Specific Oral Microbial Differences in Proteobacteria and Bacteroidetes Are Associated with Distinct Sites When Moving from Healthy Mucosa to Oral Dysplasia—A Microbiome and Gene Profiling Study and Focused Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Inclusion Criteria
2.3. Oral Swab Sample Collection
2.4. Tissue Sample Collection
2.5. DNA Extraction
2.6. 16S Amplification and Sequencing
2.7. Sequencing Data Processing
2.8. Operational Taxonomic Unit (OTU) Cluster and Taxonomic Annotation
2.9. Alpha Diversity
2.10. Beta Diversity
2.11. HSC-3 Cell Culture
2.12. Bacterial Culture
2.13. RNAseq
2.14. Statistical Analyses
3. Results
3.1. Demographics of Study Patients
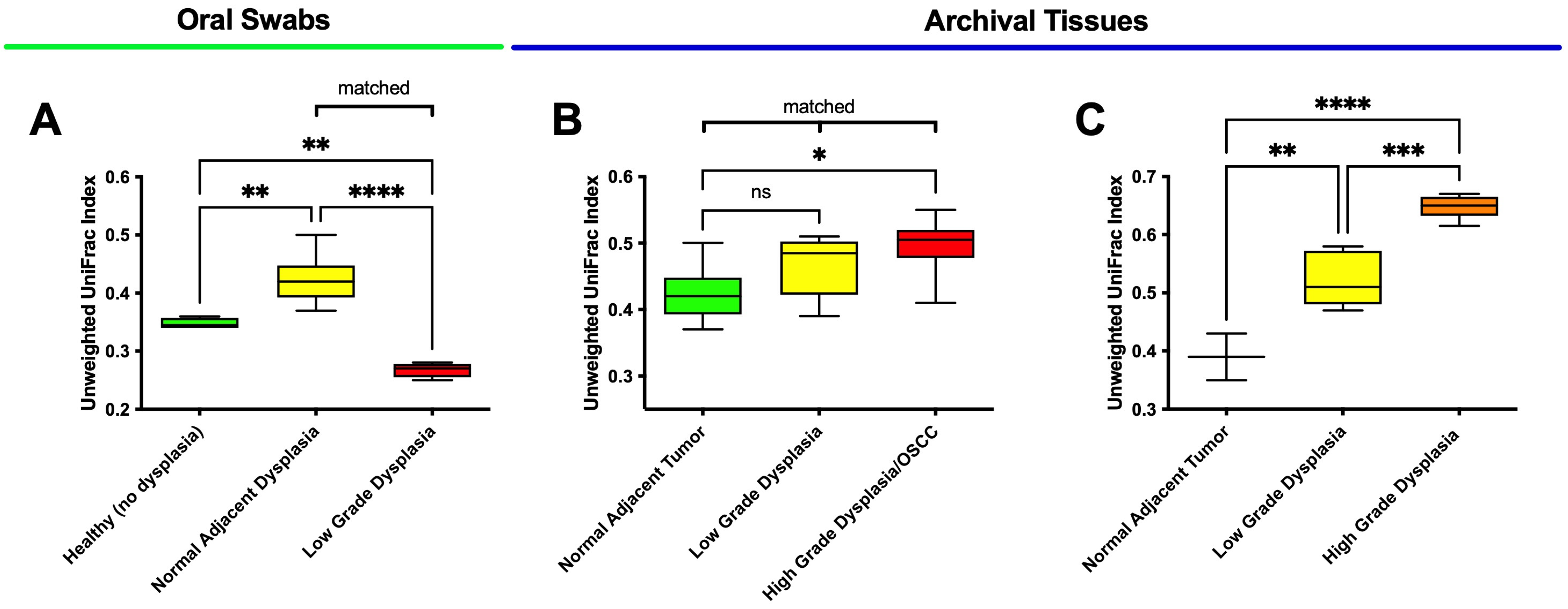
3.2. Oral Dysplasia and OSCC Microbiome Communities Are Distinct from Those in Healthy and Histologically Normal Adjacent Communities
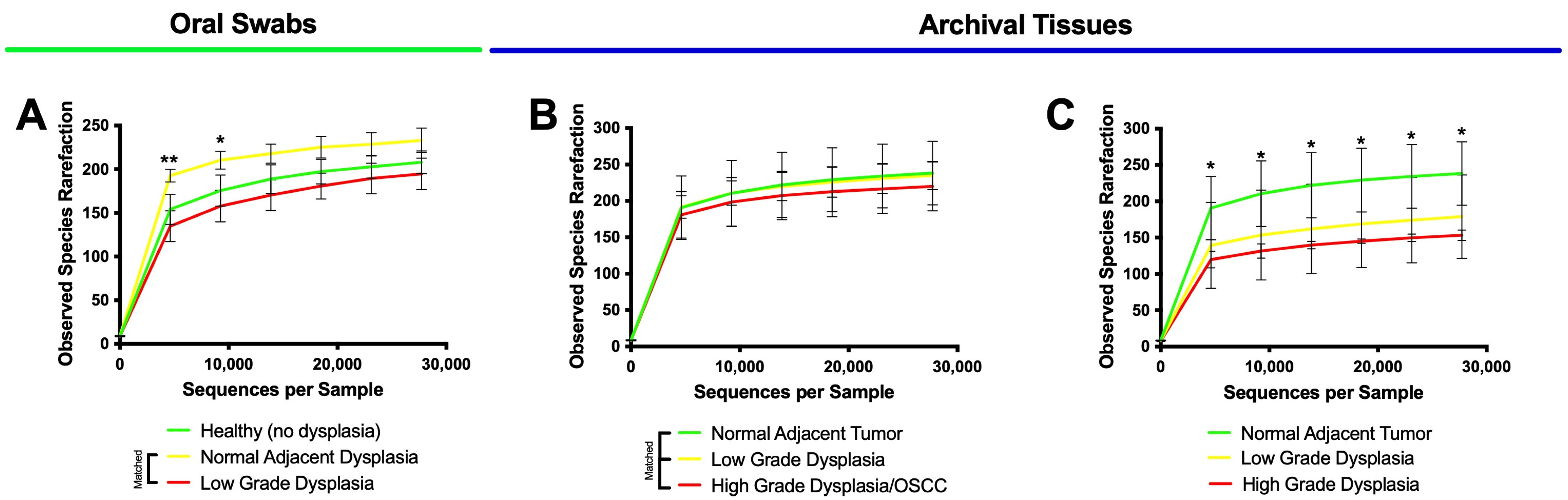
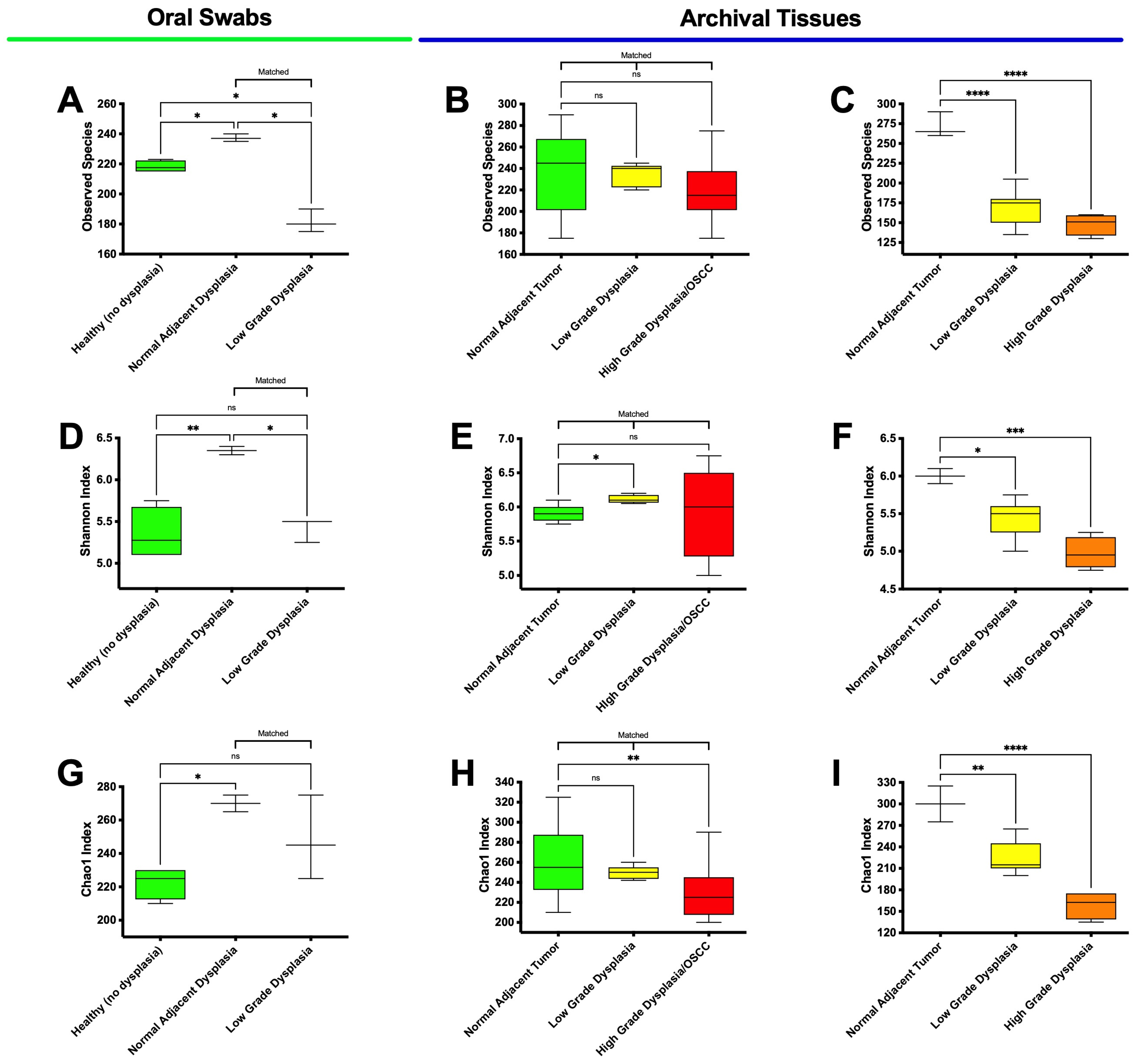
3.3. High-Grade Dysplasia and OSCC Alpha Diversities Are Significantly Different from Those in Histologically Normal Adjacent Specimens
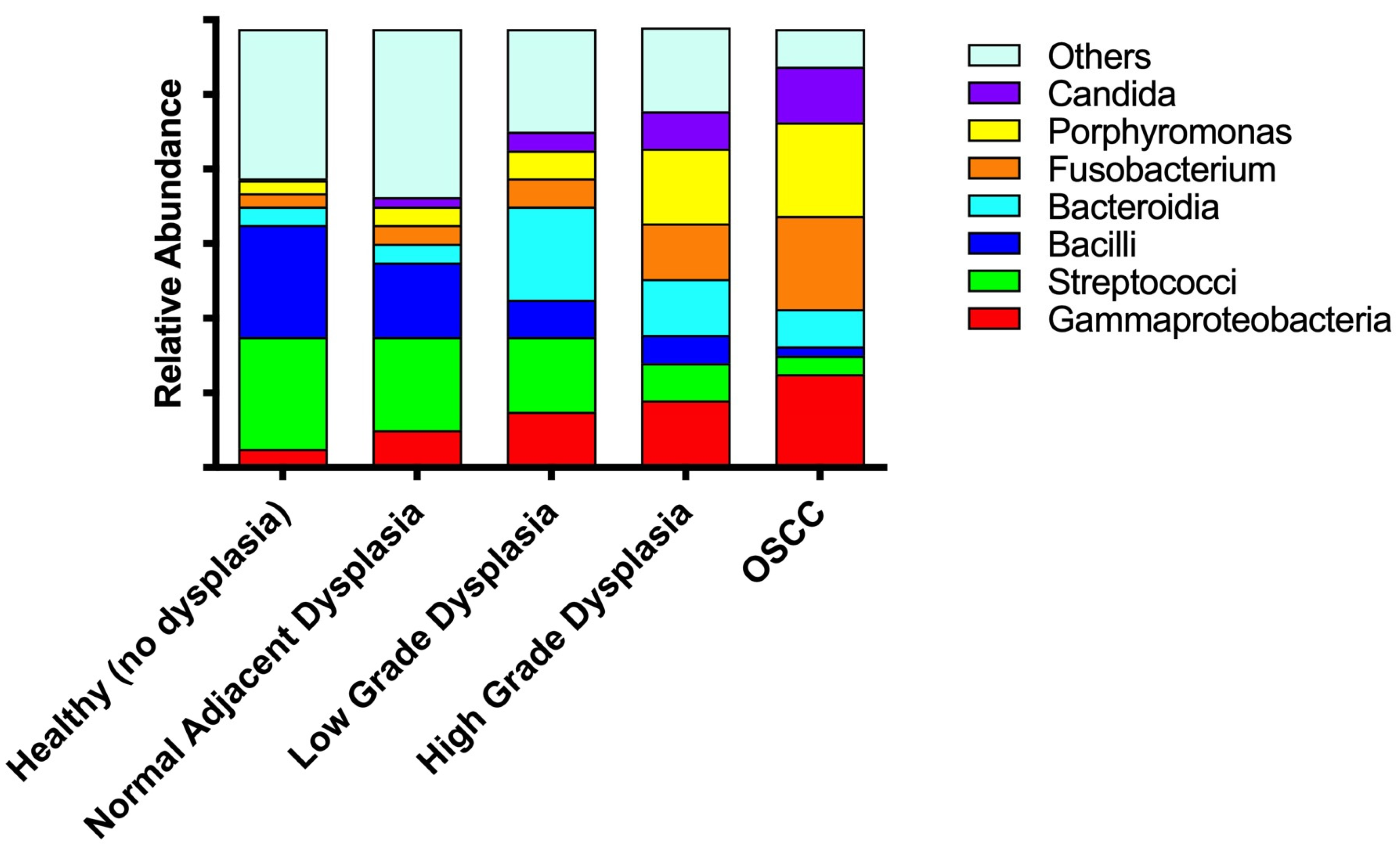
3.4. Significant Increases in Proteobacteria and Decreases in Firmicutes as well as Expansion of Fusobacteria Are Noted When Moving from the Clinically/Histologically Normal Oral Mucosa to Dysplasia and to Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Woo, S.-B. Oral Epithelial Dysplasia and Premalignancy. Head Neck Pathol. 2019, 13, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Pritzker, K.P.H.; Darling, M.R.; Hwang, J.T.-K.; Mock, D. Oral Potentially Malignant Disorders (OPMD): What Is the Clinical Utility of Dysplasia Grade? Expert Rev. Mol. Diagn. 2021, 21, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Reibel, J.; Gale, N.; Hille, J. Oral Potentially Malignant Disoders and Oral Epithelial Dysplasia. In WHO Classification of Head and Neck Tumours; IARC: Lyon, France, 2017; Volume 9, ISBN 978-92-832-2438-9. [Google Scholar]
- Tilakaratne, W.M.; Jayasooriya, P.R.; Jayasuriya, N.S.; De Silva, R.K. Oral Epithelial Dysplasia: Causes, Quantification, Prognosis, and Management Challenges. Periodontol. 2000 2019, 80, 126–147. [Google Scholar] [CrossRef] [PubMed]
- Wils, L.J.; Poell, J.B.; Evren, I.; Koopman, M.S.; Brouns, E.R.E.A.; De Visscher, J.G.A.M.; Brakenhoff, R.H.; Bloemena, E. Incorporation of Differentiated Dysplasia Improves Prediction of Oral Leukoplakia at Increased Risk of Malignant Progression. Mod. Pathol. 2020, 33, 1033–1040. [Google Scholar] [CrossRef]
- Wenig, B.M. Squamous Cell Carcinoma of the Upper Aerodigestive Tract: Dysplasia and Select Variants. Mod. Pathol. 2017, 30, S112–S128. [Google Scholar] [CrossRef]
- Speight, P.M. Update on Oral Epithelial Dysplasia and Progression to Cancer. Head Neck Pathol. 2007, 1, 61–66. [Google Scholar] [CrossRef]
- Iocca, O.; Sollecito, T.P.; Alawi, F.; Weinstein, G.S.; Newman, J.G.; De Virgilio, A.; Di Maio, P.; Spriano, G.; Pardiñas López, S.; Shanti, R.M. Potentially Malignant Disorders of the Oral Cavity and Oral Dysplasia: A Systematic Review and Meta-analysis of Malignant Transformation Rate by Subtype. Head Neck 2020, 42, 539–555. [Google Scholar] [CrossRef]
- Nevanpää, T.T.; Terävä, A.E.; Laine, H.K.; Rautava, J. Malignant Transformation of Oral Epithelial Dysplasia in Southwest Finland. Sci. Rep. 2022, 12, 8261. [Google Scholar] [CrossRef]
- Shubhasini, A.R.; Praveen, B.N.; Usha, H.; Uma, K.; Shubha, G.; Keerthi, G.; Shiladitya, S. Inter- and Intra-Observer Variability in Diagnosis of Oral Dysplasia. Asian Pac. J. Cancer Prev. 2017, 18, 3251–3254. [Google Scholar] [CrossRef]
- Smith, J.; Rattay, T.; McConkey, C.; Helliwell, T.; Mehanna, H. Biomarkers in Dysplasia of the Oral Cavity: A Systematic Review. Oral Oncol. 2009, 45, 647–653. [Google Scholar] [CrossRef]
- Radaic, A.; Ganther, S.; Kamarajan, P.; Grandis, J.; Yom, S.S.; Kapila, Y.L. Paradigm Shift in the Pathogenesis and Treatment of Oral Cancer and Other Cancers Focused on the Oralome and Antimicrobial-Based Therapeutics. Periodontol. 2000 2021, 87, 76–93. [Google Scholar] [CrossRef] [PubMed]
- Radaic, A.; Kapila, Y. The Oralome and Its Dysbiosis: New Insights into Oral Microbiome-Host Interactions. Comput. Struct. Biotechnol. J. 2021, 19, 1335–1360. [Google Scholar] [CrossRef] [PubMed]
- Van Dyke, T.E.; Bartold, P.M.; Reynolds, E.C. The Nexus between Periodontal Inflammation and Dysbiosis. Front. Immunol. 2020, 11, 511. [Google Scholar] [CrossRef]
- Sarkar, P.; Malik, S.; Laha, S.; Das, S.; Bunk, S.; Ray, J.G.; Chatterjee, R.; Saha, A. Dysbiosis of Oral Microbiota during Oral Squamous Cell Carcinoma Development. Front. Oncol. 2021, 11, 614448. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.M.; Luo, T.; Kamarajan, P.; Fenno, J.C.; Rickard, A.H.; Kapila, Y.L. Microbial Communities Associated with Primary and Metastatic Head and Neck Squamous Cell Carcinoma—A High Fusobacterial and Low Streptococcal Signature. Sci. Rep. 2017, 7, 9934. [Google Scholar] [CrossRef] [PubMed]
- Kamarajan, P.; Ateia, I.; Shin, J.M.; Fenno, J.C.; Le, C.; Zhan, L.; Chang, A.; Darveau, R.; Kapila, Y.L. Periodontal Pathogens Promote Cancer Aggressivity via TLR/MyD88 Triggered Activation of Integrin/FAK Signaling That Is Therapeutically Reversible by a Probiotic Bacteriocin. PLoS Pathog. 2020, 16, e1008881. [Google Scholar] [CrossRef]
- La Rosa, G.R.M.; Gattuso, G.; Pedullà, E.; Rapisarda, E.; Nicolosi, D.; Salmeri, M. Association of Oral Dysbiosis with Oral Cancer Development. Oncol. Lett. 2020, 19, 3045–3058. [Google Scholar] [CrossRef]
- Geng, F.; Zhang, Y.; Lu, Z.; Zhang, S.; Pan, Y. Fusobacterium nucleatum Caused DNA Damage and Promoted Cell Proliferation by the Ku70/P53 Pathway in Oral Cancer Cells. DNA Cell Biol. 2020, 39, 144–151. [Google Scholar] [CrossRef]
- Chang, C.; Geng, F.; Shi, X.; Li, Y.; Zhang, X.; Zhao, X.; Pan, Y. The Prevalence Rate of Periodontal Pathogens and Its Association with Oral Squamous Cell Carcinoma. Appl. Microbiol. Biotechnol. 2019, 103, 1393–1404. [Google Scholar] [CrossRef]
- Olsen, I.; Yilmaz, Ö. Possible Role of Porphyromonas Gingivalis in Orodigestive Cancers. J. Oral Microbiol. 2019, 11, 1563410. [Google Scholar] [CrossRef]
- Rai, A.K.; Panda, M.; Das, A.K.; Rahman, T.; Das, R.; Das, K.; Sarma, A.; Kataki, A.C.; Chattopadhyay, I. Dysbiosis of Salivary Microbiome and Cytokines Influence Oral Squamous Cell Carcinoma through Inflammation. Arch. Microbiol. 2021, 203, 137–152. [Google Scholar] [CrossRef]
- He, J.; Li, Y.; Cao, Y.; Xue, J.; Zhou, X. The Oral Microbiome Diversity and Its Relation to Human Diseases. Folia Microbiol. 2015, 60, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-L.; Wang, S.-S.; Wang, H.-F.; Tang, Y.-J.; Tang, Y.-L.; Liang, X.-H. Who Is Who in Oral Cancer? Exp. Cell Res. 2019, 384, 111634. [Google Scholar] [CrossRef] [PubMed]
- Costalonga, M.; Herzberg, M.C. The Oral Microbiome and the Immunobiology of Periodontal Disease and Caries. Immunol. Lett. 2014, 162, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, Y.; Zhu, J.; Xu, B. Oral Dysbiosis in the Onset and Carcinogenesis of Oral Epithelial Dysplasia: A Systematic Review. Arch. Oral Biol. 2023, 147, 105630. [Google Scholar] [CrossRef]
- Ganly, I.; Yang, L.; Giese, R.A.; Hao, Y.; Nossa, C.W.; Morris, L.G.T.; Rosenthal, M.; Migliacci, J.; Kelly, D.; Tseng, W.; et al. Periodontal Pathogens Are a Risk Factor of Oral Cavity Squamous Cell Carcinoma, Independent of Tobacco and Alcohol and Human Papillomavirus. Int. J. Cancer 2019, 145, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Hujoel, P.P.; Drangsholt, M.; Spiekerman, C.; Weiss, N.S. An Exploration of the Periodontitis-Cancer Association. Ann. Epidemiol. 2003, 13, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Nwizu, N.; Wactawski-Wende, J.; Genco, R.J. Periodontal Disease and Cancer: Epidemiologic Studies and Possible Mechanisms. Periodontol. 2000 2020, 83, 213–233. [Google Scholar] [CrossRef]
- Guerrero-Preston, R.; Godoy-Vitorino, F.; Jedlicka, A.; Rodríguez-Hilario, A.; González, H.; Bondy, J.; Lawson, F.; Folawiyo, O.; Michailidi, C.; Dziedzic, A.; et al. 16S RRNA Amplicon Sequencing Identifies Microbiota Associated with Oral Cancer, Human Papilloma Virus Infection and Surgical Treatment. Oncotarget 2016, 7, 51320–51334. [Google Scholar] [CrossRef]
- Yang, K.; Wang, Y.; Zhang, S.; Zhang, D.; Hu, L.; Zhao, T.; Zheng, H. Oral Microbiota Analysis of Tissue Pairs and Saliva Samples from Patients with Oral Squamous Cell Carcinoma–A Pilot Study. Front. Microbiol. 2021, 12, 719601. [Google Scholar] [CrossRef]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of Oral Microbiota in Tumor and Non-Tumor Tissues of Patients with Oral Squamous Cell Carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.-W.; Deng, W.-W.; Song, W.-F.; Wu, C.-C.; Liu, J.; Hong, S.; Zhuang, Z.-N.; Cheng, H.; Sun, Z.-J.; Zhang, X.-Z. Biomaterial-Mediated Modulation of Oral Microbiota Synergizes with PD-1 Blockade in Mice with Oral Squamous Cell Carcinoma. Nat. Biomed. Eng. 2022, 6, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hao, Y.; Peng, X.; Li, B.; Han, Q.; Ren, B.; Li, M.; Li, L.; Li, Y.; Cheng, G.; et al. The Clinical Potential of Oral Microbiota as a Screening Tool for Oral Squamous Cell Carcinomas. Front. Cell. Infect. Microbiol. 2021, 11, 728933. [Google Scholar] [CrossRef]
- Zhao, H.; Chu, M.; Huang, Z.; Yang, X.; Ran, S.; Hu, B.; Zhang, C.; Liang, J. Variations in Oral Microbiota Associated with Oral Cancer. Sci. Rep. 2017, 7, 11773. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zheng, H.J.; Zhang, C.P. The Oral Microbiota May Have Influence on Oral Cancer. Front. Cell. Infect. Microbiol. 2020, 9, 476. [Google Scholar] [CrossRef] [PubMed]
- Yost, S.; Stashenko, P.; Choi, Y.; Kukuruzinska, M.; Genco, C.A.; Salama, A.; Weinberg, E.O.; Kramer, C.D.; Frias-Lopez, J. Increased Virulence of the Oral Microbiome in Oral Squamous Cell Carcinoma Revealed by Metatranscriptome Analyses. Int. J. Oral Sci. 2018, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Panda, M.; Rai, A.K.; Rahman, T.; Das, A.; Das, R.; Sarma, A.; Kataki, A.C.; Chattopadhyay, I. Alterations of Salivary Microbial Community Associated with Oropharyngeal and Hypopharyngeal Squamous Cell Carcinoma Patients. Arch. Microbiol. 2020, 202, 785–805. [Google Scholar] [CrossRef]
- Amer, A.; Galvin, S.; Healy, C.M.; Moran, G.P. The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species. Front. Microbiol. 2017, 8, 2391. [Google Scholar] [CrossRef]
- Wang, H.; Funchain, P.; Bebek, G.; Altemus, J.; Zhang, H.; Niazi, F.; Peterson, C.; Lee, W.T.; Burkey, B.B.; Eng, C. Microbiomic Differences in Tumor and Paired-Normal Tissue in Head and Neck Squamous Cell Carcinomas. Genome Med. 2017, 9, 14. [Google Scholar] [CrossRef]
- Gopinath, D.; Kunnath Menon, R.; Chun Wie, C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; George Botelho, M.; et al. Salivary Bacterial Shifts in Oral Leukoplakia Resemble the Dysbiotic Oral Cancer Bacteriome. J. Oral Microbiol. 2020, 13, 1857998. [Google Scholar] [CrossRef]
- Al-Hebshi, N.N.; Borgnakke, W.S.; Johnson, N.W. The Microbiome of Oral Squamous Cell Carcinomas: A Functional Perspective. Curr. Oral Health Rep. 2019, 6, 145–160. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Q.; Hua, H.; Chen, F. Changes in the Salivary Microbiota of Oral Leukoplakia and Oral Cancer. Oral Oncol. 2016, 56, e6–e8. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-Filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S RRNA Sequence Formation and Detection in Sanger and 454-Pyrosequenced PCR Amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of RRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Willis, A.D. Rarefaction, Alpha Diversity, and Statistics. Front. Microbiol. 2019, 10, 2407. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to Analyze 16S RRNA Gene Sequences from Microbial Communities. Curr. Protoc. Microbiol. 2012, 27, 1E-5. [Google Scholar] [CrossRef] [PubMed]
- Koleff, P.; Gaston, K.J.; Lennon, J.J. Measuring Beta Diversity for Presence–Absence Data. J. Anim. Ecol. 2003, 72, 367–382. [Google Scholar] [CrossRef]
- Tuomisto, H. A Diversity of Beta Diversities: Straightening up a Concept Gone Awry. Part 1. Defining Beta Diversity as a Function of Alpha and Gamma Diversity. Ecography 2010, 33, 2–22. [Google Scholar] [CrossRef]
- Radaic, A.; Malone, E.; Kamarajan, P.; Kapila, Y.L. Solid Lipid Nanoparticles Loaded with Nisin (SLN-Nisin) Are More Effective Than Free Nisin as Antimicrobial, Antibiofilm, and Anticancer Agents. J. Biomed. Nanotechnol. 2022, 18, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Radaic, A.; Joo, N.E.; Jeong, S.-H.; Yoo, S.-I.; Kotov, N.; Kapila, Y.L. Phosphatidylserine-Gold Nanoparticles (PS-AuNP) Induce Prostate and Breast Cancer Cell Apoptosis. Pharmaceutics 2021, 13, 1094. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Kang, M.; Zhang, M.J.; Reza Sailani, M.; Kuraji, R.; Martinez, A.; Ye, C.; Kamarajan, P.; Le, C.; Zhan, L.; et al. Polymicrobial Periodontal Disease Triggers a Wide Radius of Effect and Unique Virome. NPJ Biofilms Microbiomes 2020, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Kuraji, R.; Zhang, M.J.; Martinez, A.; Radaic, A.; Kamarajan, P.; Le, C.; Zhan, L.; Ye, C.; Rangé, H.; et al. Nisin Probiotic Prevents Inflammatory Bone Loss While Promoting Reparative Proliferation and a Healthy Microbiome. NPJ Biofilms Microbiomes 2022, 8, 45. [Google Scholar] [CrossRef]
- Radaic, A.; Ye, C.; Parks, B.; Gao, L.; Kuraji, R.; Malone, E.; Kamarajan, P.; Zhan, L.; Kapila, Y.L. Modulation of Pathogenic Oral Biofilms towards Health with Nisin Probiotic. J. Oral Microbiol. 2020, 12, 1809302. [Google Scholar] [CrossRef]
- Radaic, A.; Brody, H.; Contreras, F.; Hajfathalian, M.; Lucido, L.; Kamarajan, P.; Kapila, Y.L. Nisin and Nisin Probiotic Disrupt Oral Pathogenic Biofilms and Restore Their Microbiome Composition towards Healthy Control Levels in a Peri-Implantitis Setting. Microorganisms 2022, 10, 1336. [Google Scholar] [CrossRef]
- De Abreu, P.M.; Có, A.C.G.; Azevedo, P.L.; Do Valle, I.B.; De Oliveira, K.G.; Gouvea, S.A.; Cordeiro-Silva, M.F.; Louro, I.D.; De Podestá, J.R.V.; Lenzi, J.; et al. Frequency of HPV in Oral Cavity Squamous Cell Carcinoma. BMC Cancer 2018, 18, 324. [Google Scholar] [CrossRef] [PubMed]
- Mirghani, H.; Amen, F.; Moreau, F.; Lacau St Guily, J. Do High-Risk Human Papillomaviruses Cause Oral Cavity Squamous Cell Carcinoma? Oral Oncol. 2015, 51, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Pietrobon, G.; Tagliabue, M.; Stringa, L.M.; De Berardinis, R.; Chu, F.; Zocchi, J.; Carlotto, E.; Chiocca, S.; Ansarin, M. Leukoplakia in the Oral Cavity and Oral Microbiota: A Comprehensive Review. Cancers 2021, 13, 4439. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, D.; Menon, R.K.; Wie, C.C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; Botelho, M.G.; et al. Differences in the Bacteriome of Swab, Saliva, and Tissue Biopsies in Oral Cancer. Sci. Rep. 2021, 11, 1181. [Google Scholar] [CrossRef]
- Villa, A.; Gohel, A. Oral Potentially Malignant Disorders in a Large Dental Population. J. Appl. Oral Sci. 2014, 22, 473–476. [Google Scholar] [CrossRef]
- Peng, R.T.; Sun, Y.; Zhou, X.D.; Liu, S.Y.; Han, Q.; Cheng, L.; Peng, X. Treponema denticola Promotes OSCC Development via the TGF-β Signaling Pathway. J. Dent. Res. 2022, 101, 704–713. [Google Scholar] [CrossRef]
- Jaber, M.A.; Elameen, E.M. Long-Term Follow-up of Oral Epithelial Dysplasia: A Hospital Based Cross-Sectional Study. J. Dent. Sci. 2021, 16, 304–310. [Google Scholar] [CrossRef]
- Krogh, P.; Hald, B.; Holmstrup, P. Possible Mycological Etiology of Oral Mucosal Cancer: Catalytic Potential of Infecting Candida aibicans and Other Yeasts in Production of N-Nitrosobenzylmethylamine. Carcinogenesis 1987, 8, 1543–1548. [Google Scholar] [CrossRef] [PubMed]
- Johnson, N.W.; Jayasekara, P.; Amarasinghe, A.A.H.K. Squamous Cell Carcinoma and Precursor Lesions of the Oral Cavity: Epidemiology and Aetiology. Periodontol. 2000 2011, 57, 19–37. [Google Scholar] [CrossRef]
- Wright, R.J.; Pewarchuk, M.E.; Marshall, E.A.; Murrary, B.; Rosin, M.P.; Laronde, D.M.; Zhang, L.; Lam, W.L.; Langille, M.G.I.; Rock, L.D. Exploring the Microbiome of Oral Epithelial Dysplasia as a Predictor of Malignant Progression. BMC Oral Health 2023, 23, 206. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Pomares, A.; Hervás, D.; Bagan-Debon, L.; Proaño, A.; Garcia, D.; Sandoval, J.; Bagan, J. Oral Cancers Preceded by Proliferative Verrucous Leukoplakia Exhibit Distinctive Molecular Features. Oral Dis. 2023. [CrossRef] [PubMed]
- Sami, A.; Elimairi, I.; Stanton, C.; Ross, R.P.; Ryan, C.A. The Role of the Microbiome in Oral Squamous Cell Carcinoma with Insight into the Microbiome–Treatment Axis. Int. J. Mol. Sci. 2020, 21, 8061. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-H.; Chen, H.-M.; Yang, S.-F.; Liang, C.; Peng, C.-Y.; Lin, F.-M.; Tsai, L.-L.; Wu, B.-C.; Hsin, C.-H.; Chuang, C.-Y.; et al. Bacterial Alterations in Salivary Microbiota and Their Association in Oral Cancer. Sci. Rep. 2017, 7, 16540. [Google Scholar] [CrossRef] [PubMed]
- Mok, S.F.; Karuthan, C.; Cheah, Y.K.; Ngeow, W.C.; Rosnah, Z.; Yap, S.F.; Ong, H.K.A. The Oral Microbiome Community Variations Associated with Normal, Potentially Malignant Disorders and Malignant Lesions of the Oral Cavity. Malays. J. Pathol. 2017, 39, 1–15. [Google Scholar] [PubMed]
- Hebbar, P.B.; Pai, A. Mycological and Histological Associations of Candida in Oral Mucosal Lesions. J. Oral Sci. 2013, 55, 157–160. [Google Scholar] [CrossRef][Green Version]
- Spolidorio, L.C.; Martins, V.R.G.; Nogueira, R.D.; Spolidorio, D.M.P. Freqüência de Candida Sp. Em Biópsias de Lesões Da Mucosa Bucal. Pesqui. Odontol. Bras. 2003, 17, 89–93. [Google Scholar] [CrossRef]
- McCullough, M.; Jaber, M.; Barrett, A.W.; Bain, L.; Speight, P.M.; Porter, S.R. Oral Yeast Carriage Correlates with Presence of Oral Epithelial Dysplasia. Oral Oncol. 2002, 38, 391–393. [Google Scholar] [CrossRef]
- Barrett, A.; Kingsmill, V.; Speight, P. The Frequency of Fungal Infection in Biopsies of Oral Mucosal Lesions. Oral Dis. 2008, 4, 26–31. [Google Scholar] [CrossRef]
- Rindum, J.L.; Stenderup, A.; Holmstrup, P. Identification of Candida Albicans Types Related to Healthy and Pathological Oral Mucosa. J. Oral Pathol. Med. 1994, 23, 406–412. [Google Scholar] [CrossRef]
- Conway, C.; Graham, J.L.; Chengot, P.; Daly, C.; Chalkley, R.; Ross, L.; Droop, A.; Rabbitts, P.; Stead, L.F. Elucidating Drivers of Oral Epithelial Dysplasia Formation and Malignant Transformation to Cancer Using RNAseq. Oncotarget 2015, 6, 40186–40201. [Google Scholar] [CrossRef]
- Abdalla, Z.; Walsh, T.; Thakker, N.; Ward, C.M. Loss of Epithelial Markers Is an Early Event in Oral Dysplasia and Is Observed within the Safety Margin of Dysplastic and T1 OSCC Biopsies. PLoS ONE 2017, 12, e0187449. [Google Scholar] [CrossRef]
- Babji, D.V.; Kale, A.D.; Hallikerimath, S.R.; Kotrashetti, V.S. Histomorphometric Study to Compare Histological Changes Between Oral Squamous Cell Carcinoma and Apparently Normal Adjacent Oral Mucosa. Indian J. Otolaryngol. Head Neck Surg. 2015, 67, 21–28. [Google Scholar] [CrossRef]
- Han, X.Y.; Hong, T.; Falsen, E. Neisseria Bacilliformis Sp. Nov. Isolated from Human Infections. J. Clin. Microbiol. 2006, 44, 474–479. [Google Scholar] [CrossRef]
- Fang, J.; Long, L.; Maeda, H. Enhancement of Tumor-Targeted Delivery of Bacteria with Nitroglycerin Involving Augmentation of the EPR Effect. Methods Mol. Biol. 2016, 1409, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Gaonkar, P.P.; Patankar, S.R.; Tripathi, N.; Sridharan, G. Oral Bacterial Flora and Oral Cancer: The Possible Link? J. Oral Maxillofac. Pathol. 2018, 22, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.L. Bacteria and Cancer: Cause, Coincidence or Cure? A Review. J. Transl. Med. 2006, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Groeger, S.; Meyle, J. Oral Mucosal Epithelial Cells. Front. Immunol. 2019, 10, 208. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, K.; Kavitha, L. Oral Epithelial Dysplasia: Classifications and Clinical Relevance in Risk Assessment of Oral Potentially Malignant Disorders. J. Oral Maxillofac. Pathol. 2019, 23, 19. [Google Scholar] [CrossRef]
- Sarode, S.C.; Sarode, G.S.; Tupkari, J.V. Oral Potentially Malignant Disorders: A Proposal for Terminology and Definition with Review of Literature. J. Oral Maxillofac. Pathol. 2014, 18, S77–S80. [Google Scholar] [CrossRef]
- Warnakulasuriya, S.; Johnson, N.W.; Van Der Waal, I. Nomenclature and Classification of Potentially Malignant Disorders of the Oral Mucosa: Potentially Malignant Disorders. J. Oral Pathol. Med. 2007, 36, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, Y.; Yang, X.; Li, C.; Song, Z. The Oral Microbiota: Community Composition, Influencing Factors, Pathogenesis, and Interventions. Front. Microbiol. 2022, 13, 895537. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Li, H.; Ni, C.; Du, Z.; Yan, F. Human Oral Microbiota and Its Modulation for Oral Health. Biomed. Pharmacother. 2018, 99, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Dabney, J.; Meyer, M.; Pääbo, S. Ancient DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012567. [Google Scholar] [CrossRef]
- Berrino, E.; Annaratone, L.; Miglio, U.; Maldi, E.; Piccinelli, C.; Peano, E.; Balmativola, D.; Cassoni, P.; Pisacane, A.; Sarotto, I.; et al. Cold Formalin Fixation Guarantees DNA Integrity in Formalin Fixed Paraffin Embedded Tissues: Premises for a Better Quality of Diagnostic and Experimental Pathology with a Specific Impact on Breast Cancer. Front. Oncol. 2020, 10, 173. [Google Scholar] [CrossRef]
- Love, C.J.; Gubert, C.; Kodikara, S.; Kong, G.; Lê Cao, K.-A.; Hannan, A.J. Microbiota DNA Isolation, 16S RRNA Amplicon Sequencing, and Bioinformatic Analysis for Bacterial Microbiome Profiling of Rodent Fecal Samples. STAR Protoc. 2022, 3, 101772. [Google Scholar] [CrossRef]





| Variable | Healthy Mucosa (No Dysplasia) | Histologically Normal Adjacent/Low Grade Dysplasia (Matched) |
|---|---|---|
| Age (Years ± SD) | 50.67 ± 13.58 | 67.50 ± 9.98 |
| Sex | ||
| Male | 2 | 1 |
| Female | 2 | 3 |
| Swab Collection Location | ||
| Left Tongue | 0 | 1 |
| Right Lingual Gingiva | 0 | 0 |
| Left Lingual Gingiva | 0 | 1 |
| Mandibular Gingiva | 4 | 2 |
| Dysplasia diagnosis | - | Mild to moderate |
| Smoking Status | ||
| Current | 0 | 0 |
| Past | 0 | 0 |
| Never | 4 | 2 |
| N/A | 0 | 2 |
| Year | Site | Diagnosis | Age | Sex | HPV Status | Smoking Status | Alcohol Use | Other Relevant History | |
|---|---|---|---|---|---|---|---|---|---|
| Unmatched Tissue Samples | 2015 | Left posterior floor of mouth | Mild dysplasia | 60 | M | Negative | Never smoker | One standard drink per week | History of dysplasia and squamous cell carcinoma in the left posterolateral tongue |
| 2010 | Gingiva, between 1st and 2nd premolars | Squamous cell carcinoma in situ | 82 | F | Negative | n/a | n/a | History of squamous cell carcinoma in Gingiva, #7–9 | |
| 2013 | Left tongue, posterior dorsal mucosal margin | Histologically normal adjacent tumor | 76 | M | Negative | Never smoker | 2 standard drinks per week | Negative margin in a patient with left tongue squamous cell carcinoma | |
| 2005 | Left lateral tongue | Low-grade dysplasia | 33 | F | Negative | Never smoker | None | ||
| 2005 | Left lateral tongue | High-grade dysplasia | 33 | ||||||
| 2012 | Left lateral tongue | Mild dysplasia | 60 | M | Negative | Former smoker (0.25 packs per day, 1.5 pack-year) | 12 standard drinks per week | History of left tongue squamous cell carcinoma in situ | |
| 2015 | Left soft palate | Moderate dysplasia | 63 | ||||||
| 2012 | Right base of tongue mucosal margin | Carcinoma in situ | 54 | F | Negative | Former smoker (1 pack-year, 15 pack-years, quit 25 years prior) | <1 standard drink per day | Margin in a patient with oral tongue multifocal squamous cell carcinoma | |
| 2009 | Right tongue | Moderate dysplasia | 53 | M | Negative | n/a | n/a | History of right lateral tongue squamous cell carcinoma and HIV+ | |
| 2017 | Right tongue | Severe dysplasia | 60 | M | Negative | Daily tobacco chew user for 22 years | Longtime drinker | Adjacent to right tongue squamous cell carcinoma in resection specimen | |
| 2013 | Gingiva, lower left 2nd premolar region | Atypical papillary verrucous proliferation | 70 | F | Negative | Former smoker (1 packs per day, 24 pack-years, quit 26 years prior) | None | History of proliferative verrucous leukoplakia | |
| 2015 | Right ventral lateral tongue | Moderate dysplasia | 95 | F | Negative | Former smoker (0.25 packs per day, 10 pack-years, quit 35 years prior) | None | History of right tongue leukoplakia | |
| 2014 | Left tongue | Histologically normal adjacent tumor | 66 | F | Negative | Never smoker | None | Adjacent to left tongue squamous cell carcinoma in resection specimen | |
| Matched Samples | 2011 | Anterior dorsal tongue | Histologically normal adjacent (margin in resection) | 60 | M | Negative | Former smoker (2 packs per day, 68 pack-years) | ≥15 standard drinks per week | History of other recreational drug use |
| Mild to moderate dysplasia (margin in resection) | |||||||||
| Moderately differentiated SCC | |||||||||
| 2011 | Left posterior ventral tongue | Histologically normal adjacent (margin in resection) | 45 | M | Negative | Never smoker | One standard drink per week | History of non-Hodgkin lymphoma, esophageal squamous cell carcinoma, and thyroid cancer | |
| Mild to moderate dysplasia (margin in resection) | |||||||||
| Moderately differentiated SCC | |||||||||
| 2009 | Anterior dorsal tongue | Hyperkeratosis, no dysplasia (margin in SCC resection) | 41 | M | Negative | n/a | n/a | History of proliferative verrucous leukoplakia | |
| Mild dysplasia (adjacent to SCC) | |||||||||
| Moderately differentiated SCC | |||||||||
| 2009 | Ventral tongue, anterior margin | Histologically normal adjacent (margin in resection) | 57 | M | Negative | Former smoker (60 pack-years) | One standard drink per day | ||
| Moderate dysplasia (margin in SCC resection) | |||||||||
| Moderately differentiated SCC | |||||||||
| 1999 | Left lateral tongue | Histologically normal adjacent (to SCCIS) | 76 | M | Negative | Former smoker (25 years prior) | <2 standard drink per day | ||
| Squamous cell carcinoma in situ with possible superficial microinvasion | |||||||||
| 2010 | Left lateral tongue | Histologically normal adjacent (to SCCIS) | 77 | M | Negative | None | None | ||
| Mild to moderate dysplasia | |||||||||
| Squamous cell carcinoma in situ |
| Author/Year | Specimen Type | Method | Sample Size | Group Comparison | Microbiome | Other Remarks |
|---|---|---|---|---|---|---|
| Shen et al. (2023) [26] | - | Systematic Review and Metanalysis | 802 | - | Dysplasia—increase in phylum Bacteroidetes Dysplasia and OSCC—increase in Fusobacterium and decrease of Streptococcus | Analyzed studies presented a high risk of bias due to non-negligible heterogeneity in the type and size of the sample and inconsistent oral microbiome composition, strongly limiting the analysis. However, only 6 out of 11 analyzed studies histopathologically diagnosed their OPMDs as dysplasia, which could account for the discrepancies found. |
| Wright et al. (2023) [72] | Oral Swabs | 16S Sequencing | 90 | Progressing vs. non-progressing dysplasia | Increase in Campylobacter in progressing dysplasia compared to non-progressing ones | No significant differences between progressing vs. non-progressing dysplasia |
| Herreros-Pomares et al. (2021) [73] | Tissue | 16S Sequencing | 10 | Healthy vs. Leukoplakia + Dysplasia | Leukoplakia + dysplasia—increase in Oribacterium sp. oral taxon 108, Campylobacter jejuni, uncultured Eubacterium sp., Tannerella, and Porphyromonas | Authors have not controlled for dysplasia and mixed no dysplasia samples with mild, moderate, and severe dysplasia samples. |
| Sami et al. (2020) [74] | - | Reveiw | - | - | Fusobacterium nucleatum and Candida species has been associated with high-grade dysplasia and its severity | - |
| Gopinath et al. (2020) [41] | Whole Mouth Fluid | 16S Sequencing | 74 | Healthy vs. Leukoplakia + Dysplasia vs. OSCC | Leukoplakia + dysplasia—increase in Bacteroidetes and decrease in Firmicutes Leuloplakia + dysplasia and OSCC—increase in Actinobacteria | Shift in bacterial communities of leukoplakia + dysplasia and oral cancer patients; no significant difference in richness and diversity |
| Al-Hebshi et al. (2019) [42] | - | Review | - | - | Significantly increased frequency and yeast colony counts (predominantly Candida) in dysplasia and OSCC; significant yeast colony increase correlated with dysplasia severity | Some of the studies did not have healthy controls, rather compared to other OPMD |
| Ganly et al. (2019) [27] | Saliva | 16S Sequencing | 38 | Healthy vs. Leukoplakia + Dysplasia vs. OSCC | Leukoplakia + dysplasia—increase in Fusobacterium and Veillonella OSCC—increase in Fusobacterium, Prevotella, Alloprevotella; decrease in Streptococcus | Significantly increase in HSP90 gene and ligands for TLRs 1, 2, and 4 along the healthy → leukoplakia/dysplasia → OSCC progression |
| Lee et al. (2017) [75] | Saliva | 16S sequencing | 376 | Healthy vs. Dysplasia vs. OSCC | Significantly different levels of Bacillus, Enterococcus, Parvimonas, Peptostreptococcus, and Slackia in dysplasia compared to cancer | - |
| Mok et al. (2017) [76] | Oral Swabs | 16S Sequencing | 27 | Healthy vs. Dysplasia vs. OSCC | Dysplasia—increase in Neisseria and Granulicatella; decrease in Streptococcus | Analysis of Molecular Variance (AMOVA) showed no significant difference between dysplasia and other groups |
| Amer et al. (2017) [39] | Oral Swabs | 16S Sequencing | 6 | Healthy vs. Leukoplakia + Dysplasia | Severe dysplasia was associated with elevated levels of Leptotrichia spp. and Campylobacter concisus | - |
| Hebbar et al. (2013) [77] | Tissue and Oral Rinse | Periodic Acid–Schiff Staining | 50 | Healthy vs. Dysplasia vs. OSCC | Significant increase in yeast infection and colony number with higher dysplasia grades and OSCC | - |
| Spolidorio et al. (2003) [78] | Tissue | Periodic Acid–Schiff Staining | 832 | Healthy vs. Dysplasia | 27.2% of dysplasia samples were PAS-positive; significant association of yeast infection and dysplasia | Tongue was the significant most affected site by yeast infection |
| McCullough et al. (2002) [79] | Tissue and Oral Rinse | 223 | Healthy vs. Dysplasia vs. OSCC | Dysplasia and OSCC—significantly higher frequency of oral yeast carriage and higher number of yeast (>1000 cfu/mL) than control. Correlation between dysplasia degree and yeast amount in oral cavity | ||
| Barrett et al. (1998) [80] | Tissue | Periodic Acid–Schiff Staining | 4724 | Healthy vs. Dysplasia | 4.7% of the biopsies contained PAS-positive fungi; significant positive association of fungal infection with moderate and severe dysplasia | Significantly higher number of males infected compared to females; 21.9% of fungi-infected dysplasia worsened in histological severity, compared to 7.6% of non-infected dysplasia |
| Rindum et al. (1994) [81] | Tissue | Periodic Acid–Schiff Staining and Smear Culture | 153 | Healthy vs. Leukoplakia/erythroleukoplakia + Dysplasia | 4 Candida albicans strains were found in moderate and severe dysplasia, but none on mild dysplasia | - |
| Krogh et al. (1987) [70] | Swab over biopsy | Yeast culture | 12 | Healthy vs. leukoplakia/erythroleukoplakia +Dysplasia | 4 different strains of Candida albicans, one strain of Candida parapsilosis found in dysplasia samples | Samples positive for dysplasia showed yeast strains with lower nitrosation potential compared to the ones on samples negative for dysplasia (no statistical analysis) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radaic, A.; Shamir, E.R.; Jones, K.; Villa, A.; Garud, N.R.; Tward, A.D.; Kamarajan, P.; Kapila, Y.L. Specific Oral Microbial Differences in Proteobacteria and Bacteroidetes Are Associated with Distinct Sites When Moving from Healthy Mucosa to Oral Dysplasia—A Microbiome and Gene Profiling Study and Focused Review. Microorganisms 2023, 11, 2250. https://doi.org/10.3390/microorganisms11092250
Radaic A, Shamir ER, Jones K, Villa A, Garud NR, Tward AD, Kamarajan P, Kapila YL. Specific Oral Microbial Differences in Proteobacteria and Bacteroidetes Are Associated with Distinct Sites When Moving from Healthy Mucosa to Oral Dysplasia—A Microbiome and Gene Profiling Study and Focused Review. Microorganisms. 2023; 11(9):2250. https://doi.org/10.3390/microorganisms11092250
Chicago/Turabian StyleRadaic, Allan, Eliah R. Shamir, Kyle Jones, Alessandro Villa, Nandita R. Garud, Aaron D. Tward, Pachiyappan Kamarajan, and Yvonne L. Kapila. 2023. "Specific Oral Microbial Differences in Proteobacteria and Bacteroidetes Are Associated with Distinct Sites When Moving from Healthy Mucosa to Oral Dysplasia—A Microbiome and Gene Profiling Study and Focused Review" Microorganisms 11, no. 9: 2250. https://doi.org/10.3390/microorganisms11092250
APA StyleRadaic, A., Shamir, E. R., Jones, K., Villa, A., Garud, N. R., Tward, A. D., Kamarajan, P., & Kapila, Y. L. (2023). Specific Oral Microbial Differences in Proteobacteria and Bacteroidetes Are Associated with Distinct Sites When Moving from Healthy Mucosa to Oral Dysplasia—A Microbiome and Gene Profiling Study and Focused Review. Microorganisms, 11(9), 2250. https://doi.org/10.3390/microorganisms11092250






