The Prevalence of Metallo-Beta-Lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Search Strategy and Eligibility Criteria
2.3. Data Extraction and Methodological Quality Assessment
2.4. Statistical Analysis
3. Results
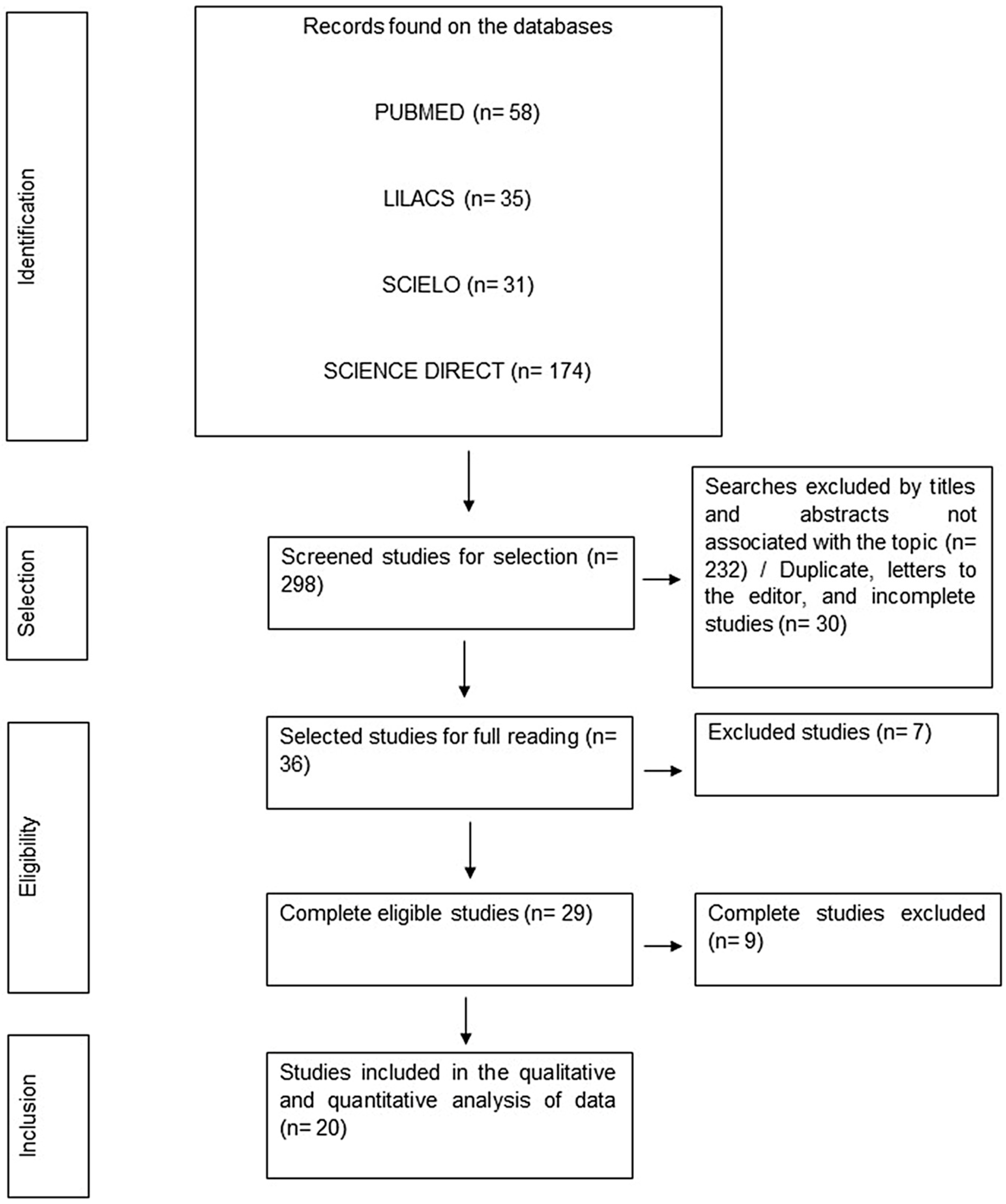
3.1. Literature Search
3.2. Characterization of Included Studies
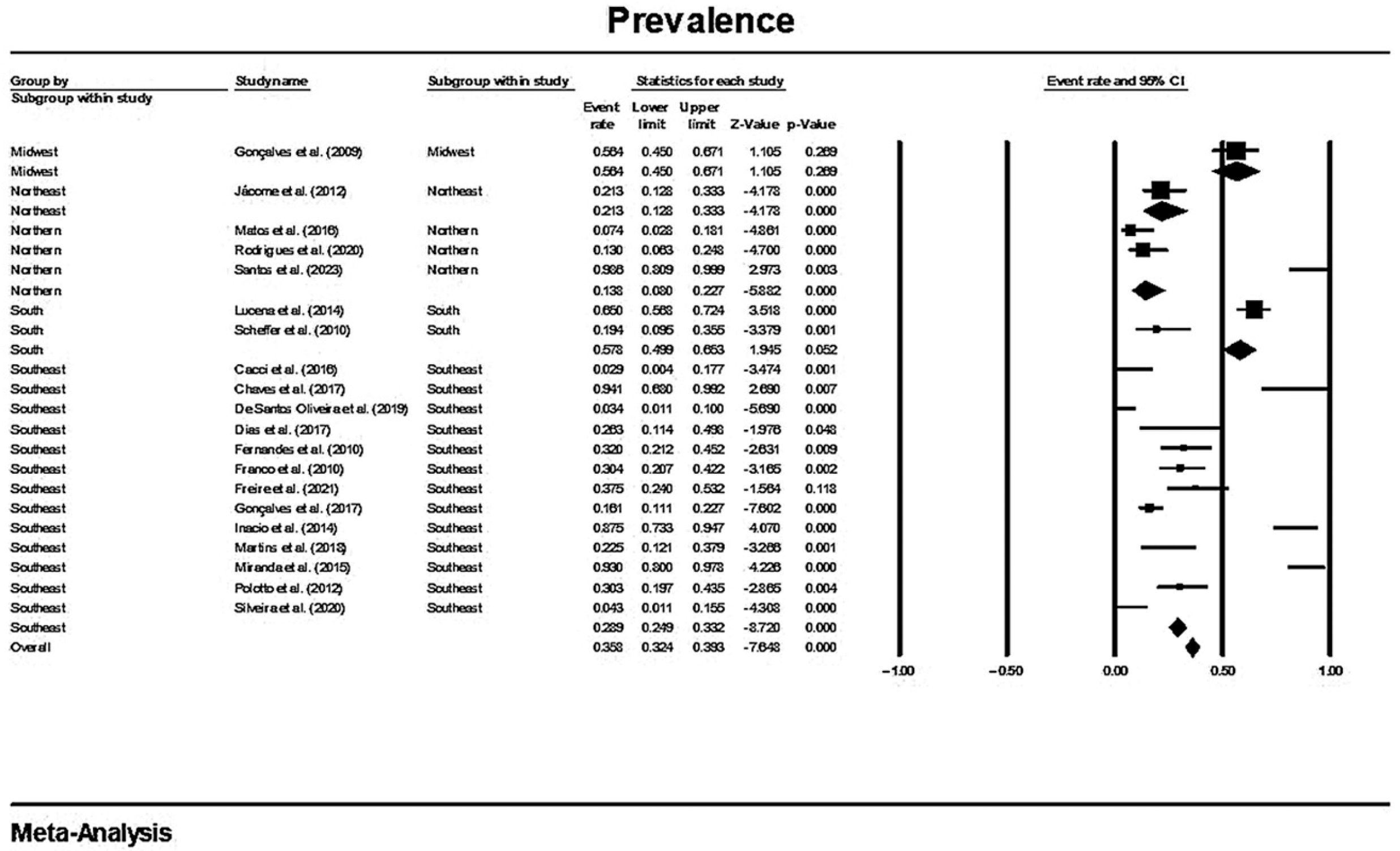
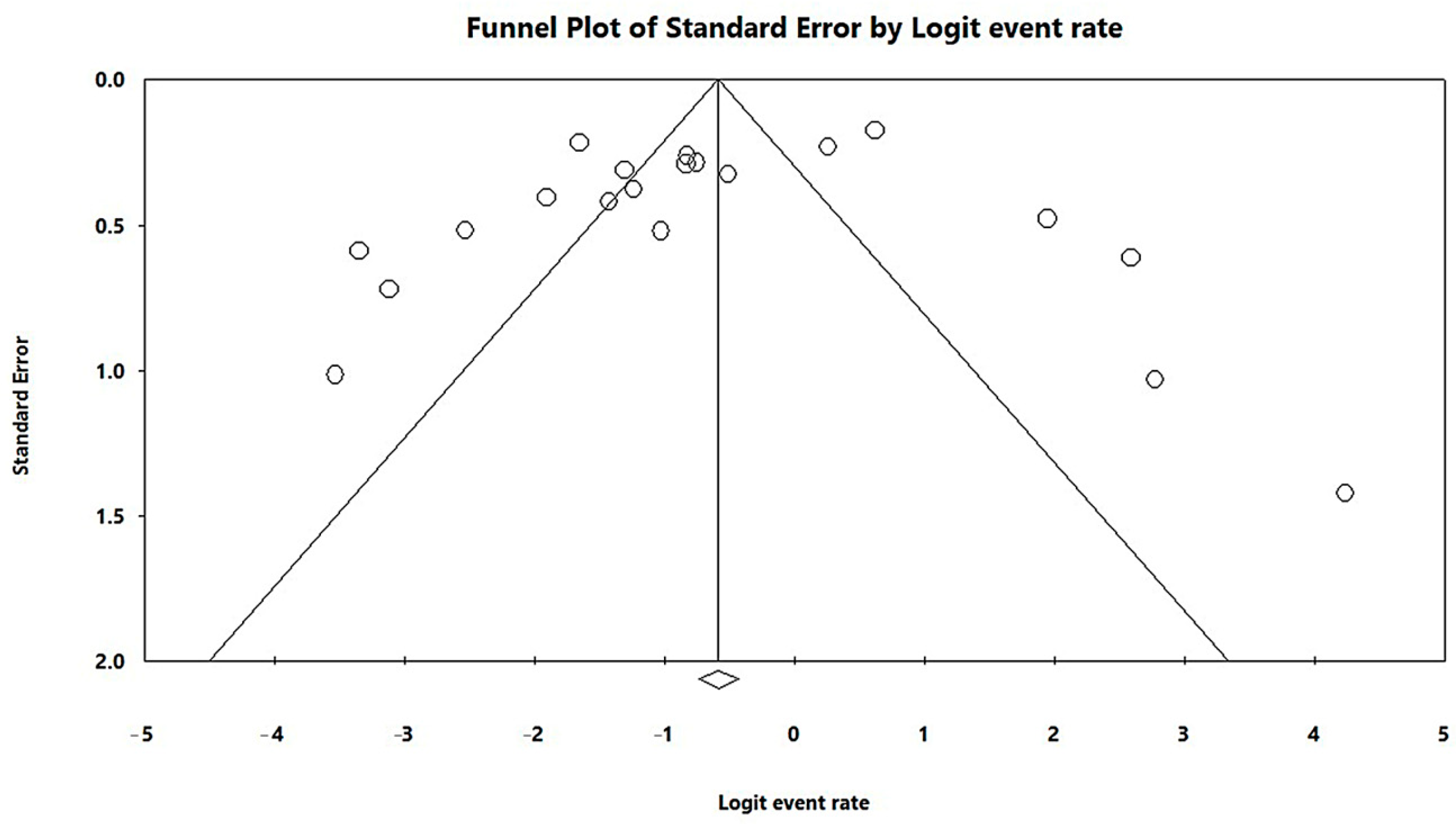
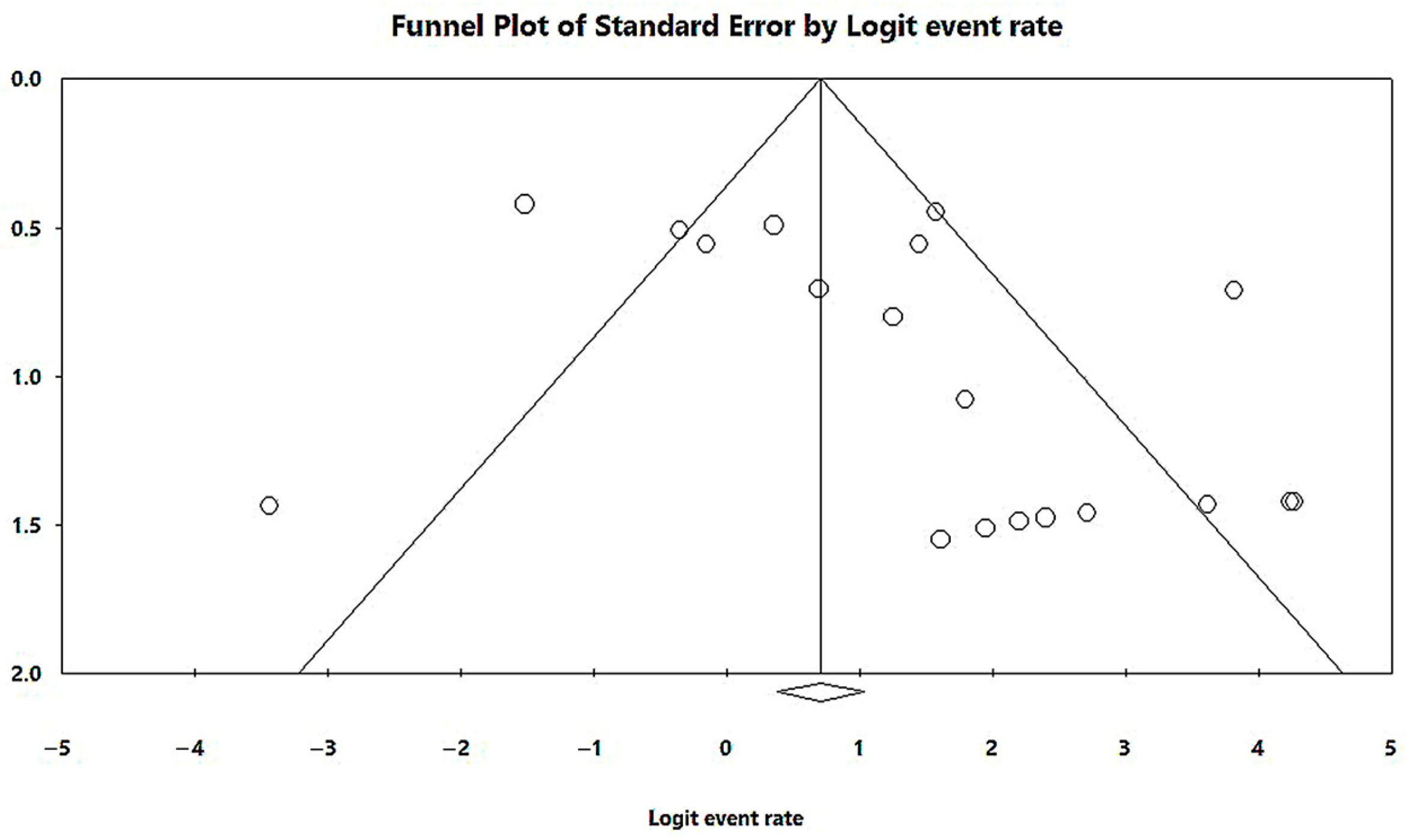
3.3. Results and Publication Bias of Meta-Analysis of Proportion of MβL-PA
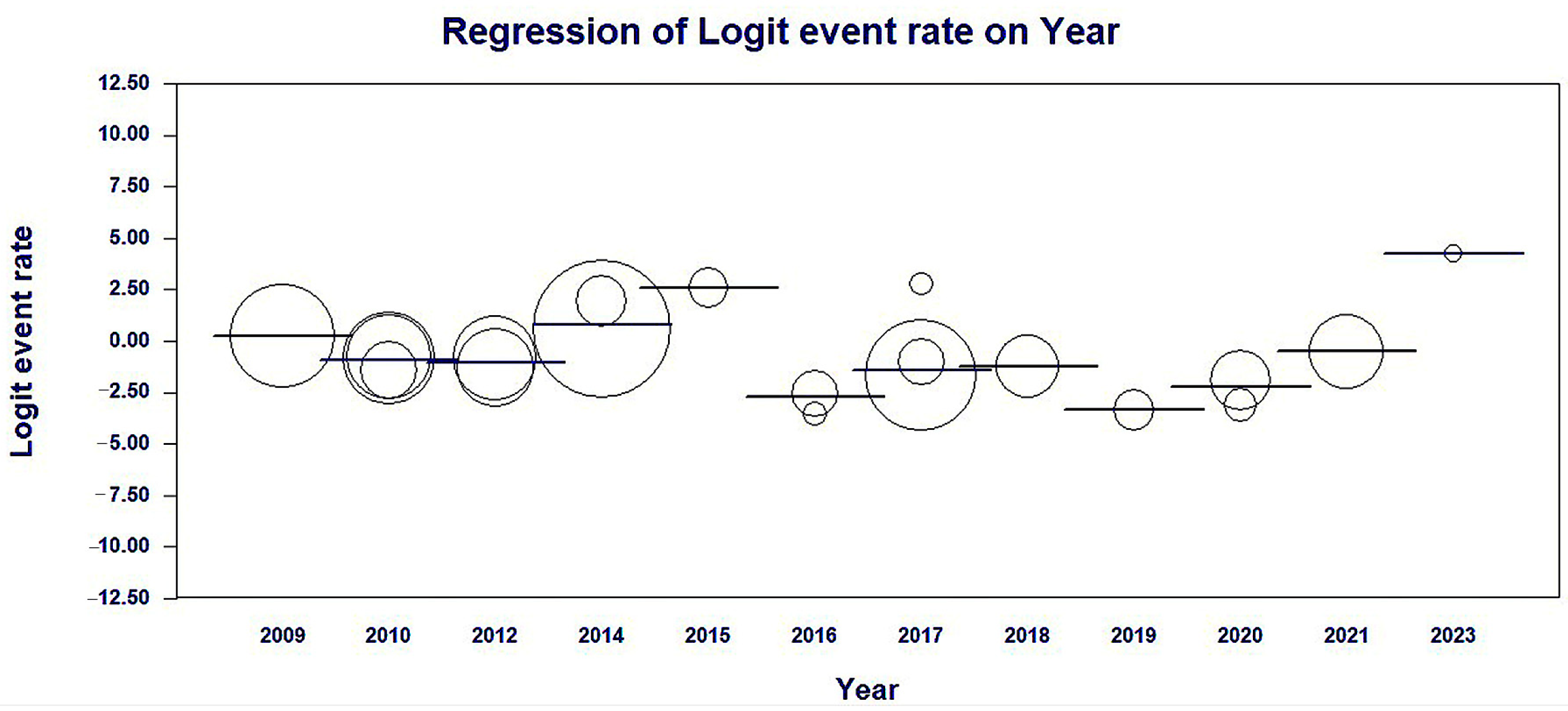
3.4. Meta-Regression of the Studies Included by Year of Publication
3.5. Results and Publication Bias of the Meta-Analysis of blaSPM-1 within MβL-PA Isolates
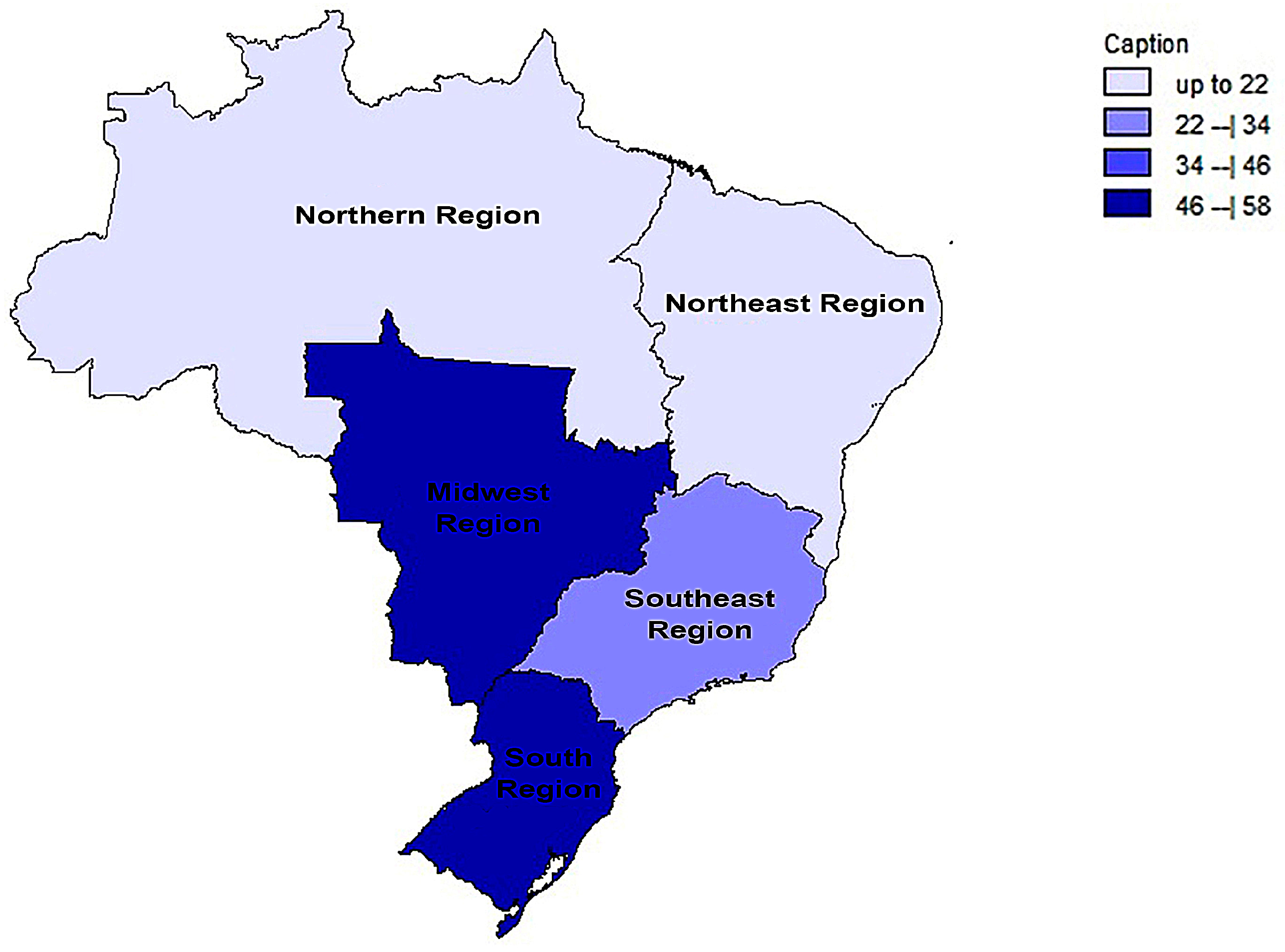
3.6. Comparison between the Results of this Brazilian Meta-Analysis and Prevalence Worldwide
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mustafai, M.M.; Hafeez, M.; Munawar, S.; Basha, S.; Rabaan, A.A.; Halwani, M.A.; Alawfi, A.; Alshengeti, A.; Najim, M.A.; Alwarthan, S.; et al. Prevalence of Carbapenemase and Extended-Spectrum β-Lactamase Producing Enterobacteriaceae: A Cross-Sectional Study. Antibiotics 2023, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Allel, K.; Day, L.; Hamilton, A.; Lin, L.; Furuya-Kanamori, L.; Moore, C.E.; Van Boeckel, T.; Laxminarayan, R.; Yakob, L. Global Antimicrobial-Resistance Drivers: An Ecological Country-Level Study at the Human-Animal Interface. Lancet Planet. Health 2023, 7, e291–e303. [Google Scholar] [CrossRef] [PubMed]
- Büchler, A.C.; Shahab, S.N.; Severin, J.A.; Vos, M.C.; Voor in ‘t holt, A.F. Outbreak Investigations after Identifying Carbapenem-Resistant Pseudomonas aeruginosa: A Systematic Review. Antimicrob. Resist. Infect. Control 2023, 12, 28. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, L.; Li, M.; Liang, H.; He, Y.; Li, S. Risk Factors and Outcomes of Patients with Carbapenem-Resistant Pseudomonas aeruginosa Bloodstream Infection. Infect. Drug Resist. 2023, 16, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Falcone, M.; Tiseo, G.; Carbonara, S.; Marino, A.; Di Caprio, G.; Carretta, A.; Mularoni, A.; Mariani, M.F.; Maraolo, A.E.; Scotto, R.; et al. Mortality Attributable to Bloodstream Infections Caused by Different Carbapenem-Resistant Gram-Negative Bacilli: Results From a Nationwide Study in Italy (ALARICO Network). Clin. Infect. Dis. 2023, 76, 2059–2069. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Wei, X.; Xu, G.; Zhang, X.; Wang, X. Carbapenem-Resistant Pseudomonas aeruginosa Infections in Critically Ill Children: Prevalence, Risk Factors, and Impact on Outcome in a Large Tertiary Pediatric Hospital of China. Front. Public Health 2023, 11, 1088262. [Google Scholar] [CrossRef] [PubMed]
- de Matos, E.C.O.; Andriolo, R.B.; Rodrigues, Y.C.; de Lima, P.D.L.; Carneiro, I.C.d.R.S.; Lima, K.V.B. Mortality in Patients with Multidrug-Resistant Pseudomonas aeruginosa Infections: A Meta-Analysis. Rev. Soc. Bras. Med. Trop. 2018, 51, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.-J.; Jeong, S.H. Mobile Carbapenemase Genes in Pseudomonas aeruginosa. Front. Microbiol. 2021, 12, 614058. [Google Scholar] [CrossRef]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.-J.; Cheng, Z. Antibiotic Resistance in Pseudomonas aeruginosa: Mechanisms and Alternative Therapeutic Strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Meletis, G. Carbapenem Resistance: Overview of the Problem and Future Perspectives. Ther. Adv. Infect. Dis. 2016, 3, 15–21. [Google Scholar] [CrossRef]
- Codjoe, F.S.; Donkor, E.S. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Aurilio, C.; Sansone, P.; Barbarisi, M.; Pota, V.; Giaccari, L.G.; Coppolino, F.; Barbarisi, A.; Passavanti, M.B.; Pace, M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics 2022, 11, 421. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Walsh, T.R.; Livermore, D.M. The Emerging NDM Carbapenemases. Trends Microbiol. 2011, 19, 588–595. [Google Scholar] [CrossRef] [PubMed]
- Elshamy, A.A.; Aboshanab, K.M. A Review on Bacterial Resistance to Carbapenems: Epidemiology, Detection and Treatment Options. Future Sci. OA 2020, 6, FSO438. [Google Scholar] [CrossRef] [PubMed]
- Bahr, G.; González, L.J.; Vila, A.J. Metallo-β-Lactamases in the Age of Multidrug Resistance: From Structure and Mechanism to Evolution, Dissemination and Inhibitor Design. Chem. Rev. 2021, 121, 7957–8094. [Google Scholar] [CrossRef] [PubMed]
- Hammoudi Halat, D.; Ayoub Moubareck, C. The Intriguing Carbapenemases of Pseudomonas aeruginosa: Current Status, Genetic Profile, and Global Epidemiology. Yale J. Biol. Med. 2022, 95, 507–515. [Google Scholar] [PubMed]
- Hong, D.J.; Bae, I.K.; Jang, I.-H.; Jeong, S.H.; Kang, H.-K.; Lee, K. Epidemiology and Characteristics of Metallo-β-Lactamase-Producing Pseudomonas aeruginosa. Infect. Chemother. 2015, 47, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Iyobe, S.; Inoue, M.; Mitsuhashi, S. Transferable Imipenem Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1991, 35, 147–151. [Google Scholar] [CrossRef]
- Toleman, M.A. Molecular Characterization of SPM-1, a Novel Metallo-Beta-Lactamase Isolated in Latin America: Report from the SENTRY Antimicrobial Surveillance Programme. J. Antimicrob. Chemother. 2002, 50, 673–679. [Google Scholar] [CrossRef]
- Fonseca, E.L.; Marin, M.A.; Encinas, F.; Vicente, A.C.P. Full Characterization of the Integrative and Conjugative Element Carrying the Metallo-β-Lactamase Bla SPM-1 and Bicyclomycin Bcr1 Resistance Genes Found in the Pandemic Pseudomonas aeruginosa Clone SP/ST277. J. Antimicrob. Chemother. 2015, 70, 2547–2550. [Google Scholar] [CrossRef][Green Version]
- Murphy, T.A.; Simm, A.M.; Toleman, M.A.; Jones, R.N.; Walsh, T.R. Biochemical Characterization of the Acquired Metallo-β-Lactamase SPM-1 from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2003, 47, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Brem, J.; Struwe, W.B.; Rydzik, A.M.; Tarhonskaya, H.; Pfeffer, I.; Flashman, E.; van Berkel, S.S.; Spencer, J.; Claridge, T.D.; McDonough, M.A. Studying the Active-Site Loop Movement of the São Paolo Metallo-β-Lactamase-1. Chem. Sci. 2015, 6, 956–963. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Murphy, T.A.; Catto, L.E.; Halford, S.E.; Hadfield, A.T.; Minor, W.; Walsh, T.R.; Spencer, J. Crystal Structure of Pseudomonas aeruginosa SPM-1 Provides Insights into Variable Zinc Affinity of Metallo-β-Lactamases. J. Mol. Biol. 2006, 357, 890–903. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Santos, J.V.; da Costa Júnior, S.D.; de Fátima Ramos dos Santos Medeiros, S.M.; Cavalcanti, I.D.L.; de Souza, J.B.; Coriolano, D.L.; da Silva, W.R.C.; Alves, M.H.M.E.; Cavalcanti, I.M.F. Panorama of Bacterial Infections Caused by Epidemic Resistant Strains. Curr. Microbiol. 2022, 79, 175. [Google Scholar] [CrossRef] [PubMed]
- García-Betancur, J.C.; Appel, T.M.; Esparza, G.; Gales, A.C.; Levy-Hara, G.; Cornistein, W.; Vega, S.; Nuñez, D.; Cuellar, L.; Bavestrello, L.; et al. Update on the Epidemiology of Carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infect. Ther. 2021, 19, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Martins, A.F.; Zavascki, A.P.; Gaspareto, P.B.; Barth, A.L. Dissemination of Pseudomonas aeruginosa Producing SPM-1-like and IMP-1-like Metallo-β-Lactamases in Hospitals from Southern Brazil. Infection 2007, 35, 457–460. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, M.R.; Sellera, F.P.; Moura, Q.; Carvalho, M.P.N.; Rosato, P.N.; Cerdeira, L.; Lincopan, N. Zooanthroponotic Transmission of Drug-Resistant Pseudomonas aeruginosa, Brazil. Emerg. Infect. Dis. 2018, 24, 1160–1162. [Google Scholar] [CrossRef]
- Neves, P.R.; Perdigão Neto, L.V.; Ruedas Martins, R.C.; Ramos, J.F.; Leite, G.; Rossi, F.; Sanabani, S.S.; Rocha, V.; Batista, M.V.; Guimaraes, T.; et al. Carbapenem-Resistant Pseudomonas aeruginosa Carrying BlaVIM-36 Assigned to ST308: Indicated Non-Virulence in a Galleria Mellonella Model. J. Glob. Antimicrob. Resist. 2019, 16, 92–97. [Google Scholar] [CrossRef]
- Scavuzzi, A.M.L.; Beltrão, E.M.B.; Firmo, E.F.; de Oliveira, É.M.; Beserra, F.G.; de Souza Lopes, A.C. Emergence of BlaVIM-2, BlaNDM-1, BlaIMP-7 and BlaGES-1 in BlaKPC-2-Harbouring Pseudomonas aeruginosa Isolates in Brazil. J. Glob. Antimicrob. Resist. 2019, 19, 181–182. [Google Scholar] [CrossRef]
- de Bertoncheli, C.M.; Hörner, R. Uma revisão sobre metalo-β-lactamases. Rev. Bras. Cienc. Farm. 2008, 44, 577–599. [Google Scholar] [CrossRef]
- Inacio, H.S.; Bomfim, M.R.Q.; França, R.O.; Farias, L.M.; Carvalho, M.A.R.; Serufo, J.C.; Santos, S.G. Phenotypic and Genotypic Diversity of Multidrug-Resistant Pseudomonas aeruginosa Isolates from Bloodstream Infections Recovered in the Hospitals of Belo Horizonte, Brazil. Chemotherapy 2014, 60, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, Y.C.; Furlaneto, I.P.; Maciel, A.H.P.; Quaresma, A.J.P.G.; de Matos, E.C.O.; Conceição, M.L.; da Vieira, M.C.S.; da Brabo, G.L.C.; do Sarges, E.S.N.F.; Lima, L.N.G.C.; et al. High Prevalence of Atypical Virulotype and Genetically Diverse Background among Pseudomonas aeruginosa Isolates from a Referral Hospital in the Brazilian Amazon. PLoS ONE 2020, 15, e0238741. [Google Scholar] [CrossRef] [PubMed]
- Franco, M.R.G.; Caiaffa-Filho, H.H.; Burattini, M.N.; Rossi, F. Metallo-Beta-Lactamases among Imipenem-Resistant Pseudomonas aeruginosa in a Brazilian University Hospital. Clinics 2010, 65, 825–829. [Google Scholar] [CrossRef] [PubMed]
- de Jácome, P.R.L.A.; Alves, L.R.; Cabral, A.B.; Lopes, A.C.S.; Maciel, M.A.V. Phenotypic and Molecular Characterization of Antimicrobial Resistance and Virulence Factors in Pseudomonas aeruginosa Clinical Isolates from Recife, State of Pernambuco, Brazil. Rev. Soc. Bras. Med. Trop. 2012, 45, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Gräf, T.; Fuentefria, D.B.; Corção, G. Ocorrência de Cepas de Pseudomonas aeruginosa Multirresistentes Produtoras de Metalo-β-Lactamase BlaSPM-1 Em Amostras Clínicas. Rev. Soc. Bras. Med. Trop. 2008, 41, 306–308. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- da Santos, C.M.C.; de Pimenta, C.A.M.; Nobre, M.R.C. The PICO Strategy for the Research Question Construction and Evidence Search. Rev. Lat.-Am. Enferm. 2007, 15, 508–511. [Google Scholar] [CrossRef]
- Kar, B.; Sharma, M.; Peter, A.; Chetia, P.; Neog, B.; Borah, A.; Pati, S.; Bhattacharya, D. Prevalence and Molecular Characterization of β-Lactamase Producers and Fluoroquinolone Resistant Clinical Isolates from North East India. J. Infect. Public Health 2021, 14, 628–637. [Google Scholar] [CrossRef]
- Aromataris, E.; Munn, Z. Joanna Briggs Institute Reviewer’s Manual. Joanna Briggs Inst. 2017, 2017. [Google Scholar] [CrossRef]
- Munn, Z.; Aromataris, E.; Tufanaru, C.; Stern, C.; Porritt, K.; Farrow, J.; Lockwood, C.; Stephenson, M.; Moola, S.; Lizarondo, L.; et al. The Development of Software to Support Multiple Systematic Review Types: The Joanna Briggs Institute System for the Unified Management, Assessment and Review of Information (JBI SUMARI). Int. J. Evid. Based Healthc. 2019, 17, 36–43. [Google Scholar] [CrossRef]
- Foumani, A.A.; Kalurazi, T.Y.; Rostami, F.M.; Ebrahim-Saraie, H.S.; Nazari-Alam, A.; Halaji, M. Epidemiology of Pseudomonas aeruginosa in Cystic Fibrosis Patients in Iran: A Systematic Review and Meta-Analysis. Infez. Med. 2020, 28, 314–321. [Google Scholar]
- Gonçalves, D.C.P.S.; Lima, A.B.M.; de Leão, L.S.N.O.; do Carmo Filho, J.R.; Pimenta, F.C.; Vieira, J.D.G. Detecção de Metalo-Beta-Lactamase Em Pseudomonas aeruginosa Isoladas de Pacientes Hospitalizados Em Goiânia, Estado de Goiás. Rev. Soc. Bras. Med. Trop. 2009, 42, 411–414. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernandes, T.Á.; Pereira, C.A.P.; Petrili, A.S.; Pignatari, A.C.C. Caracterização Molecular de Pseudomonas aeruginosa Resistentes a Carbapenêmicos e Produtoras de Metalo-β-Lactamase Isoladas Em Hemoculturas de Crianças e Adolescentes Com Câncer. Rev. Soc. Bras. Med. Trop. 2010, 43, 372–376. [Google Scholar] [CrossRef]
- Scheffer, M.C.; Bazzo, M.L.; Steindel, M.; Darini, A.L.; Clímaco, E.; Dalla-Costa, L.M. Intrahospital Spread of Carbapenem-Resistant Pseudomonas aeruginosa in a University Hospital in Florianópolis, Santa Catarina, Brazil. Rev. Soc. Bras. Med. Trop. 2010, 43, 367–371. [Google Scholar] [CrossRef] [PubMed]
- Polotto, M.; Casella, T.; de Lucca Oliveira, M.G.; Rúbio, F.G.; Nogueira, M.L.; de Almeida, M.T.; Nogueira, M.C. Detection of P. aeruginosa Harboring Bla CTX-M-2, Bla GES-1 and Bla GES-5, Bla IMP-1 and Bla SPM-1 Causing Infections in Brazilian Tertiary-Care Hospital. BMC Infect. Dis. 2012, 12, 176. [Google Scholar] [CrossRef] [PubMed]
- Lucena, A.; Dalla Costa, L.M.; Nogueira, K.S.; Matos, A.P.; Gales, A.C.; Paganini, M.C.; Castro, M.E.S.; Raboni, S.M. Nosocomial Infections with Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa: Molecular Epidemiology, Risk Factors, Clinical Features and Outcomes. J. Hosp. Infect. 2014, 87, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Miranda, C.C.; De Filippis, I.; Pinto, L.H.; Coelho-Souza, T.; Bianco, K.; Cacci, L.C.; Picão, R.C.; Clementino, M.M. Genotypic Characteristics of Multidrug-Resistant Pseudomonas aeruginosa from Hospital Wastewater Treatment Plant in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2015, 118, 1276–1286. [Google Scholar] [CrossRef]
- Cacci, L.C.; Chuster, S.G.; Martins, N.; do Carmo, P.R.; de Girão, V.B.C.; Nouér, S.A.; de Freitas, W.V.; de Matos, J.A.; de Magalhães, A.C.G.; Ferreira, A.L.P. Mechanisms of Carbapenem Resistance in Endemic Pseudomonas aeruginosa Isolates after an SPM-1 Metallo-β-Lactamase Producing Strain Subsided in an Intensive Care Unit of a Teaching Hospital in Brazil. Memórias Inst. Oswaldo Cruz 2016, 111, 551–558. [Google Scholar] [CrossRef]
- De Matos, E.C.O.; de Matos, H.J.; Conceição, M.L.; Rodrigues, Y.C.; do Carneiro, I.C.R.S.; Lima, K.V.B. Clinical and Microbiological Features of Infections Caused by Pseudomonas aeruginosa in Patients Hospitalized in Intensive Care Units. Rev. Soc. Bras. Med. Trop. 2016, 49, 305–311. [Google Scholar] [CrossRef]
- Chaves, L.; Tomich, L.M.; Salomão, M.; Leite, G.C.; Ramos, J.; Martins, R.R.; Rizek, C.; Neves, P.; Batista, M.V.; Amigo, U.; et al. High Mortality of Bloodstream Infection Outbreak Caused by Carbapenem-Resistant P. aeruginosa Producing SPM-1 in a Bone Marrow Transplant Unit. J. Med. Microbiol. 2017, 66, 1722–1729. [Google Scholar] [CrossRef]
- Dias, V.C.; Resende, J.A.; Bastos, A.N.; De Andrade Bastos, L.Q.; De Andrade Bastos, V.Q.; Bastos, R.V.; Diniz, C.G.; Da Silva, V.L. Epidemiological, Physiological, and Molecular Characteristics of a Brazilian Collection of Carbapenem-Resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Microb. Drug Resist. 2017, 23, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, I.R.; Dantas, R.C.C.; Ferreira, M.L.; da Batistão, D.W.F.; Gontijo-Filho, P.P.; Ribas, R.M. Carbapenem-Resistant Pseudomonas aeruginosa: Association with Virulence Genes and Biofilm Formation. Braz. J. Microbiol. 2017, 48, 211–217. [Google Scholar] [CrossRef]
- Martins, W.M.B.S.; Narciso, A.C.; Cayô, R.; Santos, S.V.; Fehlberg, L.C.C.; Ramos, P.L.; Da Cruz, J.B.; Gales, A.C. SPM-1-Producing Pseudomonas aeruginosa ST277 Clone Recovered from Microbiota of Migratory Birds. Diagn. Microbiol. Infect. Dis. 2018, 90, 221–227. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Santos, I.C.; Pereira De Andrade, N.F.; Da Conceição Neto, O.C.; Da Costa, B.S.; De Andrade Marques, E.; Rocha-de-Souza, C.M.; Asensi, M.D.; D’Alincourt Carvalho-Assef, A.P. Epidemiology and Antibiotic Resistance Trends in Clinical Isolates of Pseudomonas aeruginosa from Rio de Janeiro—Brazil: Importance of Mutational Mechanisms over the Years (1995–2015). Infect. Genet. Evol. 2019, 73, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.C.; Rocha-de-Souza, C.M.; Albano, R.M.; De Oliveira Santos, I.C.; Carvalho-Assef, A.P.D. Exploring the Success of Brazilian Endemic Clone Pseudomonas aeruginosa ST277 and Its Association with the CRISPR-Cas System Type I-C. BMC Genom. 2020, 21, 255. [Google Scholar] [CrossRef] [PubMed]
- Freire, M.P.; Camargo, C.H.; Yamada, A.Y.; Nagamori, F.O.; Reusing Junior, J.O.; Spadão, F.; Cury, A.P.; Rossi, F.; Nahas, W.C.; David-Neto, E.; et al. Critical Points and Potential Pitfalls of Outbreak of IMP-1-Producing Carbapenem-Resistant Pseudomonas aeruginosa among Kidney Transplant Recipients: A Case–Control Study. J. Hosp. Infect. 2021, 115, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.A.S.; Rodrigues, Y.C.; Marcon, D.J.; Lobato, A.R.F.; Cazuza, T.B.; Gouveia, M.I.M.; Silva, M.J.A.; Souza, A.B.; Lima, L.N.G.C.; Quaresma, A.J.P.G.; et al. Endemic High-Risk Clone ST277 Is Related to the Spread of SPM-1-Producing Pseudomonas aeruginosa during the COVID-19 Pandemic Period in Northern Brazil. Microorganisms 2023, 11, 2069. [Google Scholar] [CrossRef]
- Ghasemian, A.; Rizi, K.S.; Vardanjani, H.R.; Nojoomi, F. Prevalence of Clinically Isolated Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa, Coding Genes, and Possible Risk Factors in Iran. Iran. J. Pathol. 2018, 13, 1. [Google Scholar]
- Dawadi, P.; Khadka, C.; Shyaula, M.; Syangtan, G.; Joshi, T.P.; Pepper, S.H.; Kanel, S.R.; Pokhrel, L.R. Prevalence of Metallo-β-Lactamases as a Correlate of Multidrug Resistance among Clinical Pseudomonas aeruginosa Isolates in Nepal. Sci. Total Environ. 2022, 850, 157975. [Google Scholar] [CrossRef]
- Partridge, S.R. Resistance Mechanisms in Enterobacteriaceae. Pathology 2015, 47, 276–284. [Google Scholar] [CrossRef]
- Edelstein, M.V.; Skleenova, E.N.; Shevchenko, O.V.; D’souza, J.W.; Tapalski, D.V.; Azizov, I.S.; Sukhorukova, M.V.; Pavlukov, R.A.; Kozlov, R.S.; Toleman, M.A.; et al. Spread of Extensively Resistant VIM-2-Positive ST235 Pseudomonas aeruginosa in Belarus, Kazakhstan, and Russia: A Longitudinal Epidemiological and Clinical Study. Lancet Infect. Dis. 2013, 13, 867–876. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Collignon, P.; Beggs, J.J.; Walsh, T.R.; Gandra, S.; Laxminarayan, R. Anthropological and Socioeconomic Factors Contributing to Global Antimicrobial Resistance: A Univariate and Multivariable Analysis. Lancet Planet. Health 2018, 2, e398–e405. [Google Scholar] [CrossRef] [PubMed]
- Bonelli, R.R.; Moreira, B.M.; Picão, R.C. Antimicrobial Resistance among Enterobacteriaceae in South America: History, Current Dissemination Status and Associated Socioeconomic Factors. Drug Resist. Updates 2014, 17, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Planta, M.B. The Role of Poverty in Antimicrobial Resistance. J. Am. Board. Fam. Med. 2007, 20, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Tenover, F.C.; Nicolau, D.P.; Gill, C.M. Carbapenemase-Producing Pseudomonas aeruginosa—An Emerging Challenge. Emerg. Microbes Infect. 2022, 11, 811–814. [Google Scholar] [CrossRef]
- Escandón-Vargas, K.; Reyes, S.; Gutiérrez, S.; Villegas, M.V. The Epidemiology of Carbapenemases in Latin America and the Caribbean. Expert Rev. Anti-Infect. Ther. 2017, 15, 277–297. [Google Scholar] [CrossRef]
- Zowawi, H.M.; Balkhy, H.H.; Walsh, T.R.; Paterson, D.L. β-Lactamase Production in Key Gram-Negative Pathogen Isolates from the Arabian Peninsula. Clin. Microbiol. Rev. 2013, 26, 361–380. [Google Scholar] [CrossRef]
- Poirel, L.; Nordmann, P.; Lagrutta, E.; Cleary, T.; Munoz-Price, L.S. Emergence of KPC-Producing Pseudomonas aeruginosa in the United States. Antimicrob. Agents Chemother. 2010, 54, 3072. [Google Scholar] [CrossRef]
- Walters, M.S.; Grass, J.E.; Bulens, S.N.; Hancock, E.B.; Phipps, E.C.; Muleta, D.; Mounsey, J.; Kainer, M.A.; Concannon, C.; Dumyati, G. Carbapenem-Resistant Pseudomonas aeruginosa at US Emerging Infections Program Sites, 2015. Emerg. Infect. Dis. 2019, 25, 1281. [Google Scholar] [CrossRef]
- Rojo-Bezares, B.; Estepa, V.; Cebollada, R.; de Toro, M.; Somalo, S.; Seral, C.; Castillo, F.J.; Torres, C.; Sáenz, Y. Carbapenem-Resistant Pseudomonas aeruginosa Strains from a Spanish Hospital: Characterization of Metallo-Beta-Lactamases, Porin OprD and Integrons. Int. J. Med. Microbiol. 2014, 304, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Pérez, J.E.; Buelvas, F.; Tovar, C.; Vanegas, N.; Stokes, H.W. Establishment and Multi Drug Resistance Evolution of ST235 Pseudomonas aeruginosa Strains in the Intensive Care Unit of a Colombian Hospital. Res. Microbiol. 2014, 165, 852–856. [Google Scholar] [CrossRef] [PubMed]
- El Salabi, A.; Toleman, M.A.; Weeks, J.; Bruderer, T.; Frei, R.; Walsh, T.R. First Report of the Metallo-β-Lactamase SPM-1 in Europe. Antimicrob. Agents Chemother. 2010, 54, 582. [Google Scholar] [CrossRef] [PubMed]
- Wendel, A.F.; Brodner, A.H.B.; Wydra, S.; Ressina, S.; Henrich, B.; Pfeffer, K.; Toleman, M.A.; MacKenzie, C.R. Genetic Characterization and Emergence of the Metallo-β-Lactamase GIM-1 in Pseudomonas Spp. and Enterobacteriaceae during a Long-Term Outbreak. Antimicrob. Agents Chemother. 2013, 57, 5162–5165. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Deshpande, L.M.; Costello, A.; Davies, T.A.; Jones, R.N. Epidemiology and Carbapenem Resistance Mechanisms of Carbapenem-Non-Susceptible Pseudomonas aeruginosa Collected during 2009-11 in 14 European and Mediterranean Countries. J. Antimicrob. Chemother. 2014, 69, 1804–1814. [Google Scholar] [CrossRef] [PubMed]
- Pollini, S.; Maradei, S.; Pecile, P.; Olivo, G.; Luzzaro, F.; Docquier, J.-D.; Rossolini, G.M. FIM-1, a New Acquired Metallo-β-Lactamase from a Pseudomonas aeruginosa Clinical Isolate from Italy. Antimicrob. Agents Chemother. 2013, 57, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.H.; De, A.S.; Baveja, S.M.; Gore, M.A. Prevalence and Risk Factors of Metallo β-Lactamase Producing Pseudomonas aeruginosa and Acinetobacter Species in Burns and Surgical Wards in a Tertiary Care Hospital. J. Lab. Physicians 2012, 4, 39–42. [Google Scholar] [CrossRef] [PubMed]
- El-Domany, R.A.; Emara, M.; El-Magd, M.A.; Moustafa, W.H.; Abdeltwab, N.M. Emergence of Imipenem-Resistant Pseudomonas aeruginosa Clinical Isolates from Egypt Coharboring VIM and IMP Carbapenemases. Microb. Drug Resist. 2017, 23, 682–686. [Google Scholar] [CrossRef]
- Khosravi, A.D.; Mihani, F. Detection of Metallo-β-Lactamase–Producing Pseudomonas aeruginosa Strains Isolated from Burn Patients in Ahwaz, Iran. Diagn. Microbiol. Infect. Dis. 2008, 60, 125–128. [Google Scholar] [CrossRef]
- Malkoçoğlu, G.; Aktaş, E.; Bayraktar, B.; Otlu, B.; Bulut, M.E. VIM-1, VIM-2, and GES-5 Carbapenemases among Pseudomonas aeruginosa Isolates at a Tertiary Hospital in Istanbul, Turkey. Microb. Drug Resist. 2017, 23, 328–334. [Google Scholar] [CrossRef]
- Ghamgosha, M.; Shahrekizahedani, S.; Kafilzadeh, F.; Bameri, Z.; Taheri, R.A.; Farnoosh, G. Metallo-Beta-Lactamase VIM-1, SPM-1, and IMP-1 Genes Among Clinical Pseudomonas aeruginosa Species Isolated in Zahedan, Iran. Jundishapur J. Microbiol. 2015, 8, e17489. [Google Scholar] [CrossRef] [PubMed]
- Moosavian, M.; Rahimzadeh, M. Molecular Detection of Metallo-β-Lactamase Genes, BlaIMP-1, BlaVIM-2 and BlaSPM-1 in Imipenem Resistant Pseudomonas aeruginosa Isolated from Clinical Specimens in Teaching Hospitals of Ahvaz, Iran. Iran. J. Microbiol. 2015, 7, 2–6. [Google Scholar] [PubMed]
- Hopkins, K.L.; Meunier, D.; Findlay, J.; Mustafa, N.; Parsons, H.; Pike, R.; Wright, L.; Woodford, N. SPM-1 Metallo-β-Lactamase-Producing Pseudomonas aeruginosa ST277 in the UK. J. Med. Microbiol. 2016, 65, 696–697. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, A.; Rodríguez, N.; Alcalde-Rico, M.; Castillo, C.; García, P. Primer Aislado de Pseudomonas aeruginosa Productora de Sao Paulo Metalo-β-Lactamasa (SPM-1) En Un Paciente Chileno. Rev. Chil. Infectol. 2021, 38, 724–726. [Google Scholar] [CrossRef]
- Abdelaziz, N.A. Phenotype-Genotype Correlations among Carbapenem-Resistant Enterobacterales Recovered from Four Egyptian Hospitals with the Report of SPM Carbapenemase. Antimicrob. Resist. Infect. Control 2022, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Figueredo, A.C.F.; Freitas, N.L.D.; Dalmolin, T.V.; Brandão, F. Pseudomonas aeruginosa: Panorama do perfil de resistência aos carbapenêmicos no brasil/Pseudomonas aeruginosa: Overview of the profile of resistance to carbapenems in brazil. BJD 2021, 7, 9661–9672. [Google Scholar] [CrossRef]
- ul Ain, N.; Abrar, S.; Sherwani, R.A.K.; Hannan, A.; Imran, N.; Riaz, S. Systematic Surveillance and Meta-Analysis on the Prevalence of Metallo-β-Lactamase Producers among Carbapenem Resistant Clinical Isolates in Pakistan. J. Glob. Antimicrob. Resist. 2020, 23, 55–63. [Google Scholar] [CrossRef]
- Osei Sekyere, J.; Reta, M.A. Genomic and Resistance Epidemiology of Gram-Negative Bacteria in Africa: A Systematic Review and Phylogenomic Analyses from a One Health Perspective. Msystems 2020, 5, e00897-20. [Google Scholar] [CrossRef]
- Vaez, H.; Salehi-Abargouei, A.; Ghalehnoo, Z.R.; Khademi, F. Multidrug Resistant Pseudomonas aeruginosa in Iran: A Systematic Review and Metaanalysis. J. Glob. Infect. Dis. 2018, 10, 212. [Google Scholar]
- Pisani, B.; Simões, M.; Prandi, M.A.G.; Rocha, M.M.; Gonçalves, C.R.; Vaz, T.M.; Irino, K. Surto de Bacteriemia Por Pseudomonas aeruginosa Na Unidade de Hemodiálise de Um Hospital de Campinas, São Paulo, Brasil. Rev. Inst. Adolfo Lutz 2000, 59, 51–56. [Google Scholar]
- Kümmerer, K.; Henninger, A. Promoting Resistance by the Emission of Antibiotics from Hospitals and Households into Effluent. Clin. Microbiol. Infect. 2003, 9, 1203–1214. [Google Scholar] [CrossRef]
- Kunin, C.M. Resistance to Antimicrobial Drugs—A Worldwide Calamity. Ann. Intern. Med. 1993, 118, 557. [Google Scholar] [CrossRef]
- Laranjeira, V.D.S.; Marchetti, D.P.; Steyer, J.R.; Corção, G.; Picoli, S.U. Pesquisa de Acinetobacter Sp e Pseudomonas aeruginosa Produtores de Metalo-β-Lactamase Em Hospital de Emergência de Porto Alegre, Estado Do Rio Grande Do Sul, Brasil. Rev. Soc. Bras. Med. Trop. 2010, 43, 462–464. [Google Scholar] [CrossRef]
- Camargo, C.H.; Bruder-Nascimento, A.; Mondelli, A.L.; Montelli, A.C.; Sadatsune, T. Detection of SPM and IMP Metallo-β-Lactamases in Clinical Specimens of Pseudomonas aeruginosa from a Brazilian Public Tertiary Hospital. Braz. J. Infect. Dis. 2011, 15, 478–481. [Google Scholar] [CrossRef]
- de Cavalcanti, F.L.S.; Almeida, A.C.S.; Vilela, M.A.; de Morais, M.M.C.; de Morais Junior, M.A. Changing the Epidemiology of Carbapenem-Resistant Pseudomonas aeruginosa in a Brazilian Teaching Hospital: The Replacement of São Paulo Metallo-β-Lactamase-Producing Isolates. Mem. Inst. Oswaldo Cruz 2012, 107, 420–423. [Google Scholar] [CrossRef]


| No | Authors and Year of Publication | Title | Database | Methodology/Sample Size | Purpose(s)/Objective(s) | Sample Type/City (Region)/Period | Results | JBI Score | Frequency of MβL Resistance Genes among P. aeruginosa Isolates |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Gonçalves et al. (2009) [42] | Detection of metallo-beta-lactamase in Pseudomonas aeruginosa isolated from hospitalized patients in Goiânia, State of Goiás | LILACS and SciELO and PUBMED | Cohort/75 isolates from patients with PA infection. | To investigate the susceptibility profile of P. aeruginosa previously isolated from patients in a hospital in Goiânia (GO). | Clinical/hospital settings/Goiânia-GO (midwest)/2005–2007 | Among the 62 isolates resistant to IMP and CAZ, 35 (56.4%) produced MβL, corresponding to 47.3% of the PA isolates, while 26 (74.3%) presented the blaSPM-1 gene. | (11/11) | The blaSPM-1 gene was detected in high frequency (26/35–74.28%). |
| 2 | Fernandes et al. (2010) [43] | Molecular characterization of carbapenem-resistant and MβL-producing Pseudomonas aeruginosa isolated from blood cultures from children and teenagers with cancer. | LILACS and SciELO and PUBMED | Cohort/56 PA isolates from 49 patients. | Assess the persistence of MβL-PA from BSIs. | Clinical/hospital settings/São Paulo-SP (southeast)/2000–2005 | A total of 32 (57.1%) CR-PA and, of this amount, 18 samples (32.14%) were positive for MΒL by a disk approximation test, equivalent to 17.85% of the total. All 18 samples were positive for the blaSPM-1 gene. The blaIMP-1, blaVIM-1, blaVIM-2 were not detected. | (11/11) | The blaSPM-1 gene was related to all CR-PA (18/18–100%). |
| 3 | Franco et al. (2010) [33] | Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian university hospital | LILACS and SciELO and PUBMED | Cohort/69 included PA isolates. | Determine the frequency of MβL-PA among imipenem-resistant isolates in this hospital and compare phenotypic and molecular methods of detection. | Clinical/hospital settings/São Paulo-SP (southeast)/2006 | The frequency of MβL-PA strains was 30.4% (n = 21). Of that number, 17 (80.95%) were positive for blaSPM-1 and 4 (19.05%) for blaVIM-2. The blaIMP-1, blaIMP-2, and blaVIM-1 were not detected. | (10/11) | Distinct variants were detected, with a higher frequency of the blaSPM-1 (17/21–80.95%), followed by blaVIM-2 (4–19.05%). |
| 4 | Scheffer et al. (2010) [44] | Intrahospital spread of carbapenem-resistant Pseudomonas aeruginosa in a University Hospital in Florianópolis, Santa Catarina, Brazil | LILACS and SciELO and PUBMED | Cohort/36 CR-PA isolates. | To investigate the minimum inhibitory concentration (MIC), the presence of MβL-PA and a possible clonal relationship between isolates in one teaching hospital in southern Brazil. | Clinical/hospital settings/Florianópolis–SC (south)/2003–2005 | Seven isolates (19.44%) presented positive phenotypic tests for MβL, and most carried the blaSPM-1 gene. | (11/11) | Distinct variants were detected, with a higher frequency of the blaSPM-1, (6/7–85.71%) and blaIMP-16 (1–14.29%). |
| 5 | Jácome et al. (2012) [34] | Phenotypic and molecular characterization of antimicrobial resistance and virulence factors in Pseudomonas aeruginosa clinical isolates from Recife, State of Pernambuco, Brazil | LILACS and SciELO and PUBMED | Cohort/61 PA strains from public hospitals. | Identify resistance and virulence markers and establish clonal spread of PA isolates. | Clinical/hospital settings/Recife-PE (northeast)/2006–2010 | A total of 29 isolates were resistant to imipenem and/or ceftazidime. From this total, 44.8% (13/29) were MβL-PA, of which, 46.2% (6/13) revealed the blaSPM-1 gene. | (9/11) | The blaSPM-1 gene was related to 13 (46.15%) MβL-PA. |
| 6 | Polotto et al. (2012) [45] | Detection of P. aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5, bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital | PUBMED | Cohort/56 P. aeruginosa isolates. | To investigate the carriage of genes codifying MΒLs and ESBLs by P. aeruginosa isolated from patients admitted to a Brazilian 720-bed teaching tertiary care hospital. | Clinical/hospital settings/São Paulo—SP (Sudeste)/June–December 2009 | The prevalence of isolates harboring MβL genes was 30.3% (17/56). Ten (10/17 58.82%) presented the blaSPM-1 gene, and the blaIMP-1 was detected in seven isolates (41.17). No blaVIM type was detected. | (10/11) | Distinct variants were detected, with a higher frequency of the blaSPM-1 gene. |
| 7 | Inacio et al. (2014) [31] | Phenotypic and genotypic diversity of multidrug-resistant Pseudomonas aeruginosa Isolates from bloodstream infections recovered in the Hospitals of Belo Horizonte, Brazil | PUBMED | Cohort/40 PA isolates from 5 hospitals in the city of Belo Horizonte. | To evaluate antimicrobial resistance and the production of MβL, oxacillinase, and cephalosporinases, and the genetic diversity of P. aeruginosa strains isolated from patients with BSIs. | Clinical/hospital settings/Belo Horizonte-MG (southeast)/2008–2009 | A frequency of 87.5% (n = 35/40 were MβL-producers. Of this amount, 29 isolates were exclusively detected harboring the blaSPM-1 gene (82.86%), equivalent to 72.5% of the total PA samples. Simultaneous occurrence of the blaSPM-1 and blaVIM-1 genes was observed in six MβL-PA. | (10/11) | High frequency of the blaSPM-1 (82.86%) gene. Strains co-harboring the blaSPM-1 and blaVIM-1 genes (17.14%). |
| 8 | Lucena et al. (2014) [46] | Nosocomial infections with metallo-beta-lactamase-producing Pseudomonas aeruginosa: molecular epidemiology, risk factors, clinical features and outcomes | Science Direct and PUBMED | Cohort/CR-PA were isolated from different clinical samples of hospitalized patients. | To evaluate the molecular epidemiology, risk factors, and outcomes of nosocomial infections caused by MβL-PA in a university hospital in southern Brazil. | clinical/hospital settings/Curitiba-PR (south)/2001–2008 | A total of 93/142 (65%) strains were confirmed as MβL-PA. Considering the total number of isolates, 91 isolates contained the blaSPM-1 gene (64%), followed by the blaIMP-1 (0.7%) and the blaIMP-16 (0.7%) genes in one isolate each. | (10/10) | Three distinct variants were detected among MβL-PA, with emphasis on the blaSPM-1 gene (97.84%), blaIMP-1 (1.08%), blaIMP-16 (1.08%). |
| 9 | Miranda et al. (2015) [47] | Genotypic characteristics of multidrug-resistant Pseudomonas aeruginosa from hospital wastewater treatment plant in Rio de Janeiro, Brazil | PUBMED | Cohort/41 isolates of PA. | To investigate the antimicrobial susceptibility profiles and genetic relatedness of P. aeruginosa isolates obtained from a Hospital Wastewater Treatment Plant (HWTP). | Environmental/Rio de Janeiro—RJ (southeast)/2008–2010 | A total of 38 (93%) isolates exhibited antimicrobial resistance to carbapenems. | (11/11) | Among these, 14 (37%) were blaVIM and six were (18%) blaSPM- 1-positive. Three strains were found co-harboring the blaVIM and blaSPM-1 genes. |
| 10 | Cacci et al. (2016) [48] | Mechanisms of carbapenem resistance in endemic Pseudomonas aeruginosa isolates after an SPM-1 metallo-β—lactamase producing strain subsided in an intensive care unit of a teaching hospital in Brazil | LILACS and SciELO and PUBMED | Cohort/35 clinical and 1 hemodialysis isolate of PA from 234 patients admitted to the ICU. | Compare the resistance levels of clinical isolates of P. aeruginosa detected from 2007 to 2008 with those previously described in the literature. | Clinical/hospital settings/Rio de Janeiro-RJ (southeast)/2007–2008 | CR-PA was related to one blaSPM-1 positive isolate. The blaIMP-type, blaVIM-type, blaSIM-1, blaGIM-1, and blaNDM-1 were not detected. | (10/11) | One blaSPM-1 (100%) in MβL-PA |
| 11 | Matos et al. (2016) [49] | Clinical and microbiological features of infections caused by Pseudomonas aeruginosa in patients hospitalized in intensive care units | LILACS and SciELO and PUBMED | Cohort study/54 isolates from patients in beds admitted to intensive care units (ICUs). | To analyze the clinical, epidemiological, and molecular characteristics of P. aeruginosa infections in ICU patients at a university hospital. | Clinical/hospital settings/Belém -PA (northern)/2010–2012 | The frequency of MDR-PA strains was 37% (20/54). Of this amount, 20% (4/20) of the isolates were positive for the MβL blaSPM-1 gene, which is equivalent to 7.4% of the total samples. | (11/11) | The blaSPM-1 gene was found in four MβL isolates. |
| 12 | Chaves et al. (2017) [50] | High mortality of bloodstream infection outbreak caused by carbapenem-resistant P. aeruginosa producing SPM-1 in a bone marrow transplant unit | PUBMED | Case–control/29 cases and 58 controls, with 17 isolates. | To analyze the mortality and risk factors for developing BSI caused by carbapenem-resistant P. aeruginosa (CRPA). | Clinical/hospital settings/São Paulo—SP (southeast)/2011–2013 | Most isolates were MβL-PA (16/17—94.12%). | (10/11) | Seven isolates were found harboring the blaSPM-1 (41.2%) |
| 13 | Dias et al. (2017) [51] | Epidemiological, Physiological, and Molecular Characteristics of a Brazilian Collection of Carbapenem-Resistant Acinetobacter baumannii and Pseudomonas aeruginosa | PUBMED | Cross-sectional/19 PA were isolated from a private hospital. | To determine antimicrobial susceptibility and biocide tolerance patterns, hemolytic activity, biofilm formation, and oxidative stress tolerance in carbapenem-resistant and drug-sensitive bacteria, and also track genetic markers of carbapenem resistance of A. baumanni and P. aeruginosa. | Clinical/hospital settings/Juiz de Fora-MG (southeast)/January to December 2013 | About five samples showed the MβL phenotype with the blaSPM-1 genetic marker (26.32% of the total). The blaIMP, blaVIM, blaSIM, blaGIM, and blaNDM-1 were not detected. | (8/8) | The blaSPM-1 gene was found in all 5 MβL-PA samples. |
| 14 | Gonçalves et al. (2017) [52] | Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation | LILACS and SciELO and PUBMED | Cohort/56 CR-PA isolates from patients. | Identify strains of P. aeruginosa resistant to carbapenems. | Clinical/hospital settings/Uberlândia -MG (southeast)/2009–2012 | Of the total CR-PA isolates, nine were phenotypically positive for MβL. | (10/11) | Among the MβL-PA, six (66.67%) were positive for the blaSPM-1 and three (33.33%) for the blaVIM genes. |
| 15 | Martins et al. (2018) [53] | SPM-1-producing Pseudomonas aeruginosa ST277 clone recovered from microbiota of migratory birds | PUBMED | Cohort/40 strains of P. aeruginosa. | To report the detection and molecular characterization of the SPM-1-producing P. aeruginosa ST277 clone recovered from the microbiota of migratory birds (Dendrocygna viduata) in Brazil. | Animal-related/São Paulo—SP (southeast)/July–August 2012 | A total of nine CR-PA isolates (22.5%) were MβL producers. | (11/11) | The blaSPM-1 gene was found in 7 strains (77.77%). |
| 16 | De Oliveira Santos et al. (2019) [54] | Epidemiology and antibiotic resistance trends in clinical isolates of Pseudomonas aeruginosa from Rio de janeiro—Brazil: Importance of mutational mechanisms over the years (1995–2015) | PUBMED and Science Direct | Cohort/88 isolates of PA. | To determine patterns and mechanisms of antimicrobial resistance and its spread over the years in Rio de Janeiro. | Clinical/hospitalar/Rio de Janeiro—RJ (southeast)/2006–2010 | A total of three P. aeruginosa strains were MβL producers (3.4%). | (10/11) | All three MβL-PA isolates were blaSPM-1 (100%) |
| 17 | Rodrigues et al. (2020) [32] | High prevalence of atypical virulotype and genetically diverse background among Pseudomonas aeruginosa isolates from a referral hospital in the Brazilian Amazon | PUBMED | Cross-sectional study/54 PA isolates from an Amazonian referral hospital. | To report in-depth data on virulence, resistance properties, and genetic diversity of P. aeruginosa isolates from patients admitted to the ICU of a reference hospital in the Brazilian Amazon region. | Clinical/hospitalar/Belém -PA (northern)/2010–2013 | A blaSPM-1 frequency of 9.2% (n = 5/54) was found. Two isolates co-harbored the blaSPM-1/blaOXA-2 and blaSPM-1/blaOXA-10 genes, and one isolate was positive for blaSPM-1 only. The blaIMP, blaVIM, blaNDM, and blaKPC genes were not detected. | (8/8) | The blaSPM-1 was found in five MβL-PA. |
| 18 | Silveira et al. (2020) [55] | Exploring the success of Brazilian endemic clone Pseudomonas aeruginosa ST277 and its association with the CRISPR-Cas system type I-C | PUBMED | Cohort/47 strains of P. aeruginosa. | To examine the phylogenetic distribution and conservation of genetic determinants among ST277 P. aeruginosa genomes available at NCBI. | Environmental/Rio de Janeiro—RJ (southeast)/1997–2018 | A total of two SPM-1 isolates were recovered from impacted rivers (4.25%). | (9/11) | All two samples were 100% blaSPM-1. |
| 19 | Freire et al. (2021) [56] | Critical points and potential pitfalls of outbreak of IMP-1-producing carbapenem-resistant Pseudomonas aeruginosa among kidney transplant recipients: a case control study | PUBMED | Case–control/40 CRPA isolates. | To analyze an outbreak of infection/colonization with IMP-1-producing CRPA on a kidney transplantation (KT) ward. | Clinical/hospitalar/São Paulo—SP (southeast)/2019–2020 | A total of 15 P. aeruginosa strains were MβL producers (37.5%). | (10/11) | The blaSPM-1 gene was not found. All 15 strains were blaIMP-1 (100%). |
| 20 | Dos Santos et al. (2023) [57] | Endemic high-risk clone ST277 is related to the spread of SPM-1-producing Pseudomonas aeruginosa during the COVID-19 pandemic period in the Brazilian Northern region | PUBMED | Cross-sectional study/34 PA isolates. | To investigate the antimicrobial resistance, virulence, and genotypic characteristics of SPM-1-producing P. aeruginosa recovered from the pre-pandemic era in medical facilities in the states of Pará (PA) and Acre (AC), northern Brazilian Amazon area. | Clinical/hospitalar/Pará State (northern region) 2018–2022 | All samples were classified as CRPA and MβL producers (n = 34; 100%). | (8/8) | All samples were 100% blaSPM-1 (100%). |
| Set | Covariate | Coefficient | Standard Error | 95% Lower | 95% Upper | Z-Value | 2-Sided p-Value | Set |
|---|---|---|---|---|---|---|---|---|
| Intercept | 0.2574 | 0.2329 | −0.199 | 0.7138 | 1.11 | 0.269 | ||
| Year | 2010 | −1.1612 | 0.2917 | −1.7329 | −0.5895 | −3.98 | 0.0001 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2012 | −1.31 | 0.3155 | −1.9284 | −0.6915 | −4.15 | 0 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2014 | 0.5199 | 0.2855 | −0.0396 | 1.0794 | 1.82 | 0.0686 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2015 | 2.3293 | 0.6549 | 1.0457 | 3.6128 | 3.56 | 0.0004 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2016 | −2.9922 | 0.5181 | −4.0076 | −1.9768 | −5.78 | 0 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2017 | −1.6585 | 0.3048 | −2.256 | −1.061 | −5.44 | 0 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2018 | −1.4942 | 0.4445 | −2.3654 | −0.6229 | −3.36 | 0.0008 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2019 | −3.6042 | 0.6326 | −4.8441 | −2.3643 | −5.7 | 0 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2020 | −2.4511 | 0.4233 | −3.2807 | −1.6215 | −5.79 | 0 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2021 | −0.7682 | 0.4011 | −1.5544 | 0.0179 | −1.92 | 0.0555 | Q = 212.13, df = 11, p < 0.0001 |
| Year | 2023 | 3.9767 | 1.4433 | 1.1478 | 6.8056 | 2.76 | 0.0059 | Q = 212.13, df = 11, p < 0.0001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dos Santos, P.A.S.; Silva, M.J.A.; Gouveia, M.I.M.; Lima, L.N.G.C.; Quaresma, A.J.P.G.; De Lima, P.D.L.; Brasiliense, D.M.; Lima, K.V.B.; Rodrigues, Y.C. The Prevalence of Metallo-Beta-Lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: A Systematic Review and Meta-Analysis. Microorganisms 2023, 11, 2366. https://doi.org/10.3390/microorganisms11092366
Dos Santos PAS, Silva MJA, Gouveia MIM, Lima LNGC, Quaresma AJPG, De Lima PDL, Brasiliense DM, Lima KVB, Rodrigues YC. The Prevalence of Metallo-Beta-Lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: A Systematic Review and Meta-Analysis. Microorganisms. 2023; 11(9):2366. https://doi.org/10.3390/microorganisms11092366
Chicago/Turabian StyleDos Santos, Pabllo Antonny Silva, Marcos Jessé Abrahão Silva, Maria Isabel Montoril Gouveia, Luana Nepomuceno Gondim Costa Lima, Ana Judith Pires Garcia Quaresma, Patrícia Danielle Lima De Lima, Danielle Murici Brasiliense, Karla Valéria Batista Lima, and Yan Corrêa Rodrigues. 2023. "The Prevalence of Metallo-Beta-Lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: A Systematic Review and Meta-Analysis" Microorganisms 11, no. 9: 2366. https://doi.org/10.3390/microorganisms11092366
APA StyleDos Santos, P. A. S., Silva, M. J. A., Gouveia, M. I. M., Lima, L. N. G. C., Quaresma, A. J. P. G., De Lima, P. D. L., Brasiliense, D. M., Lima, K. V. B., & Rodrigues, Y. C. (2023). The Prevalence of Metallo-Beta-Lactamese-(MβL)-Producing Pseudomonas aeruginosa Isolates in Brazil: A Systematic Review and Meta-Analysis. Microorganisms, 11(9), 2366. https://doi.org/10.3390/microorganisms11092366









