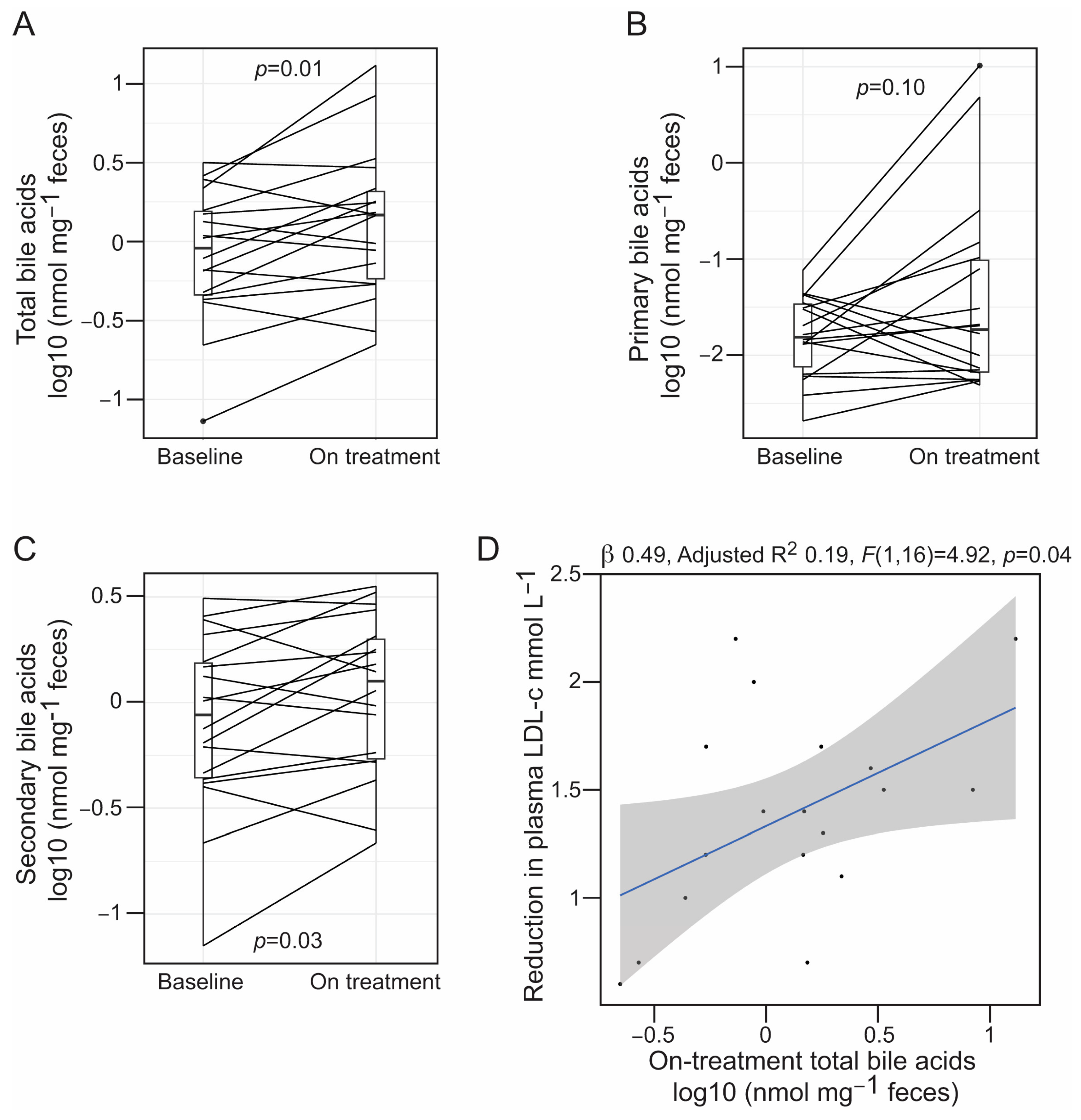
3.3. Alirocumab Is Linked to Changes in the Profiles of Gut Bacteria-Associated Metabolites
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reklou, A.; Doumas, M.; Imprialos, K.; Stavropoulos, K.; Patoulias, D.; Athyros, V.G. Reduction of Vascular Inflammation, LDL-C, or Both for the Protection from Cardiovascular Events? Open Cardiovasc. Med. J. 2018, 12, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Packard, C.; Chapman, M.J.; Sibartie, M.; Laufs, U.; Masana, L. Intensive low-density lipoprotein cholesterol lowering in cardiovascular disease prevention: Opportunities and challenges. Heart 2021, 107, 1369–1375. [Google Scholar] [CrossRef]
- Baigent, C.; Keech, A.; Kearney, P.M.; Blackwell, L.; Buck, G.; Pollicino, C.; Kirby, A.; Sourjina, T.; Peto, R.; Collins, R.; et al. Efficacy and safety of cholesterol-lowering treatment: Prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet 2005, 366, 1267–1278. [Google Scholar] [CrossRef] [PubMed]
- Cholesterol Treatment Trialists, C.; Baigent, C.; Blackwell, L.; Emberson, J.; Holland, L.E.; Reith, C.; Bhala, N.; Peto, R.; Barnes, E.H.; Keech, A.; et al. Efficacy and safety of more intensive lowering of LDL cholesterol: A meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet 2010, 376, 1670–1681. [Google Scholar] [CrossRef] [PubMed]
- Heart Protection Study Collaborative, G. MRC/BHF Heart Protection Study of cholesterol lowering with simvastatin in 20,536 high-risk individuals: A randomised placebo-controlled trial. Lancet 2002, 360, 7–22. [Google Scholar] [CrossRef]
- Cannon, C.P.; Blazing, M.A.; Giugliano, R.P.; McCagg, A.; White, J.A.; Theroux, P.; Darius, H.; Lewis, B.S.; Ophuis, T.O.; Jukema, J.W.; et al. Ezetimibe Added to Statin Therapy after Acute Coronary Syndromes. N. Engl. J. Med. 2015, 372, 2387–2397. [Google Scholar] [CrossRef] [PubMed]
- Duarte Lau, F.; Giugliano, R.P. Lipoprotein(a) and its Significance in Cardiovascular Disease: A Review. JAMA Cardiol. 2022, 7, 760–769. [Google Scholar] [CrossRef]
- Grundy, S.M.; Stone, N.J.; Bailey, A.L.; Beam, C.; Birtcher, K.K.; Blumenthal, R.S.; Braun, L.T.; de Ferranti, S.; Faiella-Tommasino, J.; Forman, D.E.; et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J. Am. Coll. Cardiol. 2019, 73, e285–e350. [Google Scholar] [CrossRef]
- Watts, G.F.; Chan, D.C.; Pang, J.; Ma, L.; Ying, Q.; Aggarwal, S.; Marcovina, S.M.; Barrett, P.H.R. PCSK9 Inhibition with alirocumab increases the catabolism of lipoprotein(a) particles in statin-treated patients with elevated lipoprotein(a). Metabolism 2020, 107, 154221. [Google Scholar] [CrossRef]
- Hess, C.N.; Low Wang, C.C.; Hiatt, W.R. PCSK9 Inhibitors: Mechanisms of Action, Metabolic Effects, and Clinical Outcomes. Annu. Rev. Med. 2018, 69, 133–145. [Google Scholar] [CrossRef]
- Ahmad, A.F.; Dwivedi, G.; O’Gara, F.; Caparros-Martin, J.; Ward, N.C. The gut microbiome and cardiovascular disease: Current knowledge and clinical potential. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H923–H938. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. [Google Scholar] [CrossRef]
- Tang, W.H.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Senthong, V.; Wang, Z.; Li, X.S.; Fan, Y.; Wu, Y.; Tang, W.H.; Hazen, S.L. Intestinal Microbiota-Generated Metabolite Trimethylamine-N-Oxide and 5-Year Mortality Risk in Stable Coronary Artery Disease: The Contributory Role of Intestinal Microbiota in a COURAGE-Like Patient Cohort. J. Am. Heart Assoc. 2016, 5, e002816. [Google Scholar] [CrossRef]
- Roberts, A.B.; Gu, X.; Buffa, J.A.; Hurd, A.G.; Wang, Z.; Zhu, W.; Gupta, N.; Skye, S.M.; Cody, D.B.; Levison, B.S.; et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 2018, 24, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Mackay, C.R.; Kaye, D.M. Beyond gut feelings: How the gut microbiota regulates blood pressure. Nat. Rev. Cardiol. 2018, 15, 20–32. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, J.; Wu, W.; Zhu, Y.; Liu, X. The role of bile acids in cardiovascular diseases: From mechanisms to clinical implications. Aging Dis. 2023, 14, 261–282. [Google Scholar] [CrossRef] [PubMed]
- van der Hee, B.; Wells, J.M. Microbial regulation of host physiology by short-chain fatty acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Chen, M.L.; Takeda, K.; Sundrud, M.S. Emerging roles of bile acids in mucosal immunity and inflammation. Mucosal Immunol. 2019, 12, 851–861. [Google Scholar] [CrossRef]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Edwards, P.A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef]
- Larabi, A.B.; Masson, H.L.; Bäumler, A.J. Bile acids as modulators of gut microbiota composition and function. Gut Microbes 2023, 15, 2172671. [Google Scholar] [CrossRef] [PubMed]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.L.; Stine, J.G.; Bisanz, J.E.; Okafor, C.D.; Patterson, A.D. Bile acids and the gut microbiota: Metabolic interactions and impacts on disease. Nat. Rev. Microbiol. 2023, 21, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Hageman, J.; Herrema, H.; Groen, A.K.; Kuipers, F. A role of the bile salt receptor FXR in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 1519–1528. [Google Scholar] [CrossRef] [PubMed]
- Haghikia, A.; Zimmermann, F.; Schumann, P.; Jasina, A.; Roessler, J.; Schmidt, D.; Heinze, P.; Kaisler, J.; Nageswaran, V.; Aigner, A.; et al. Propionate attenuates atherosclerosis by immune-dependent regulation of intestinal cholesterol metabolism. Eur. Heart J. 2022, 43, 518–533. [Google Scholar] [CrossRef]
- Kondo, T.; Kishi, M.; Fushimi, T.; Ugajin, S.; Kaga, T. Vinegar intake reduces body weight, body fat mass, and serum triglyceride levels in obese Japanese subjects. Biosci. Biotechnol. Biochem. 2009, 73, 1837–1843. [Google Scholar] [CrossRef]
- Laffitte, B.A.; Kast, H.R.; Nguyen, C.M.; Zavacki, A.M.; Moore, D.D.; Edwards, P.A. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J. Biol. Chem. 2000, 275, 10638–10647. [Google Scholar] [CrossRef]
- Langhi, C.; Le May, C.; Kourimate, S.; Caron, S.; Staels, B.; Krempf, M.; Costet, P.; Cariou, B. Activation of the farnesoid X receptor represses PCSK9 expression in human hepatocytes. FEBS Lett. 2008, 582, 949–955. [Google Scholar] [CrossRef]
- Sayin, S.I.; Wahlström, A.; Felin, J.; Jäntti, S.; Marschall, H.U.; Bamberg, K.; Angelin, B.; Hyötyläinen, T.; Orešič, M.; Bäckhed, F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013, 17, 225–235. [Google Scholar] [CrossRef]
- Caparros-Martin, J.A.; Lareu, R.R.; Ramsay, J.P.; Peplies, J.; Reen, F.J.; Headlam, H.A.; Ward, N.C.; Croft, K.D.; Newsholme, P.; Hughes, J.D.; et al. Statin therapy causes gut dysbiosis in mice through a PXR-dependent mechanism. Microbiome 2017, 5, 95. [Google Scholar] [CrossRef]
- Vieira-Silva, S.; Falony, G.; Belda, E.; Nielse, T.; Aron-Wisnewsky, J.; Chakaroun, R.; Forslund, S.K.; Assmann, K.; Valles-Colomer, M.; Nguyen, T.T.D.; et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 2020, 581, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Wang, J.; Cheng, R.; Ren, Y.; Miao, Z.; Luo, Y.; Zhou, Q.; Xue, Y.; Shen, X.; He, F.; et al. Orlistat and ezetimibe could differently alleviate the high-fat diet-induced obesity phenotype by modulating the gut microbiota. Front. Microbiol. 2022, 13, 908327. [Google Scholar] [CrossRef] [PubMed]
- Catry, E.; Pachikian, B.D.; Salazar, N.; Neyrinck, A.M.; Cani, P.D.; Delzenne, N.M. Ezetimibe and simvastatin modulate gut microbiota and expression of genes related to cholesterol metabolism. Life Sci. 2015, 132, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Kliewer, S.A.; Willson, T.M. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane X receptor. J. Lipid Res. 2002, 43, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef]
- Tan, S.; Caparros-Martin, J.A.; Matthews, V.B.; Koch, H.; O’Gara, F.; Croft, K.D.; Ward, N.C. Isoquercetin and inulin synergistically modulate the gut microbiome to prevent development of the metabolic syndrome in mice fed a high fat diet. Sci. Rep. 2018, 8, 10100. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Saladie, M.; Caparros-Martin, J.A.; Agudelo-Romero, P.; Wark, P.A.B.; Stick, S.M.; O’Gara, F. Microbiomic Analysis on Low Abundant Respiratory Biomass Samples; Improved Recovery of Microbial DNA From Bronchoalveolar Lavage Fluid. Front. Microbiol. 2020, 11, 572504. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. Peer J. 2016, 18, e2584. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Croft, K.D.; Caparros-Martin, J.; O’Gara, F.; Mori, T.A.; Ward, N.C. Beneficial effects of inorganic nitrate in non-alcoholic fatty liver disease. Arch. Biochem. Biophys. 2021, 711, 109032. [Google Scholar] [CrossRef] [PubMed]
- Blake, S.J.; James, J.; Ryan, F.J.; Caparros-Martin, J.; Eden, G.L.; Tee, Y.C.; Salamon, J.R.; Benson, S.C.; Tumes, D.J.; Sribnaia, A.; et al. The immunotoxicity, but not anti-tumor efficacy, of anti-CD40 and anti-CD137 immunotherapies is dependent on the gut microbiota. Cell Rep. Med. 2021, 2, 100464. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Peddada, S.D. Analysis of compositions of microbiomes with bias correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef] [PubMed]
- Holmes, I.; Harris, K.; Quince, C. Dirichlet multinomial mixtures: Generative models for microbial metagenomics. PLoS ONE 2012, 7, e30126. [Google Scholar] [CrossRef]
- Hofmann, A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef]
- Ohira, H.; Tsutsui, W.; Fujioka, Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J. Atheroscler. Thromb. 2017, 24, 660–672. [Google Scholar] [CrossRef]
- Jie, Z.; Xia, H.; Zhong, S.L.; Feng, Q.; Li, S.; Liang, S.; Zhong, H.; Liu, Z.; Gao, Y.; Zhao, H.; et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 2017, 8, 845. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Fak, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Backhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Gao, R.; Zhang, Y.; Pan, D.; Zhu, Y.; Zhang, X.; Yang, R.; Jiang, R.; Xu, Y.; Qin, H. Dysbiosis signatures of gut microbiota in coronary artery disease. Physiol. Genom. 2018, 50, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Zhao, T.; Hu, H.; Zhang, W.; Hua, X. Association Study of Gut Flora in Coronary Heart Disease through High-Throughput Sequencing. BioMed Res. Int. 2017, 2017, 3796359. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Raufman, J.P.; Pallone, T.L. Bile acids regulate cardiovascular function. Clin. Transl. Sci. 2011, 4, 210–218. [Google Scholar] [CrossRef] [PubMed]
- von Haehling, S.; Schefold, J.C.; Jankowska, E.A.; Springer, J.; Vazir, A.; Kalra, P.R.; Sandek, A.; Fauler, G.; Stojakovic, T.; Trauner, M.; et al. Ursodeoxycholic acid in patients with chronic heart failure: A double-blind, randomized, placebo-controlled, crossover trial. J. Am. Coll. Cardiol. 2012, 59, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Bishop-Bailey, D.; Walsh, D.T.; Warner, T.D. Expression and activation of the farnesoid X receptor in the vasculature. Proc. Natl. Acad. Sci. USA 2004, 101, 3668–3673. [Google Scholar] [CrossRef]
- Tominaga, T.; Suzuki, H.; Ogata, Y.; Imafuku, T.; Saruta, T. Bile acids are able to reduce blood pressure by attenuating the vascular reactivity in spontaneously hypertensive rats. Life Sci. 1988, 42, 1861–1868. [Google Scholar] [CrossRef]
- Jama, H.A.; Rhys-Jones, D.; Nakai, M.; Yao, C.K.; Climie, R.E.; Sata, Y.; Anderson, D.; Creek, D.J.; Head, G.A.; Kaye, D.M.; et al. Prebiotic intervention with HAMSAB in untreated essential hypertensive patients assessed in a phase II randomized trial. Nat. Cardiovasc. Res. 2023, 2, 35–43. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Kashima, H.; Rahmanto, Y.S.; Banno, K.; Yu, Y.; Matoba, Y.; Watanabe, K.; Iijima, M.; Takeda, T.; Kunitomi, H.; et al. Drug repositioning of mevalonate pathway inhibitors as antitumor agents for ovarian cancer. Oncotarget 2017, 8, 72147–72156. [Google Scholar] [CrossRef]
- Nelson, D.L.; Lehninger, A.L.; Cox, M.M. Lehninger Principles of Biochemistry, 5th ed.; W.H. Freeman: New York, NY, USA, 2005. [Google Scholar]
- Parker, R.A.; Garcia, R.; Ryan, C.S.; Liu, X.; Shipkova, P.; Livanov, V.; Patel, P.; Ho, S.P. Bile acid and sterol metabolism with combined HMG-CoA reductase and PCSK9 suppression. J. Lipid Res. 2013, 54, 2400–2409. [Google Scholar] [CrossRef] [PubMed]
- Prizment, A.E.; Staley, C.; Onyeaghala, G.C.; Vivek, S.; Thyagarajan, B.; Straka, R.J.; Demmer, R.T.; Demmer, R.T.; Knights, D.; Meyer, K.A.; et al. Randomised clinical study: Oral aspirin 325 mg daily vs placebo alters gut microbial composition and bacterial taxa associated with colorectal cancer risk. Aliment. Pharmacol. Ther. 2020, 52, 967–987. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Wang, Z.; Song, W.; Liao, Y.; Wang, X.; Chen, C.; Ming, J.; Cui, J.; Xu, K. Effects of long-term regular oral aspirin combined with atorvastatin to prevent ischemic stroke on human gut microbiota. Eur. J. Pharmacol. 2023, 951, 175800. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Liu, Y.; Zhao, Y.; Yan, R.; Yang, L.; Ma, H.; Wang, J.; Wang, T.; Li, Y.; Zhang, G.; et al. Aspirin ameliroates atherosclerotic immuno-inflammation through regulating the Treg/Th17 axis and CD39-CD73 adenosine signaling via remodeling the gut microbiota in ApoeE-/- mice. Int. Immunopharmacol. 2023, 120, 110296. [Google Scholar] [CrossRef]
| Value | |
|---|---|
| Sex (% males) | 61% |
| Age (years) | 56.5 ± 11.2 |
| BMI (kg m−2) | 28.6 ± 4.4 |
| Systolic blood pressure (mmHg) | 124.3 ± 7.3 |
| Diastolic blood pressure (mmHg) | 76.3 ± 8.5 |
| Total cholesterol (mmol L−1) | 3.8 ± 0.6 |
| Triglyceride (mmol L−1) | 1.1 (0.9, 1.4) |
| HDL-cholesterol (mmol L−1) | 1.2 ± 0.3 |
| LDL-cholesterol (mmol L−1) | 2.1 ± 0.5 |
| ApoB (g L−1) | 0.8 ± 0.1 |
| Lp(a) (g L−1) | 1.2 (0.96, 1.44) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caparrós-Martín, J.A.; Maher, P.; Ward, N.C.; Saladié, M.; Agudelo-Romero, P.; Stick, S.M.; Chan, D.C.; Watts, G.F.; O’Gara, F. An Analysis of the Gut Microbiota and Related Metabolites following PCSK9 Inhibition in Statin-Treated Patients with Elevated Levels of Lipoprotein(a). Microorganisms 2024, 12, 170. https://doi.org/10.3390/microorganisms12010170
Caparrós-Martín JA, Maher P, Ward NC, Saladié M, Agudelo-Romero P, Stick SM, Chan DC, Watts GF, O’Gara F. An Analysis of the Gut Microbiota and Related Metabolites following PCSK9 Inhibition in Statin-Treated Patients with Elevated Levels of Lipoprotein(a). Microorganisms. 2024; 12(1):170. https://doi.org/10.3390/microorganisms12010170
Chicago/Turabian StyleCaparrós-Martín, Jose A., Patrice Maher, Natalie C. Ward, Montserrat Saladié, Patricia Agudelo-Romero, Stephen M. Stick, Dick C. Chan, Gerald F. Watts, and Fergal O’Gara. 2024. "An Analysis of the Gut Microbiota and Related Metabolites following PCSK9 Inhibition in Statin-Treated Patients with Elevated Levels of Lipoprotein(a)" Microorganisms 12, no. 1: 170. https://doi.org/10.3390/microorganisms12010170
APA StyleCaparrós-Martín, J. A., Maher, P., Ward, N. C., Saladié, M., Agudelo-Romero, P., Stick, S. M., Chan, D. C., Watts, G. F., & O’Gara, F. (2024). An Analysis of the Gut Microbiota and Related Metabolites following PCSK9 Inhibition in Statin-Treated Patients with Elevated Levels of Lipoprotein(a). Microorganisms, 12(1), 170. https://doi.org/10.3390/microorganisms12010170








