Bibliometric Analysis of Global Trends in Research on Seasonal Variations in Gut Microbiota from 2012 to 2022
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Statistics and Analysis
3. Results
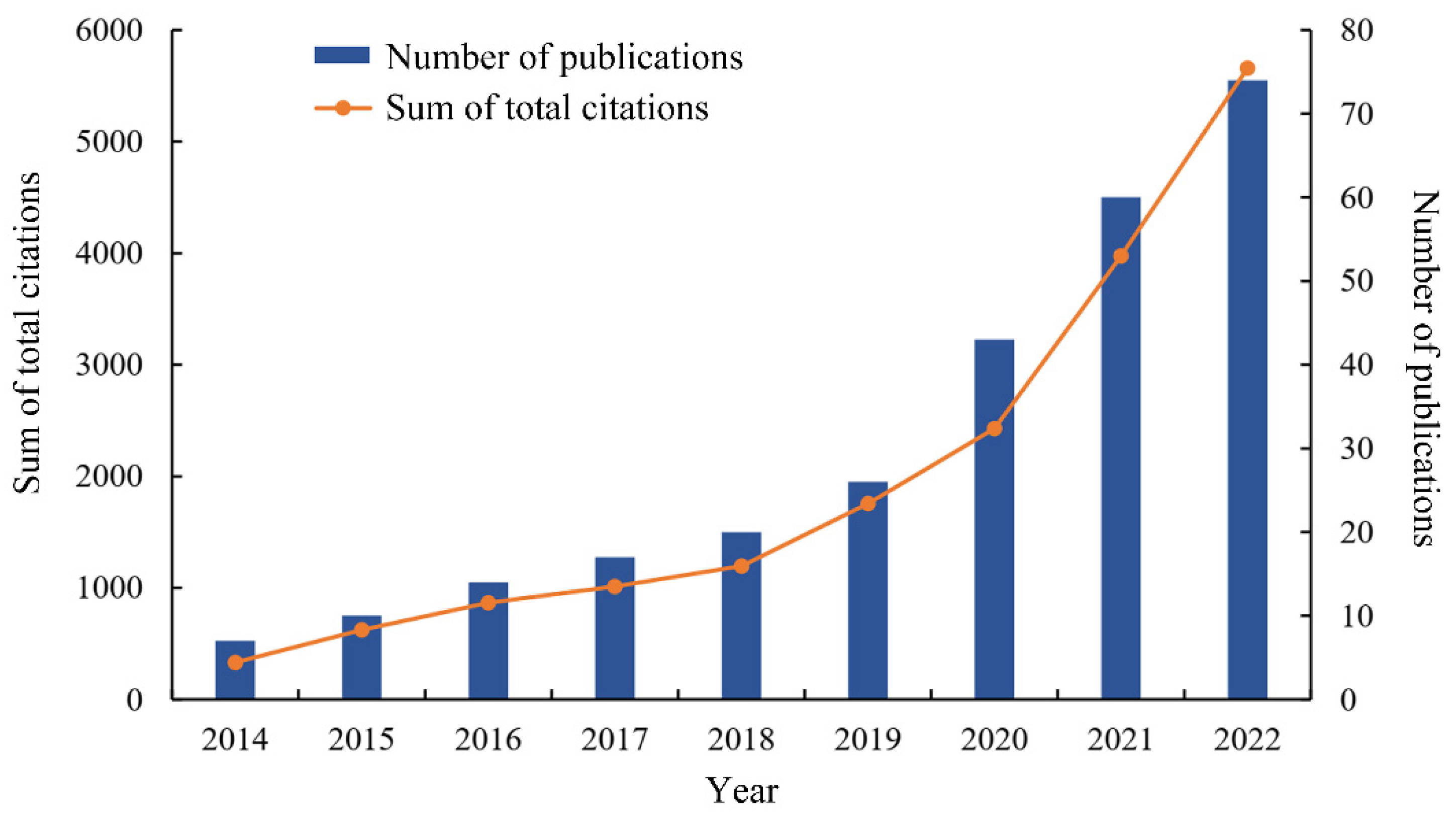
3.1. Temporal Trends of Publications and Citations
3.2. Countries and Institutions Analysis
3.3. Journal and Author Analysis
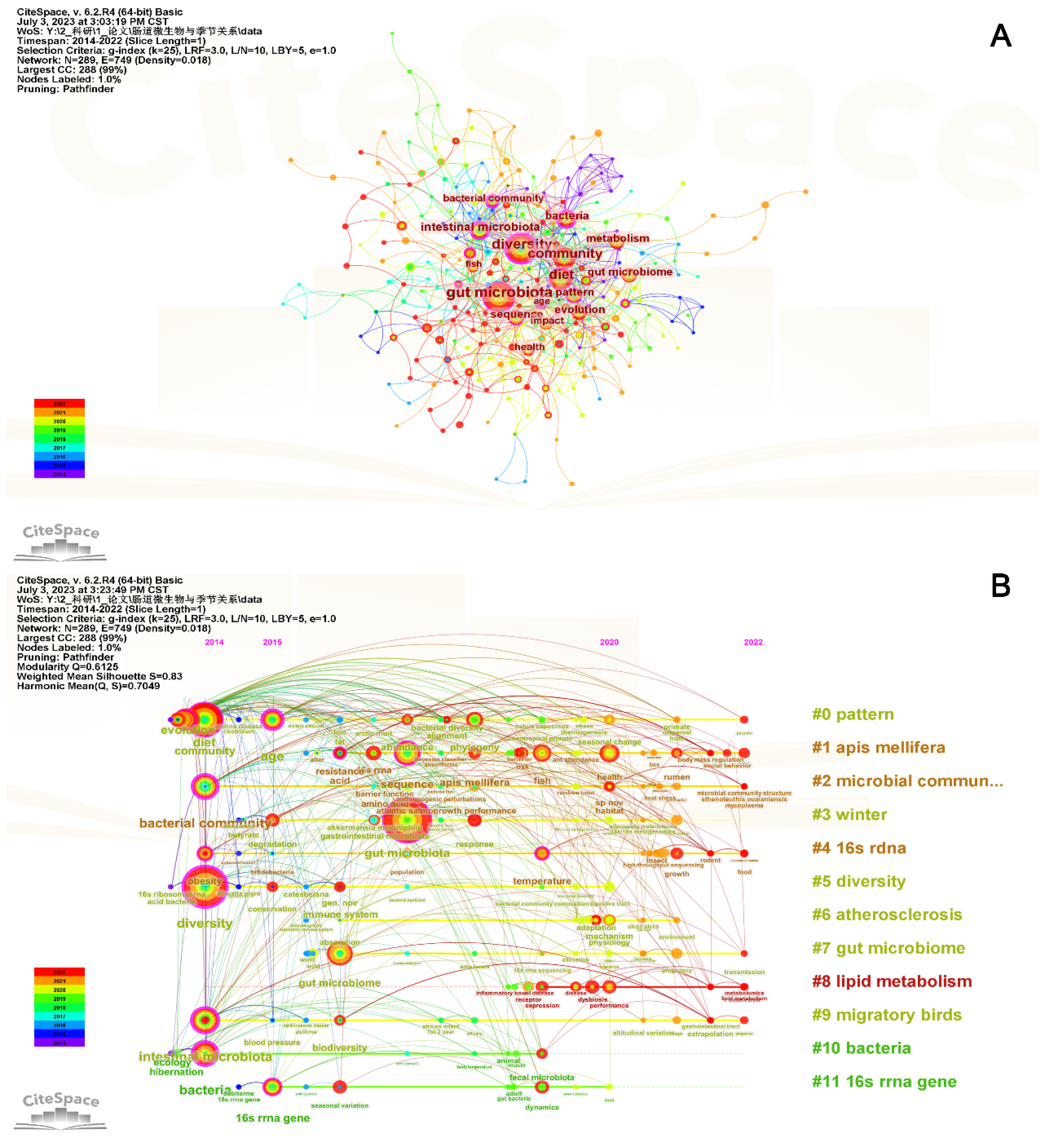
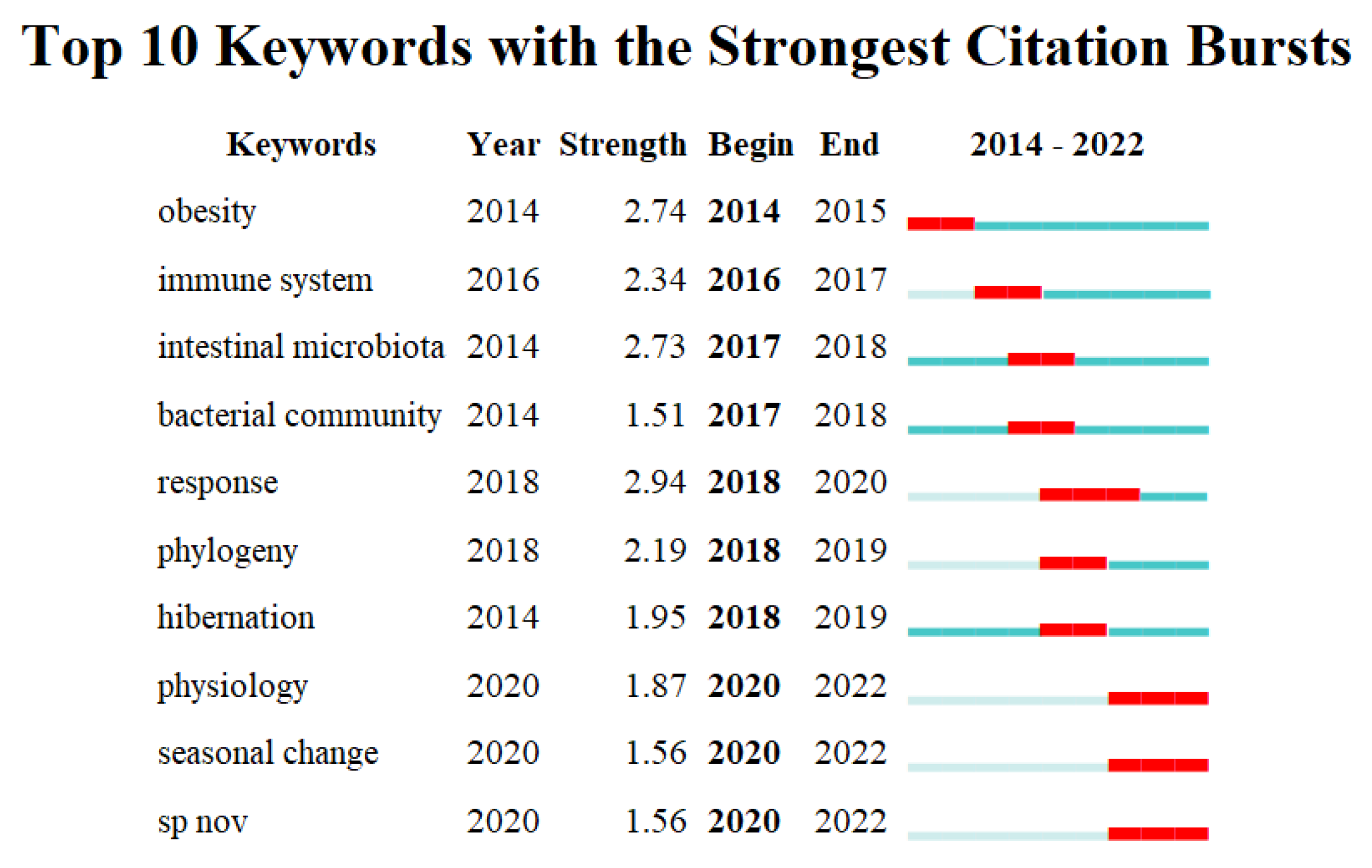
3.4. Keyword Analysis
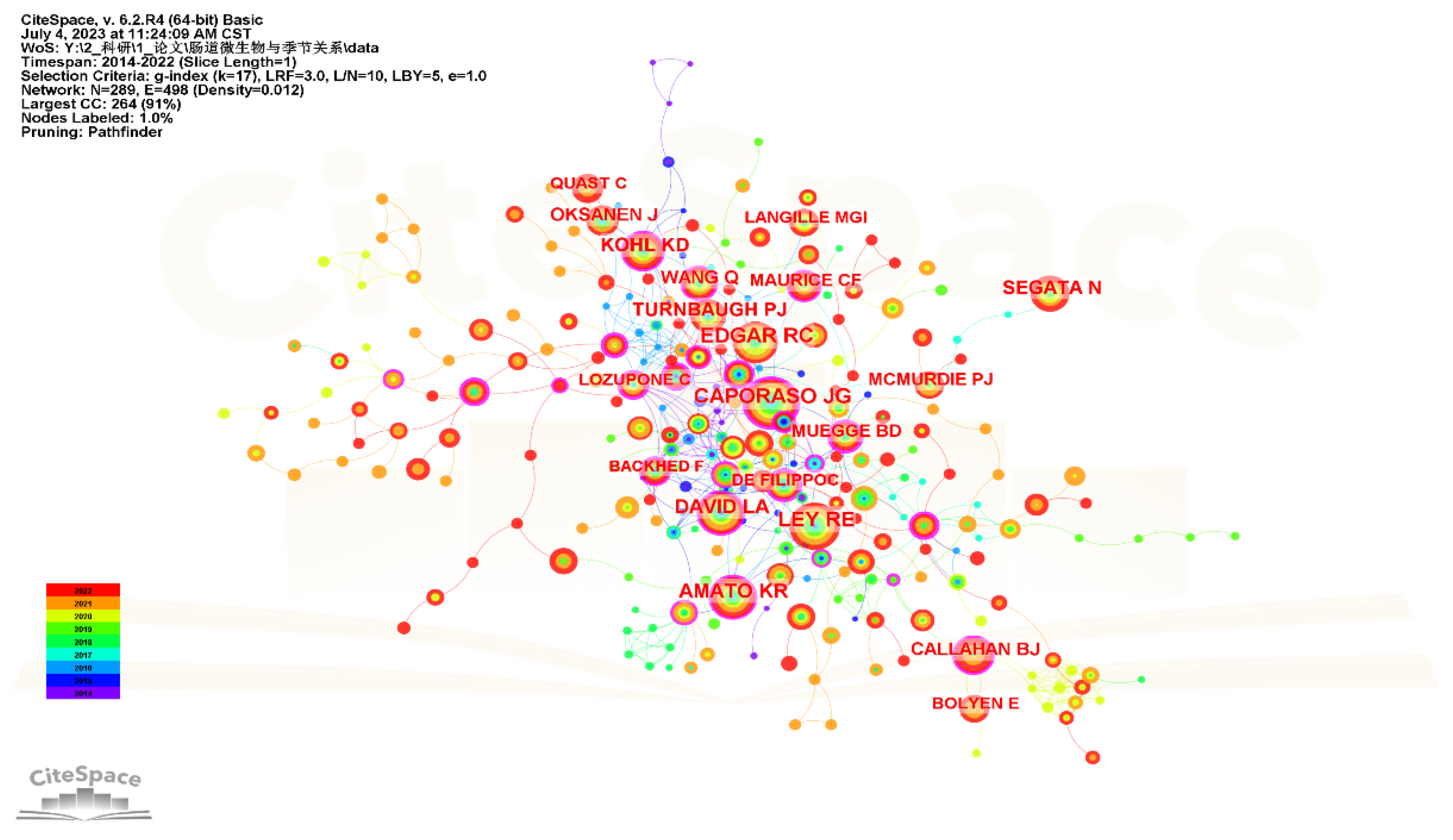
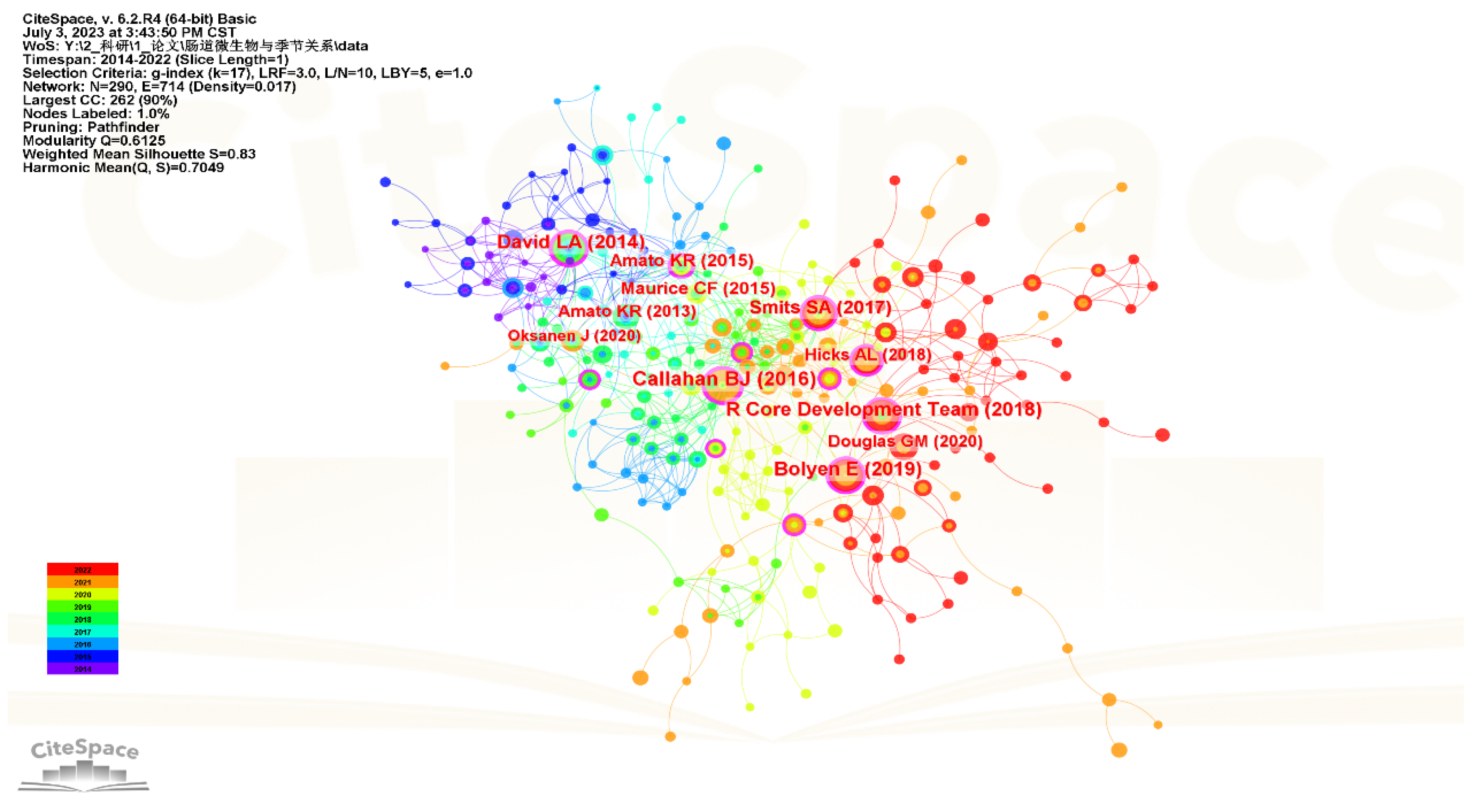
3.5. Analysis of Cited Authors and Co-Cited References
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Hao, Y.; Liu, X.; Yu, S.; Zhang, W.; Yang, S.; Yu, Z.; Ren, F. Gut microbiota from end-stage renal disease patients disrupt gut barrier function by excessive production of phenol. J. Genet. Genom. 2019, 46, 409–412. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut Microbiota and Immune System Interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef] [PubMed]
- Priadko, K.; Romano, L.; Olivieri, S.; Romeo, M.; Barone, B.; Sciorio, C.; Spirito, L.; Morelli, M.; Crocetto, F.; Arcaniolo, D.; et al. Intestinal microbiota, intestinal permeability and the urogenital tract: Is there a pathophysiological link? J. Physiol. Pharmacol. 2022, 73, 575–585. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- O’Toole, P.W.; Jeffery, I.B. Gut microbiota and aging. Science 2015, 350, 1214–1215. [Google Scholar] [CrossRef]
- Rojas, C.A.; Ramírez-Barahona, S.; Holekamp, K.E.; Theis, K.R. Host phylogeny and host ecology structure the mammalian gut microbiota at different taxonomic scales. Anim. Microbiome 2021, 3, 33. [Google Scholar] [CrossRef]
- Youngblut, N.D.; Reischer, G.H.; Walters, W.; Schuster, N.; Walzer, C.; Stalder, G.; Ley, R.E.; Farnleitner, A.H. Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat. Commun. 2019, 10, 2200. [Google Scholar] [CrossRef]
- Liu, P.Y.; Cheng, A.C.; Huang, S.W.; Chang, H.W.; Oshida, T.; Yu, H.T. Variations in Gut Microbiota of Siberian Flying Squirrels Correspond to Seasonal Phenological Changes in Their Hokkaido Subarctic Forest Ecosystem. Microb. Ecol. 2019, 78, 223–231. [Google Scholar] [CrossRef]
- Baniel, A.; Amato, K.R.; Beehner, J.C.; Bergman, T.J.; Mercer, A.; Perlman, R.F.; Petrullo, L.; Reitsema, L.; Sams, S.; Lu, A.; et al. Seasonal shifts in the gut microbiome indicate plastic responses to diet in wild geladas. Microbiome 2021, 9, 26. [Google Scholar] [CrossRef]
- Maurice, C.F.; Knowles, S.C.; Ladau, J.; Pollard, K.S.; Fenton, A.; Pedersen, A.B.; Turnbaugh, P.J. Marked seasonal variation in the wild mouse gut microbiota. ISME J. 2015, 9, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Orkin, J.D.; Campos, F.A.; Myers, M.S.; Cheves Hernandez, S.E.; Guadamuz, A.; Melin, A.D. Seasonality of the gut microbiota of free-ranging white-faced capuchins in a tropical dry forest. ISME J. 2019, 13, 183–196. [Google Scholar] [CrossRef]
- Bergmann, G.T.; Craine, J.M.; Robeson, M.S., 2nd; Fierer, N. Seasonal Shifts in Diet and Gut Microbiota of the American Bison (Bison bison). PLoS ONE 2015, 10, e0142409. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liu, G.; Li, Y.; Wei, Y.; Lin, S.; Liu, S.; Zheng, Y.; Hu, D. High-Throughput Analysis Reveals Seasonal Variation of the Gut Microbiota Composition Within Forest Musk Deer (Moschus berezovskii). Front. Microbiol. 2018, 9, 1674. [Google Scholar] [CrossRef]
- You, Z.; Deng, J.; Liu, J.; Fu, J.; Xiong, H.; Luo, W.; Xiong, J. Seasonal variations in the composition and diversity of gut microbiota in white-lipped deer (Cervus albirostris). PeerJ 2022, 10, e13753. [Google Scholar] [CrossRef]
- Wei, X.; Dong, Z.; Cheng, F.; Shi, H.; Zhou, X.; Li, B.; Wang, L.; Wang, W.; Zhang, J. Seasonal diets supersede host species in shaping the distal gut microbiota of Yaks and Tibetan sheep. Sci. Rep. 2021, 11, 22626. [Google Scholar] [CrossRef]
- Sun, B.; Wang, X.; Bernstein, S.; Huffman, M.A.; Xia, D.P.; Gu, Z.; Chen, R.; Sheeran, L.K.; Wagner, R.S.; Li, J. Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Sci. Rep. 2016, 6, 26035. [Google Scholar] [CrossRef]
- Amato, K.R.; Leigh, S.R.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The Gut Microbiota Appears to Compensate for Seasonal Diet Variation in the Wild Black Howler Monkey (Alouatta pigra). Microb. Ecol. 2015, 69, 434–443. [Google Scholar] [CrossRef]
- Chevalier, C.; Stojanović, O.; Colin, D.J.; Suarez-Zamorano, N.; Tarallo, V.; Veyrat-Durebex, C.; Rigo, D.; Fabbiano, S.; Stevanović, A.; Hagemann, S.; et al. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell 2015, 163, 1360–1374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Goerlandt, F.; van Nunen, K.; Ponnet, K.; Reniers, G. Conceptualizing the Contextual Dynamics of Safety Climate and Safety Culture Research: A Comparative Scientometric Analysis. Int. J. Environ. Res. Public Health 2022, 19, 813. [Google Scholar] [CrossRef]
- Mazov, N.A.; Gureev, V.N.; Glinskikh, V.N. The Methodological Basis of Defining Research Trends and Fronts. Sci. Tech. Inf. Process. 2020, 47, 221–231. [Google Scholar] [CrossRef]
- Chen, C. CiteSpace II: Detecting and visualizing emerging trends and transient patterns in scientific literature. J. Am. Soc. Inf. Sci. Technol. 2006, 57, 359–377. [Google Scholar] [CrossRef]
- Chen, C.; Ibekwe-Sanjuan, F.; Hou, J. The Structure and Dynamics of Co-Citation Clusters: A Multiple-Perspective Co-Citation Analysis. J. Am. Soc. Inf. Sci. Technol. 2014, 61, 1386–1409. [Google Scholar] [CrossRef]
- Garfield, E. The history and meaning of the journal impact factor. JAMA 2006, 295, 90–93. [Google Scholar] [CrossRef]
- Cipresso, P.; Giglioli, I.A.C.; Raya, M.A.; Riva, G. The Past, Present, and Future of Virtual and Augmented Reality Research: A Network and Cluster Analysis of the Literature. Front. Psychol. 2018, 9, 2086. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, K.; Weng, L.; Chen, C.; Kiartivich, S.; Jiang, X.; Su, X.; Wang, Y.; Wang, X. Bibliometric evaluation of 2000-2019 publications on functional near-infrared spectroscopy. NeuroImage 2020, 220, 117121. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Amato, K.R.; Yeoman, C.J.; Kent, A.; Righini, N.; Carbonero, F.; Estrada, A.; Gaskins, H.R.; Stumpf, R.M.; Yildirim, S.; Torralba, M.; et al. Habitat degradation impacts black howler monkey (Alouatta pigra) gastr ointestinal microbiomes. ISME J. 2013, 7, 1344–1353. [Google Scholar] [CrossRef]
- Weinstock, G.M. Genomic approaches to studying the human microbiota. Nature 2012, 489, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wang, L.; Li, J.; Hou, R.; Wang, M.; Wang, Z.; Qu, Q.; Zhou, W.; Nie, Y.; Hu, Y.; et al. Seasonal shift of the gut microbiome synchronizes host peripheral circadian rhythm for physiological adaptation to a low-fat diet in the giant panda. Cell Rep. 2022, 38, 110203. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, X.; Ding, Y.; Hu, Y.; Nie, Y.; Wei, W.; Ma, S.; Yan, L.; Zhu, L.; Wei, F. Seasonal variation in nutrient utilization shapes gut microbiome structure and function in wild giant pandas. Proc. Biol. Sci. 2017, 284, 20170955. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ding, J.; Yang, Z.; Chen, H.; Yao, R.; Dai, Q.; Ding, Y.; Zhu, L. Père David’s deer gut microbiome changes across captive and translocated populations: Implications for conservation. Evol. Appl. 2019, 12, 622–635. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Gao, H.; Qin, W.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Wang, D.; Zhang, T. Marked Seasonal Variation in Structure and Function of Gut Microbiota in Forest and Alpine Musk Deer. Front. Microbiol. 2021, 12, 699797. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Chi, X.; Li, G.; Qin, W.; Song, P.; Jiang, F.; Liu, D.; Zhang, J.; Zhou, X.; Li, S.; et al. Gut microbial diversity and stabilizing functions enhance the plateau adaptability of Tibetan wild ass (Equus kiang). MicrobiologyOpen 2020, 9, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Khakisahneh, S.; Zhang, X.Y.; Nouri, Z.; Wang, D.H. Gut Microbiota and Host Thermoregulation in Response to Ambient Temperature Fluctuations. mSystems 2020, 5, e00514-20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Xu, T.W.; Wang, X.G.; Geng, Y.Y.; Liu, H.J.; Hu, L.Y.; Zhao, N.; Kang, S.P.; Zhang, W.M.; Xu, S.X. The Effect of Transitioning between Feeding Methods on the Gut Microbiota Dynamics of Yaks on the Qinghai-Tibet Plateau. Animals 2020, 10, 1641. [Google Scholar] [CrossRef]
- Qin, W.; Huang, Y.G.; Wang, L.; Lin, G.; Zhang, T. Gut Microbiota enabled Goitered Gazelle (Gazella subgutturosa) to Adapt to Seasonal Changes. Pak. J. Zool. 2020, 52, 1637–1646. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Sukhchuluun, G.; Bo, T.B.; Chi, Q.S.; Yang, J.J.; Chen, B.; Zhang, L.; Wang, D.H. Huddling remodels gut microbiota to reduce energy requirements in a small mammal species during cold exposure. Microbiome 2018, 6, 103. [Google Scholar] [CrossRef]
- Wang, W.; Wang, F.; Li, L.; Wang, A.; Sharshov, K.; Druzyaka, A.; Lancuo, Z.; Wang, S.; Shi, Y. Characterization of the gut microbiome of black-necked cranes (Grus nigricollis) in six wintering areas in China. Arch. Microbiol. 2020, 202, 983–993. [Google Scholar] [CrossRef]
- Wang, W.; Cao, J.; Yang, F.; Wang, X.; Zheng, S.; Sharshov, K.; Li, L. High-throughput sequencing reveals the core gut microbiome of Bar-headed goose (Anser indicus) in different wintering areas in Tibet. MicrobiologyOpen 2016, 5, 287–295. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, Y.; Cao, L.; Yin, H.; Xu, M.; Wang, Z.; Liu, Y.; Wang, X.; Deng, Y. Habitat environments impacted the gut microbiome of long-distance migratory swan geese but central species conserved. Sci. Rep. 2018, 8, 13314. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Deng, Y.; Cao, L. Characterising the interspecific variations and convergence of gut microbiota in Anseriformes herbivores at wintering areas. Sci. Rep. 2016, 6, 32655. [Google Scholar] [CrossRef]
- Greene, L.K.; Andriambeloson, J.B.; Rasoanaivo, H.A.; Yoder, A.D.; Blanco, M.B. Variation in gut microbiome structure across the annual hibernation cycle in a wild primate. FEMS Microbiol. Ecol. 2022, 98, fiac070. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Arango, L.F.; Barrett, H.L.; McIntyre, H.D.; Callaway, L.K.; Morrison, M.; Dekker Nitert, M. Increased Systolic and Diastolic Blood Pressure Is Associated With Altered Gut Microbiota Composition and Butyrate Production in Early Pregnancy. Hypertension 2016, 68, 974–981. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Elias, J.E.; et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science 2017, 357, 802–806. [Google Scholar] [CrossRef]
- Hicks, A.L.; Lee, K.J.; Couto-Rodriguez, M.; Patel, J.; Sinha, R.; Guo, C.; Olson, S.H.; Seimon, A.; Seimon, T.A.; Ondzie, A.U.; et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 2018, 9, 1786. [Google Scholar] [CrossRef]
- Lin, X.; Zhou, R.; Liang, D.; Xia, L.; Zeng, L.; Chen, X. The role of microbiota in autism spectrum disorder: A bibliometric analysis based on original articles. Front. Psychiatry 2022, 13, 976827. [Google Scholar] [CrossRef] [PubMed]
- Davenport, E.R.; Mizrahi-Man, O.; Michelini, K.; Barreiro, L.B.; Ober, C.; Gilad, Y. Seasonal variation in human gut microbiome composition. PLoS ONE 2014, 9, e90731. [Google Scholar] [CrossRef]
- Zhang, J.; Guo, Z.; Lim, A.A.; Zheng, Y.; Koh, E.Y.; Ho, D.; Qiao, J.; Huo, D.; Hou, Q.; Huang, W.; et al. Mongolians core gut microbiota and its correlation with seasonal dietary changes. Sci. Rep. 2014, 4, 5001. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Qi, W.; Dai, J.; Zhang, S.; Wei, F. Evidence of cellulose metabolism by the giant panda gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 17714–17719. [Google Scholar] [CrossRef] [PubMed]
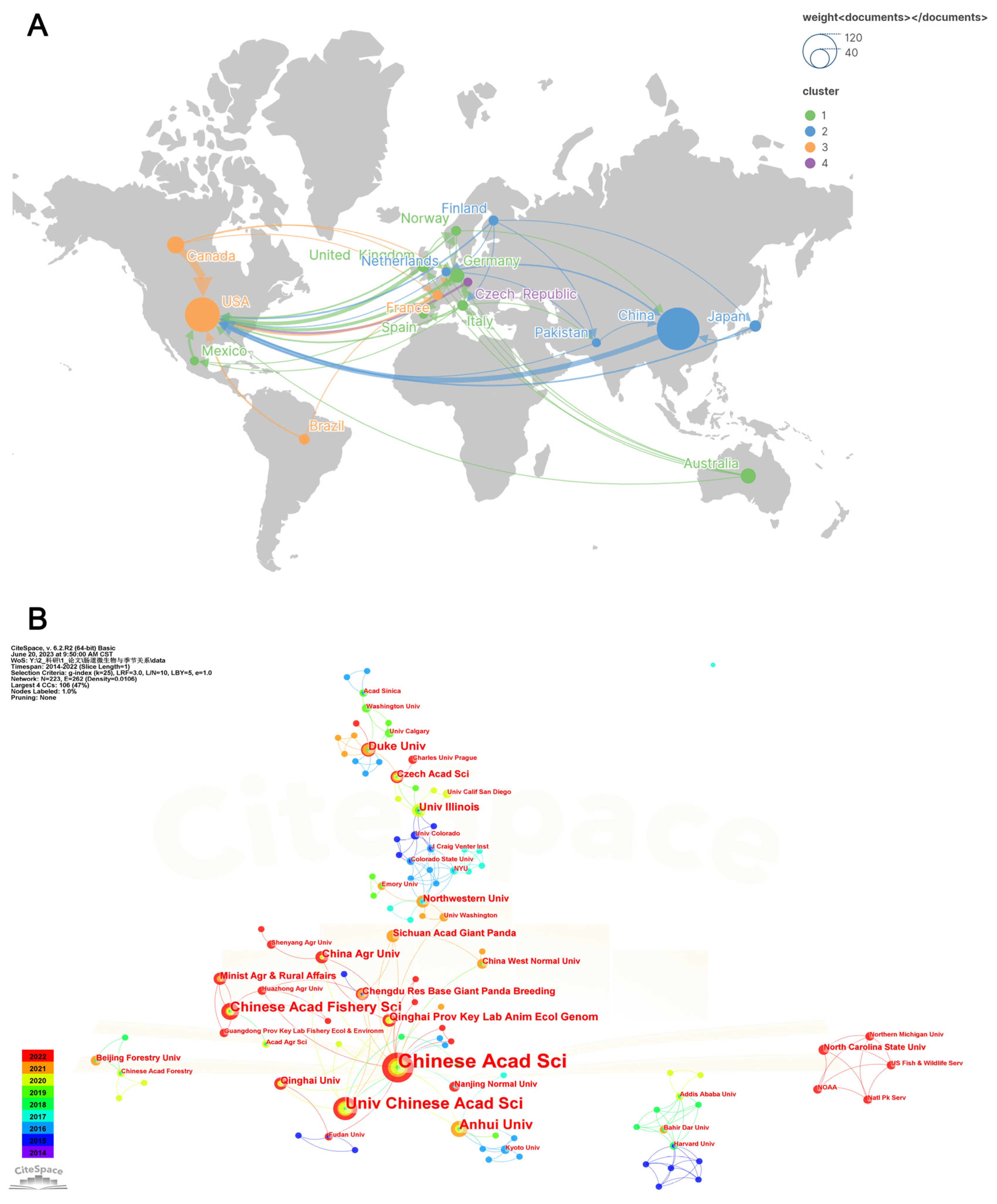

| Rank | Freq | Centrality | Country | Rank | Freq | Centrality | Institution |
|---|---|---|---|---|---|---|---|
| 1 | 112 | 0.07 | PEOPLES R CHINA | 1 | 30 | 0.17 | Chinese Acad Sci |
| 2 | 74 | 0.93 | USA | 2 | 15 | 0.01 | Univ Chinese Acad Sci |
| 3 | 18 | 0.02 | CANADA | 3 | 9 | 0.04 | Anhui Univ |
| 4 | 14 | 0.08 | AUSTRALIA | 4 | 9 | 0.02 | Chinese Acad Fishery Sci |
| 5 | 12 | 0.11 | GERMANY | 5 | 6 | 0.08 | Duke Univ |
| 6 | 9 | 0.15 | ENGLAND | 6 | 6 | 0 | Univ Queensland |
| 7 | 8 | 0.08 | JAPAN | 7 | 6 | 0 | Royal Brisbane and Womens Hosp |
| 8 | 7 | 0.06 | ITALY | 8 | 5 | 0.1 | Univ Illinois |
| 9 | 7 | 0 | BRAZIL | 9 | 5 | 0 | Qinghai Prov Key Lab Anim Ecol Genom |
| 10 | 6 | 0.04 | FINLAND | 10 | 5 | 0.01 | China Agr Univ |
| 11 | 6 | 0.04 | NORWAY | 11 | 4 | 0.13 | Northwestern Univ |
| 12 | 6 | 0.03 | FRANCE | 12 | 4 | 0.08 | Czech Acad Sci |
| 13 | 5 | 0.02 | NETHERLANDS | 13 | 4 | 0.01 | Chengdu Res Base Giant Panda Breeding |
| 14 | 5 | 0.06 | PAKISTAN | 14 | 4 | 0 | Sichuan Acad Giant Panda |
| 15 | 5 | 0 | MEXICO | 15 | 4 | 0 | Qinghai Univ |
| 16 | 5 | 0 | CZECH REPUBLIC | 16 | 4 | 0 | Chinese Acad Agr Sci |
| 17 | 5 | 0 | SPAIN | 17 | 4 | 0 | Northeast Agr Univ |
| 18 | 4 | 0.03 | SWEDEN | 18 | 4 | 0 | Minist Agr and Rural Affairs |
| 19 | 4 | 0 | ISRAEL | 19 | 3 | 0 | North Carolina State Univ |
| 20 | 4 | 0 | NEW ZEALAND | 20 | 3 | 0.01 | China West Normal Univ |
| Rank | Freq | Centrality | Journal | IF 1 | Quartile in Category |
|---|---|---|---|---|---|
| 1 | 211 | 0.02 | PLoS One | 3.7 | Q2 |
| 2 | 188 | 0.02 | Appl Environ Microb | 4.4 | Q2 |
| 3 | 186 | 0.03 | P Natl Acad Sci Usa | 11.1 | Q1 |
| 4 | 179 | 0.02 | ISME J | 11 | Q1 |
| 5 | 178 | 0.02 | Science | 56.9 | Q1 |
| 6 | 178 | 0.01 | Nature | 64.8 | Q1 |
| 7 | 161 | 0.02 | Front Microbiol | 5.2 | Q2 |
| 8 | 153 | 0.03 | Sci Rep-UK | 4.6 | Q2 |
| 9 | 145 | 0.03 | Nat Methods | 48.0 | Q1 |
| 10 | 130 | 0.01 | Bioinformatics | 5.8 | Q1 |
| 11 | 118 | 0.02 | Microb Ecol | 3.6 | Q1 |
| 12 | 115 | 0.02 | Microbiome | 15.1 | Q1 |
| 13 | 114 | 0.09 | Mol Ecol | 4.9 | Q1 |
| 14 | 114 | 0.02 | Nucleic Acids Res | 14.9 | Q1 |
| 15 | 112 | 0.02 | Nat Commun | 16.6 | Q1 |
| 16 | 96 | 0.03 | Environ Microbiol | 5.1 | Q2 |
| 17 | 94 | 0.06 | Genome Biol | 12.3 | Q1 |
| 18 | 91 | 0.04 | Nat Rev Microbiol | 88.1 | Q1 |
| 19 | 90 | 0.03 | Fems Microbiol Ecol | 4.2 | Q2 |
| 20 | 88 | 0.14 | Cell | 64.5 | Q1 |
| Rank | Count | Centrality | Cited Authors | Year |
|---|---|---|---|---|
| 1 | 90 | 0.23 | Caporaso JG | 2014 |
| 2 | 86 | 0.08 | Ley RE | 2014 |
| 3 | 82 | 0.06 | Edgar RC | 2014 |
| 4 | 63 | 0.11 | Amato KR | 2015 |
| 5 | 60 | 0.11 | David LA | 2014 |
| 6 | 52 | 0 | Segata N | 2017 |
| 7 | 50 | 0.12 | Kohl KD | 2015 |
| 8 | 49 | 0.04 | Turnbaugh PJ | 2014 |
| 9 | 46 | 0.11 | Wang Q | 2015 |
| 10 | 46 | 0.16 | Callahan BJ | 2018 |
| Rank | Count | Centrality | Author | Year | Title | Journal |
|---|---|---|---|---|---|---|
| 1 | 29 | 0.23 | Callahan BJ | 2016 | DADA2: High-resolution sample inference from Illumina amplicon data | Nat Methods |
| 2 | 28 | 0.15 | Bolyen E | 2019 | Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 | Nat Biotechnol |
| 3 | 27 | 0.25 | David LA | 2014 | Diet rapidly and reproducibly alters the human gut microbiome | Nature |
| 4 | 26 | 0.14 | R Core Development Team | 2018 | R: A language and environment for statistical computing | R Lang Env Stat Comp |
| 5 | 23 | 0.12 | Smits SA | 2017 | Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania | Science |
| 6 | 17 | 0.35 | Amato KR | 2015 | The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra) | Microb Ecol |
| 7 | 17 | 0.07 | Maurice CF | 2015 | Marked seasonal variation in the wild mouse gut microbiota | ISME J |
| 8 | 16 | 0.11 | Hicks AL | 2018 | Gut microbiomes of wild great apes fluctuate seasonally in response to diet | Nat Commun |
| 9 | 16 | 0.01 | Douglas GM | 2020 | PICRUSt2 for prediction of metagenome functions | Nat Biotechnol |
| 10 | 15 | 0.08 | Amato KR | 2013 | Habitat degradation impacts black howler monkey (Alouatta pigra) gastrointestinal microbiomes | ISME J |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, J.; Sun, X.; Lu, R.; Hu, X.; Huang, Z. Bibliometric Analysis of Global Trends in Research on Seasonal Variations in Gut Microbiota from 2012 to 2022. Microorganisms 2023, 11, 2125. https://doi.org/10.3390/microorganisms11082125
Zhai J, Sun X, Lu R, Hu X, Huang Z. Bibliometric Analysis of Global Trends in Research on Seasonal Variations in Gut Microbiota from 2012 to 2022. Microorganisms. 2023; 11(8):2125. https://doi.org/10.3390/microorganisms11082125
Chicago/Turabian StyleZhai, Jiancheng, Xiao Sun, Rui Lu, Xueqin Hu, and Zhiqiang Huang. 2023. "Bibliometric Analysis of Global Trends in Research on Seasonal Variations in Gut Microbiota from 2012 to 2022" Microorganisms 11, no. 8: 2125. https://doi.org/10.3390/microorganisms11082125
APA StyleZhai, J., Sun, X., Lu, R., Hu, X., & Huang, Z. (2023). Bibliometric Analysis of Global Trends in Research on Seasonal Variations in Gut Microbiota from 2012 to 2022. Microorganisms, 11(8), 2125. https://doi.org/10.3390/microorganisms11082125






