Abstract
Atopic dermatitis (AD) is a common chronic inflammatory skin disease with a significant association with various type-2 inflammation-related comorbidities. Ongoing research suggests the crucial involvement of gut microbiome, especially in childhood onset AD, and hence, probiotics have emerged as a potential non-steroid-based therapeutics option to complement existing AD management plans. In order to delineate the impact of probiotics in the gut microbiome of pediatric AD patients from southern China, targeted 16S rRNA sequencing and thorough bioinformatic analysis were performed to analyze the gut microbiome profiles of 24 AD children after taking an orally administered novel synbiotics formula with triple prebiotics for 8 weeks. A notable improvement in Eczema Area and Severity Index (EASI) (p = 0.008) was observed after taking an 8-week course of probiotics, with no adverse effects observed. The relative abundances of key microbial drivers including Bacteroides fragilis and Lactobacillus acidophilus were significantly increased at week 8. We also found that the positive responsiveness towards an 8-week course of probiotics was associated with improvements in the gut microbiome profile with a higher relative abundance of probiotic species. Over-represented functional abundance pathways related to vitamin B synthesis and peptidoglycan recycling may imply the underlying mechanism. In summary, our study suggests how the gut microbial landscape shifts upon probiotic supplementation in AD children, and provides preliminary evidence to support targeted probiotic supplementation for the management of childhood AD.
1. Introduction
Atopic dermatitis (AD) is a common chronic inflammatory skin disease with both atopic and nonatopic comorbidities [1] including asthma, rhinitis; and psychiatric, autoimmune, and cardiovascular diseases [2,3,4,5,6,7,8,9,10,11,12,13,14]. The incidence of childhood eczema has been continuously rising, especially in developed countries [14,15,16]; the lifetime eczema prevalence among children could be as high as 30% in China [17,18,19,20,21]. The “hygiene hypothesis” postulates that the lifestyles of people in industrialized countries, together with a decline in the infectious burden, have been associated with an increase in allergic and autoimmune diseases [22,23]. In recent years, this hypothesis has gained increasing attention from the scientific community to explain the time trend. Although the exact cause of AD is not fully understood, it is known to result from a complex interplay of genetic, environmental, and immunological factors [24]. With advances in sequencing technology, researchers have begun to investigate the relationship between the gut microbiome and pediatric AD, and it is clear that children with AD have a distinct gut microbiome composition compared to healthy children [25,26,27,28,29,30,31,32,33]. Specifically, children with AD have been found to harbor gut dysbiosis with altered levels of beneficial bacteria, such as Bifidobacterium and Lactobacillus, and enriched subspecies of Faecalibacterium prausnitzii [28,31,32,34]. These alterations could lead to dysfunction of the intestinal barrier [35] and dysregulation of the immune system [36], e.g., inhibiting Th1 and/or Th2 maturation [37,38,39]. Hence, current evidence suggests that appropriate microbial colonization in early life may lower the risk of developing allergic diseases [40] as described in the “hygiene hypothesis”, and the gut microbiome may contribute to the underlying mechanism to some extent.
Modulating the gut microbiome as a therapeutic strategy for AD has therefore been explored in the form of prebiotics, probiotics, fecal microbiota transplantation [41], and dietary interventions [42]. In particular, the World Allergy Organization has recommended probiotic use in infants at high risk of developing allergies [43]. However, study results are inconsistent and mixed, with some groups reporting the positive beneficial effect of probiotics in alleviating AD symptoms and mitigating the risk of AD, but others reporting no effect [44,45]. Despite the ongoing debate on the best practices of probiotic use in treating AD, a number of recent meta-analyses have pooled several clinical trials [46,47,48,49,50,51] and shown that certain probiotic strains [52,53,54], such as Lactobacillus rhamnosus and Bifidobacterium lactis, may be effective in reducing AD symptoms in children. In addition, mixed probiotic strains with a longer treatment duration have apparently demonstrated better effects in children aged > 1 year [46]. However, the specific mechanisms by which probiotics exert their effects are not yet fully understood, and further investigation is warranted to elucidate the optimal bacterial strains, administration dosages, and treatment durations for achieving the desired therapeutic outcomes.
Geography, lifestyle, and diet are some of the other well-known factors influencing gut microbiota composition [55,56]. Recent studies have also suggested that the efficacy of probiotics may differ based on the host’s gut microbiome composition [57]. In view of these, in our study, we investigated the efficacy of a novel synbiotics formula in southern Chinese children with AD, and the gut microbiome dynamics upon probiotics intervention. We believe that our pilot study could augment the current knowledge in respect of the role of gut microbiota in the management of AD in early childhood and provide insights into the underlying mechanism.
2. Materials and Methods
2.1. Subject Recruitment and Study Design
(1–10 years old) with mild to moderate AD were recruited prospectively from a community health survey through local skin patient association. All participants’ legal guardians provided informed consent. Exclusion criteria of the study were as follows: (1) history of adverse reaction to probiotics; (2) known overt bacterial infections in the skin; (3) premorbid medical conditions such as cardiovascular, liver, or renal dysfunction; or diabetes mellitus; (4) prior use of oral corticosteroids, oral antibiotics, other immunosuppressive drugs, or any preparation of oral herbal medicines for the treatment of AD in the past month; (5) prior diagnosis of AD, scabies, allergic contact dermatitis, or seborrheic dermatitis; and (7) prior use of anti-coagulant or anti-platelet drugs in the past month. All study participants underwent an initial screening by a board-certified dermatologist (S.K.F.L) to evaluate the severity of their atopic dermatitis. Stool specimens were obtained for subsequent sequencing analysis. Participants were permitted to continue their customary pharmacological or topical maintenance therapy for atopic dermatitis during the course of the study. Participants were followed up to confirm their compliance and collect fecal samples after 8 weeks. Age- and sex-matched historical control of deidentified AD patients from previous longitudinal studies without the use of probiotics were extracted from the database and used for comparison. The study was conducted in compliance with the ethical principles outlined in the Declaration of Helsinki and received approval from the Research Ethics Committee of the Hong Kong Doctors Union (protocol number HKSGM-2020AD-Study-protocol-vl-20220211).
2.2. Library Preparation and 16S rRNA Sequencing
Fecal specimens were homogenized using a proprietary DNA preservative and subjected to bead beating with 425–600 μm glass beads (Sigma-Aldrich, Burlington, MA, USA) for 1 h in accordance with the manufacturer’s instructions. Microbial DNA was isolated from the fecal specimens using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). The DNA concentration of each sample was quantified using a Qubit™ dsDNA HS Assay Kit (Life Technologies, Waltham, MA, USA) and a Qubit 3 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). An amplicon library was constructed using the 515F(5’-GTGCCAGCMGCCGCGG-3’)/907R(5’-CCGTCAATTTCMTTTRAGTTT-3’) primer pair, targeting the V4-V5 hypervariable region of the 16S rRNA genes in conjunction with adapter sequences and multiplex identifier tags. The 16S rRNA gene sequencing was performed using the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA) following the original Earth Microbiome Project Protocols [58]. Index barcodes and adapter sequences were removed from demultiplexed paired-end reads for downstream analysis.
2.3. Probiotic Mixture
All AD patients received one sachet daily of a novel synbiotics formula developed by BioMed Microbiome Research Centre (Kid Allergy Formula, BioMed Laboratory Company Limited, Hong Kong) containing a mixture of 6 types of highly effective gastro-resistant probiotics (not less than 1.5 × 1010 CFU/sachet at the time of production), and triple prebiotics containing inulin, isomalto-oligosaccharides (IMOs), and fructo-oligosaccharides (FOSs) for 8 weeks. The probiotic mixture was composed of Lactobacillus rhamnosus GG, Lactobacillus acidophilus GKA7, Bifidobacterium longum GKL7, Lactobacillus plantarum GKM3, Bifidobacterium bifidum GKB2, and Lactobacillus paracasei GKS6. L. rhamnosus GG and B. bifidum formula were shown to reduce the occurrence and recurrence risks of allergies in previous studies. Prebiotics serve as a substrate for probiotics, thereby enhancing their functionality [59].
2.4. Quantitative Real-Time PCR
Leftover fecal microbial DNA from 16S rRNA sequencing was retrieved for further analysis. Real-time PCR was carried out with a total volume of 10 μL, containing 5 μL of GoTaq qPCR master mix (Promega Corporation, Madison, WI, USA), 2 μL of DNA template, and 3 μL of primer pair solution (300 nM/reaction). For each run, nuclease-free water (Promega Corporation, Madison, WI, USA) was used as the negative control and melting peaks were used to determine the specificity of the PCR. qPCR was performed in a DNA thermal cycler (QuantStudio 1 Real-Time PCR System, Thermo Fisher Scientific, Waltham, MA, USA).The PCR protocol included an initial denaturation step at 95 °C for 2 min, followed by 40 cycles of 95°C for 15 s and 60 °C for 1 min, and a final dissociation step (95 °C for 15 s, 60 °C for 1 min, followed by a slow ramp to 95 °C). Primer sequences are detailed in Table S1. ∆Ct was calculated as the Ct difference between the respective target and universal bacterial primer.
2.5. Bioinformatics Analysis
16S rRNA demultiplexed sequencing data were analyzed using QIIME 2-2023.2 [60], a plugin-based system that integrates various microbiome analysis algorithms and tools. Quality control and denoising of reads were conducted with DADA2 [61] using the q2-dada2 plugin to obtain exact amplicon sequence variants (ASVs) [62]. ASVs were aligned using mafft [63] and a phylogenetic tree was constructed using fastree2 [64] via the q2-phylogeny plugin. Taxonomic annotation of ASVs was performed using the q2-feature-classifier plugin [65] with a pre-trained naive Bayes classifier based on the SILVA v138 taxonomic reference database with 99% similarity [66,67]. Alpha diversity was assessed using six metrics: observed features, Chao1 index (Chao1), ACE index (ACE), Shannon diversity index (Shannon), Simpson index (Simpson), and Faith’s phylogenetic diversity (PD). Beta diversity was assessed based on the Bray–Curtis, cosine, Hamming, Jaccard distance metrics, and the generalized, weighted, weighted normalized, and unweighted UniFrac distance metrics. The difference in gut microbial composition across groups was computed using PERMANOVA test (999 permutations) [68], while the Adonis test was used to investigate the microbial community dissimilarity across responders and time points [69]. Differential abundance analysis was performed using ANCOM with bias correction and repeated measures (ANCOM-BC2) [70]. The co-occurrence/co-exclusion network was derived using the Sparse and Compositionally Robust Inference of Microbial Ecological Networks (SPIEC-EASI) framework [71] using the neighborhood selection framework introduced by Meinshausen and Bühlmann [72].
2.6. Statistical Analysis
Statistical analyses and visualization of the results were performed using Python 3.8.13 with the following package versions: numpy 1.22.3, scipy 1.8.0, statsmodels 0.13.2, skbio 0.5.6, matplotlib 3.5.1, and seaborn 0.11.2. Normality assumptions were assessed using D’Agostino and Pearson’s test (implemented in the scipy.stats.normaltest function) and the Shapiro–Wilk test (implemented in the scipy.stats.shapiro function) for parametric tests. Demographic characteristics were evaluated using the non-parametric Mann–Whitney U rank test (implemented in the scipy.stats.mannwhitneyu function) for continuous variables and the Fisher exact test (implemented in the scipy.stats.fisher_exact function) for categorical variables. p-value correction was performed using the Benjamini/Hochberg (non-negative) procedure, implemented in the statsmodels.stats.multitest.multipletests function. A p-value of less than 0.05 was considered statistically significant unless otherwise specified.
3. Results
3.1. Study Cohort
A total of 24 subjects aged between 1 and 10 years old with atopic dermatitis were prospectively recruited into the cohort. A summary of the participants’ demographic and disease-related characteristics is presented in Table 1. In brief, the cohort consisted of 58.3% males, and 41.6% of the participants were delivered by Caesarean section. Over 80% of caregivers of the participants reported type-1 hypersensitivity-related comorbidities including allergic rhinitis, asthma, or allergic conjunctivitis. Three participants reported having comorbid constipation, while one participant reported having comorbid diarrhea at baseline. There was one participant aged under 3 years, which is younger than the reported age of gut microbiome maturation [73]. However, we did not exclude the participant from the analysis owing to the small sample size.

Table 1.
Baseline demographic of participants.
The cohort was stratified into subgroups according to the response to the 8-week oral administration of probiotics. There was no statistically significant difference in age (mean age—responders: 9 years, non-responders: 4 years; p = 0.0524), sex, or mode of delivery between responders and non-responders. However, age and mode of delivery are well-known confounding factors of gut microbiome analysis; we controlled for age and mode of delivery during subsequent analysis. No adverse effects or discontinuations were reported or recorded throughout the study period.
3.2. Effect of 8-Week Probiotic Intake on Gut Microbiome Composition
Among all participants, there was a modest yet significant improvement in EASI score (∆EASI = −2.3 ± 3.9, p = 0.008, Figure 1A) after taking an 8-week course of orally administered probiotics. Our hypothesis posited that the observed improvement in EASI scores could be attributed to the restoration of gut dysbiosis mediated by the synbiotics formula. Therefore, we compared the gut microbiota composition of participants after 8 weeks of probiotic supplementation with their baseline profiles (Figure 1B,C). No significant differences in alpha diversities (Figure S1) and beta diversities (Figure S2) were found. Even though there was an increase in the relative abundance of Firmicutes, together with a decrease in the relative abundances of Actinobacteriota and Desulfobacterota, neither the phylum after multiple testing corrections (Wilcoxon Signed Rank, Benjamini/Hochberg correction) nor the F/B ratio were observed to be significantly different (Figure 1D and Table S2).
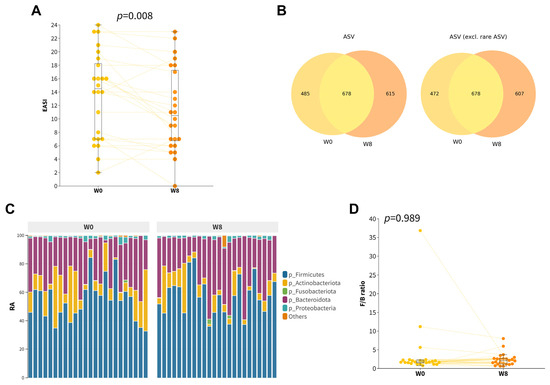
Figure 1.
Gut composition profiles of childhood AD participants at baseline and at week 8. (A) Boxplot of the EASI scores of the participants. (B) Venn diagram of all ASVs (left) and with rare ASVs excluded (right) at baseline and week 8. Rare ASVs were defined as ASVs that occurred in only one of the samples. (C) Relative abundance of top five main phyla. (D) Boxplot of Firmicutes/Bacteroidetes (F/B) ratio.
An exploratory analysis of differentially abundant units was conducted using ANCOM-BC2 at the ASV level. This analysis identified three differentially abundant ASVs, including two named species (DA−ASV1 and DA−ASV2) and one unnamed species (DA−ASV3), as detailed in Table 2. DA−ASV1 and DA−ASV2 were classified as Bacteroides fragilis and Lactobacillus acidophilus, respectively (Table S3). Higher (center-log-transformed, clr) relative abundances of DA−ASV1 (Figure 2B; q < 0.001) and DA−ASV2 (Figure 2C; q = 0.015) were observed at week eight, while a lower relative abundance of DA−ASV3 (Figure 2D) was observed after 8 weeks. MOLE-BLAST was employed to classify the DA−ASV3 unnamed Bifidobacterium species as closely related to Bifidobacterium adolescentis (Figure 2E). The increase in Lactobacillus acidophilus abundance was very likely due to the intake of synbiotics rich in Lactobacillus.

Table 2.
Differentially abundant ASVs (taxon assigned by q2-feature-classifier) identified by ANCOM-BC2 after the 8-week course of orally administered probiotics.
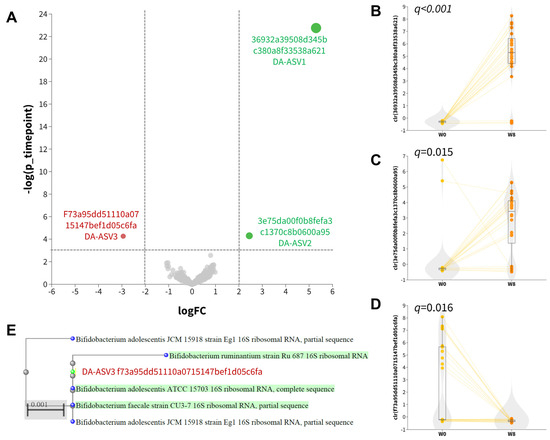
Figure 2.
Differentially abundant ASVs. (A) Volcano plot of ANCOM-BC2 results. Differentially abundant ASVs indicated in red (downregulated at week 8) and green (upregulated at week 8). (B–D) Boxplot of center-log-ratio (clr) transformed abundance of differentially abundant ASVs. (E) Phylogenetic tree of DA−ASV3 computed by MOLE−BLAST using 16S rRNA sequences as search set with default parameter settings.
3.3. Distinctive Changes in the Gut Microbiota among Responders
To investigate the differences in the changes in gut microbiota after 8 weeks of oral probiotic intake in the responder subgroup, we further examined the changes in alpha diversities within the subgroup (Figures S3–S5). Although no significant changes were observed in any of the six alpha diversity metrics tested, the Adonis test identified a significant difference in five of the eight beta-diversity metrics by taking responsiveness and timepoint into account (Figure 3A), which indicated the differences in the microbial profiles of responders compared with non-responders (Figure 3B).
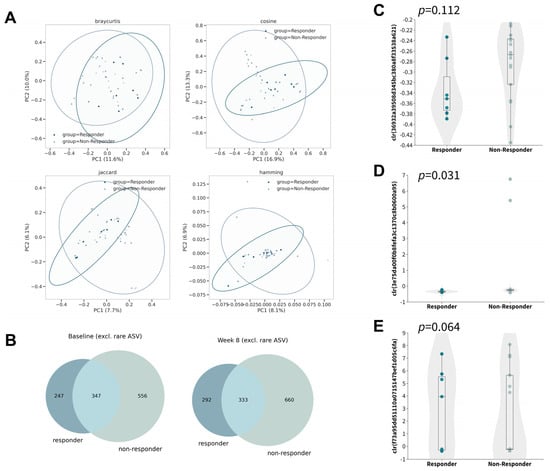
Figure 3.
Gut microbiome compositions of responders and non-responders after 8 weeks of synbiotics. (A) Principal coordinate analysis based on Bray–Curtis (upper left), cosine (upper right), Jaccard (lower left), and Hamming distance (lower right). (B) Venn diagram of ASVs (excluding rare ASVs) at baseline (left) and week 8 (right). (C–E) Boxplot of center-log ratio (clr)-transformed abundance of DA−ASV at baseline (Mann–Whitney U).
Owing to the relatively small sample size in the subgroup, we explored the dissimilarity in the abovementioned differential abundant ASVs without proceeding to subsequent analysis so as to minimize false discovery. In contrast to that of non-responders, the gut microbiota of responders was found to contain less DA−ASV1 (Figure 3C; p = 0.112, Mann–Whitney U) and DA−ASV2 (Figure 3D; p = 0.031, Mann–Whitney U) and more DA−ASV3 (Figure 3E; p = 0.064, Mann–Whitney U) at baseline with marginal statistical significance, while no statistical significance was discovered at week eight (Figure S6). Interestingly, the observation was opposite to the direction of the respective log fold change. This may imply that people who have a lower abundance of DA−ASV1 and DA−ASV2, and a higher abundance of DA−ASV3, would be more likely to respond to probiotics for the control of atopic dermatitis.
3.4. The Dynamics of Probiotic Species in Relation to Probiotic Intake Responsiveness
With the encouraging results with respect to the increased relative abundance of L. acidophilus after 8 weeks of probiotic intake, we further verified the findings using qPCR (Table S4) because it is well known that 16S targeted sequencing has a limited resolution at the species level. All probiotic species contained in the mixture were tested using the remaining DNA extract. It is evident that the ∆Ct values of L. acidophilus (p = 0.001, Wilcoxon signed rank), L. paracasei (p = 0.001, Wilcoxon signed rank), L. plantarum (p = 0.001, Wilcoxon signed rank), and L. rhamnosus (p = 0.001, Wilcoxon signed rank) were decreased at week eight regardless of responsiveness (Figure 4A), which corresponded to an increase in relative abundance. The results for B. bifidum and B. longum were mixed. Overall, responders were found to have a relatively higher relative abundance of probiotic species (Figure 4B). We also incorporated a historical placebo cohort as a control for comparison. The synbiotics formula boosted the relative abundance of probiotic species to a greater extent compared to the control group.
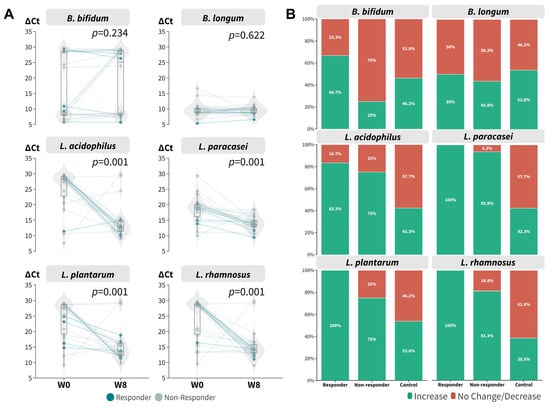
Figure 4.
Real-time PCR of beneficial bacteria contained in the synbiotics mixture. (A) ∆Ct at baseline and week 8 (signed rank test regardless of responsiveness). (B) Proportion of participants within responsiveness subgroup who achieved increase in relative abundance of respective bacterial targets.
3.5. Distinctive Functional Abundance Profile among Responders
To investigate the biological implications of the gut microbiome profile, functional abundance was inferred using PICRUSt2 in conjunction with LefSe. This analysis identified 18 discriminative features with an absolute LDA score greater than 2, as depicted in Figure 5A,B and detailed in Table 3. Most of the features over-represented in the responder subgroup (Figure S7) were related to vitamin B synthesis, such as biotin (Figure 5C,D; PWY-6519 and BIOTIN-BIOSYNTHESIS) and colonic acid (COLANSYN-PWY). However, vitamin K or menaquinol-related pathways (including PWY-5840, PWY-5838, PWY-5897, PWY-5898, and PWY-5899) and L-tryptophan synthesis were disproportionately abundant in non-responders (TRPSYN-PWY and PWY-6163).
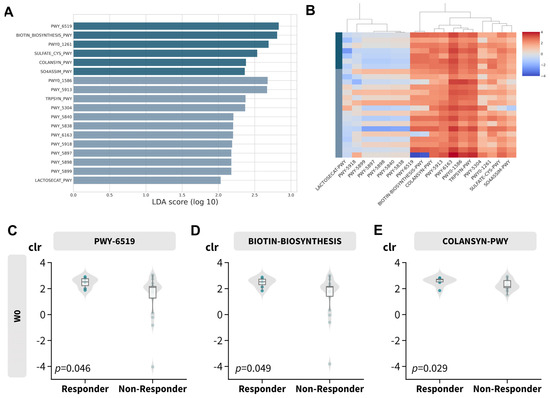
Figure 5.
Predicted MetaCyc pathway abundance. (A) Log LDA score of differentially abundant MetaCyc pathways. (B) Heatmap of center−log−ratio (clr) transformed abundance of differentially abundant MetaCyc pathways. (C–E) Boxplot of center−log−ratio (clr) transformed abundance of selected differential abundant MetaCyc pathways.

Table 3.
Differentially abundant MetaCyc pathways and corresponding LDA scores; and p-value inferred using PICRUSt2 and LefSe.
Notably, peptidoglycan recycling (PWY0-1261; leading to decreased expression of peptidoglycan) was over-represented in responders, while peptidoglycan maturation (PWY0-1586; leading to increased expression of peptidoglycan) was over-represented in non-responders. Signature fragments of peptidoglycan are known to trigger pro-inflammatory responses [74], which may explain the responsiveness in relation to the gut microbial profile.
4. Discussion
Despite the recent advancements in therapeutic options in the management of atopic dermatitis, including the use of biologics and small molecules [75,76], recurrent disease is commonplace in AD patients, yielding poor treatment satisfaction, frustration, and anxiety in patients and their caregivers [77]. In addition, the journey of children with AD is often complicated by their reluctance to use corticosteroids [78]. Therefore, it is crucial to seek alternative options to complement existing management plans. Probiotic supplementation has emerged as an attractive choice with the substantial recognition of the gut–skin axis and the pivotal role of the gut microbiome in immunity maturation [79]. In our study, we explored the gut microbial dynamics upon probiotic intake and clarified the taxonomical and functional alteration of the gut microbiome profile among responders in a pediatric atopic dermatitis cohort. To our knowledge, this represents the first study to investigate the dynamics of the gut microbiome in response to probiotic supplementation in a pediatric atopic dermatitis cohort in Hong Kong. We posit that the inclusion of gut microbiome data from southern Chinese patients could enhance our understanding of the convoluted reciprocity between the gut microbiome and host health.
We reported three significant differentially abundant ASVs (DA−ASVs) after 8 weeks of oral probiotic administration without significant changes in conventional alpha diversities, beta diversities, and taxonomic analysis. It is expected that the impact of probiotics on the gut microbiota would be reflected to a larger extent on the ASV level rather than structural changes, because the gut microbiota is reported to be resilient [80,81,82,83], and this pilot cohort may not be suitable for illustrating the subtle differences in the gut flora. Among the DA−ASVs identified, B. fragilis is reportedly associated with childhood eczema [30,32]. The bacterial polysaccharide produced by B. fragilis is known to exert anti-inflammatory properties via inducing IL-10 secretion, balancing Th1/Th2 populations, and modulating systemic T cell deficiencies [84,85,86]. L. acidophilus has also been repeatedly shown to suppress inflammation and improve atopic dermatitis symptoms [47,87,88,89,90]. However, one of the unnamed DA−ASVs that is closely related to B. adolescentis was depleted after probiotic intake. This observation was also made in a recent 4-month prospective study of infant eczema in Hong Kong [30], and in contrast to a previous study in Swedish infants with atopic dermatitis [33]. This further reinforces and justifies the need to conduct more studies with diverse ethnicities and across a wide range of geographical locations to gain a better understanding of the complex interplay between microbes and hosts.
We also noted that participants with a lower relative abundance of B. fragilis or L. acidophilus and a higher relative abundance of DA−ASV3 (B. adolescentis) may be more likely to benefit from the synbiotics formula. As L. acidophilus is one of the components of the synbiotics mixture, we decided to conduct real-time PCR to examine the relative abundances of the probiotic species contained in the mixture. It is evident that probiotic intake increased the relative abundance of beneficial bacteria reaching statistical significance in four out of six bacterial targets tested. Specifically, no effect on B. longum was observed, which could likely result from its high abundance at baseline compared to other bacterial targets. This echoes previous landmark findings showing that the baseline gut microbiome profile may play a decisive role in determining a host’s response towards probiotics [57,91]. Responders were found to have a higher relative abundance of probiotic species when compared to non-responders. It is possible that attaining adequate levels of intestinal probiotic species is necessary for an optimal clinical response. This may explain the inconsistent response to the use of oral probiotics in AD in previous clinical studies. Monitoring the adequacy of probiotic therapy with microbiome-based testing with a defined level endpoint may be a potential option to optimize the therapeutic outcome when using probiotic therapy in AD.
To gain physiological insights from the gut microbiome profile, functional abundances were investigated. Vitamin B7 (biotin) and B12 (cobalamin) synthesis was predicted to be over-represented in responders, which could imply that more vitamin B7 and vitamin B12 have been absorbed and circulated in responders’ bodies [92,93,94]. Several studies suggested that vitamin B might alleviate eczema symptoms [95,96,97,98]. It is also important to highlight that responders exhibited a high prevalence of peptidoglycan recycling, whereas peptidoglycan maturation was predominantly observed in non-responsive participants in line with the pro-inflammatory properties of the signature fragments of peptidoglycan [74]. The abovementioned observation may provide valuable insights into the correlation between the responsiveness of subjects and their respective gut microbial profiles. Vitamin B deficiency has long been known to have a dermatological manifestation [99]. For example, biotin deficiency can lead to skin rash or eczema [100,101,102]. The serum vitamin B7 level has been shown to be associated with cedar pollinosis [103], while topical vitamin B12 has been shown to reduce the severity of atopic dermatitis [104,105,106]. Although blood samples were not included in the current study design, serum vitamin B7 and B12 levels would be valuable measurements in future investigations to gain a deeper understanding of the mechanism of probiotics in pediatric AD.
In summary, our findings show the alteration of the gut microbiome due to probiotic intake in southern Chinese pediatric atopic dermatitis patients. B. fragilis and L. acidopilus may be crucial microbial drivers in atopic dermatitis development, and probiotics could potentially enhance their impact. We have also provided additional evidence to support the notion of differential probiotic colonization in a personalized manner. Our study was constrained by a limited sample size and the inclusion of only patients with mild to moderate disease severity. Well-designed and powered prospective clinical trials would be warranted to explore the efficacy of probiotics as long-term adjuvant maintenance therapy in AD patients with different disease severities. In spite of this, our study suggests the possibility and provides a scientific basis for the incorporation of probiotic treatment in the management of childhood AD, with the use of microbiome-based testing to assess the therapeutic adequacy endpoint.
5. Conclusions
We revealed significant alterations in the composition of the gut microbiome of (1) children with AD taking 8 weeks of probiotics and (2) responders consisting of a local cohort of southern Chinese patients. The crucial microbial drivers B. fragilis and Lactobacillus were restored at week eight. The gut microbiome profile before and after treatment may predict probiotic responsiveness, and this study could provide an important clue regarding the implication of gut-microbiome-based testing and therapies in the management of children with atopic dermatitis.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/microorganisms11092175/s1, Supplementary Figure S1 Boxplot of alpha diversity metrics at baseline and week 8 (Wilcoxon signed rank): (A) ACE, (B) Chao1, (C) Faith’s phylogenetic diversity (Faith PD), (D) observed features, (E) Shannon’s index, and (F) inverse Simpson; Supplementary Figure S2. Gut composition profile at baseline and week 8. Principal coordinate analysis biplot based on (A) Bray–Curtis, (B) Jaccard, (C) cosine, (D) Hamming, (E) unweighted UniFrac, (F) weighted UniFrac, (G) generalized UniFrac, and (H) weighted normalized UniFrac (Adonis); Supplementary Figure S3. Boxplot of alpha diversity metrics at baseline and week 8 stratified by responsiveness (Wilcoxon signed rank): (A) ACE, (B) Chao1, (C) Faith’s phylogenetic diversity (Faith PD), (D) observed features, (E) Shannon’s index, and (F) inverse Simpson; Supplementary Figure S4. Boxplot of alpha diversity metrics at baseline stratified by responsiveness (Mann–Whitney U): (A) ACE, (B) Chao1, (C) Faith’s phylogenetic diversity (Faith PD), (D) observed features, (E) Shannon’s index, and (F) inverse Simpson; Supplementary Figure S5. Boxplot of alpha diversity metrics at week 8 stratified by responsiveness (Mann–Whitney U): (A) ACE, (B) Chao1, (C) Faith’s phylogenetic diversity (Faith PD), (D) observed features, (E) Shannon’s index, and (F) inverse Simpson; Supplementary Figure S6. Boxplot of clr-transformed abundance of the differentially abundant ASVs identified by ANCOM-BC2 at week 8. (A) DA−ASV1, (B) DA−ASV2, and (C) DA−ASV3; Supplementary Figure S7. Over-represented functional MetaCyc pathways. (A) Violin plot of over-represented pathways stratified by responsiveness. Boxplots of clr-transformed predicted abundances of (B) PWY-6519, (C) BIOTIN-BIOSYNTHESIS, and (D) COLANSYN-PWY. Supplementary Table S1. qPCR primer sequences of beneficial bacteria; Supplementary Table S2. Adjusted p-value of the relative abundance at phylum level; Supplementary Table S3. Confidence of taxonomic assignment of differentially abundant ASVs identified by ANCOM-BC2; Supplementary Table S4. Fisher’s exact test result of the change in abundance stratified by responsiveness.
Author Contributions
Conceptualization, S.K.F.L. and S.K.W.T.; formal analysis, C.T.C.; investigation, C.J.Y.L., J.C.C.T., J.Z., C.H.W., Y.W.L. and H.W.C.; data curation, P.L.K.S. and S.K.F.L.; writing—original draft preparation, C.T.C. and S.K.F.L.; visualization, C.T.C.; supervision, S.K.F.L. and S.K.W.T.; project administration, C.J.Y.L. and P.L.K.S.; funding acquisition, S.K.F.L. and S.K.W.T. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by the Hong Kong Society of Gut Microbiome (HKSGM).
Institutional Review Board Statement
The study was conducted in compliance with the ethical principles outlined in the Declaration of Helsinki and received approval from the Research Ethics Committee of the Hong Kong Doctors Union (protocol number HKSGM-2020AD-Study-protocol-vl-20220211).
Informed Consent Statement
Informed consent was obtained from all participants and/or their legal guardians involved in the study.
Data Availability Statement
The raw sequence data are available in NCBI (BioProject PRJNA982573). Due to limitations of the consent and concerns regarding sensitive data, access to the metadata and qPCR data is restricted and available only upon reasonable request to the corresponding authors.
Conflicts of Interest
C.T.C., P.L.K.S., J.Z., C.H.W., Y.W.L., H.W.C., C.J.Y.L. and J.C.C.T. are employees of BioMed Laboratory Company Limited but the relationship did not constitute a conflict in this study. S.K.F.L. and S.K.W.T. are consultants of the BioMed Laboratory Company Limited but the relationship did not constitute a conflict in this study.
References
- Weidinger, S.; Beck, L.A.; Bieber, T.; Kabashima, K.; Irvine, A.D. Atopic Dermatitis. Nat. Rev. Dis. Primers 2018, 4, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Sorrell, J.; Petukhova, L.; Reingold, R.; Christiano, A.; Garzon, M. Shedding Light on Alopecia Areata in Pediatrics: A Retrospective Analysis of Comorbidities in Children in the National Alopecia Areata Registry. Pediatr. Dermatol. 2017, 34, e271–e272. [Google Scholar] [CrossRef]
- Schmitt, J.; Schwarz, K.; Baurecht, H.; Hotze, M.; Fölster-Holst, R.; Rodríguez, E.; Lee, Y.A.E.; Franke, A.; Degenhardt, F.; Lieb, W.; et al. Atopic Dermatitis Is Associated with an Increased Risk for Rheumatoid Arthritis and Inflammatory Bowel Disease, and a Decreased Risk for Type 1 Diabetes. J. Allergy Clin. Immunol. 2016, 137, 130–136. [Google Scholar] [CrossRef]
- Thyssen, J.P.; Hamann, C.R.; Linneberg, A.; Dantoft, T.M.; Skov, L.; Gislason, G.H.; Wu, J.J.; Egeberg, A. Atopic Dermatitis Is Associated with Anxiety, Depression, and Suicidal Ideation, but Not with Psychiatric Hospitalization or Suicide. Allergy 2018, 73, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Yaghmaie, P.; Koudelka, C.W.; Simpson, E.L. Mental Health Comorbidity in Patients with Atopic Dermatitis. J. Allergy Clin. Immunol. 2013, 131, 428–433. [Google Scholar] [CrossRef]
- Pinart, M.; Benet, M.; Annesi-Maesano, I.; von Berg, A.; Berdel, D.; Carlsen, K.C.L.; Carlsen, K.H.; Bindslev-Jensen, C.; Eller, E.; Fantini, M.P.; et al. Comorbidity of Eczema, Rhinitis, and Asthma in IgE-Sensitised and Non-IgE-Sensitised Children in MeDALL: A Population-Based Cohort Study. Lancet Respir. Med. 2014, 2, 131–140. [Google Scholar] [CrossRef]
- Kapoor, R.; Menon, C.; Hoffstad, O.; Bilker, W.; Leclerc, P.; Margolis, D.J. The Prevalence of Atopic Triad in Children with Physician-Confirmed Atopic Dermatitis. J. Am. Acad. Dermatol. 2008, 58, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Van der Hulst, A.E.; Klip, H.; Brand, P.L.P. Risk of Developing Asthma in Young Children with Atopic Eczema: A Systematic Review. J. Allergy Clin. Immunol. 2007, 120, 565–569. [Google Scholar] [CrossRef]
- Werfel, T.; Heratizadeh, A.; Niebuhr, M.; Kapp, A.; Roesner, L.M.; Karch, A.; Erpenbeck, V.J.; Losche, C.; Jung, T.; Krug, N.; et al. Exacerbation of Atopic Dermatitis on Grass Pollen Exposure in an Environmental Challenge Chamber. J. Allergy Clin. Immunol. 2015, 136, 96–103.e9. [Google Scholar] [CrossRef]
- Longo, G.; Berti, I.; Burks, A.W.; Krauss, B.; Barbi, E. IgE-Mediated Food Allergy in Children. Lancet 2013, 382, 1656–1664. [Google Scholar] [CrossRef]
- Bergmann, M.M.; Caubet, J.C.; Boguniewicz, M.; Eigenmann, P.A. Evaluation of Food Allergy in Patients with Atopic Dermatitis. J. Allergy Clin. Immunol. Pract. 2013, 1, 22–28. [Google Scholar] [CrossRef]
- Manam, S.; Tsakok, T.; Till, S.; Flohr, C. The Association between Atopic Dermatitis and Food Allergy in Adults. Curr. Opin. Allergy Clin. Immunol. 2014, 14, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Flohr, C.; Johansson, S.G.O.; Wahlgren, C.F.; Williams, H. How Atopic Is Atopic Dermatitis? J. Allergy Clin. Immunol. 2004, 114, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Asher, M.I.; Montefort, S.; Björkstén, B.; Lai, C.K.; Strachan, D.P.; Weiland, S.K.; Williams, H. Worldwide Time Trends in the Prevalence of Symptoms of Asthma, Allergic Rhinoconjunctivitis, and Eczema in Childhood: ISAAC Phases One and Three Repeat Multicountry Cross-Sectional Surveys. Lancet 2006, 368, 733–743. [Google Scholar] [CrossRef]
- Mallol, J.; Crane, J.; von Mutius, E.; Odhiambo, J.; Keil, U.; Stewart, A. The International Study of Asthma and Allergies in Childhood (ISAAC) Phase Three: A Global Synthesis. Allergol. Immunopathol. 2013, 41, 73–85. [Google Scholar] [CrossRef]
- Williams, H.; Stewart, A.; von Mutius, E.; Cookson, W.; Anderson, H.R. Is Eczema Really on the Increase Worldwide? J. Allergy Clin. Immunol. 2008, 121, 947–954.e15. [Google Scholar] [CrossRef]
- Liu, W.; Cai, J.; Sun, C.; Zou, Z.; Zhang, J.; Huang, C. Time-Trends for Eczema Prevalences among Children and Adults from 1985 to 2015 in China: A Systematic Review. BMC Public Health 2022, 22, 1294. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.L.; Wong, W.; Lau, Y.L. Increasing Prevalence of Allergic Rhinitis but Not Asthma among Children in Hong Kong from 1995 to 2001 (Phase 3 International Study of Asthma and Allergies in Childhood). Pediatr. Allergy Immunol. 2004, 15, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Survey Findings on Prevalence of Allergic Diseases among Hong Kong Primary and Secondary Schoolchildren—All News—Media—HKU. Available online: https://www.hku.hk/press/news_detail_23934.html (accessed on 10 May 2023).
- Leung, R.; Wong, G.; Lau, J.; Ho, A.; Chan, J.K.W.; Choy, D.; Douglass, C.; Lai, C.K.W. Prevalence of Asthma and Allergy in Hong Kong Schoolchildren: An ISAAC Study. Eur. Respir. J. 1997, 10, 354–360. [Google Scholar] [CrossRef]
- Hong Kong Journal of Paediatrics [HK J Paediatr (New Series) 2021, 26, 42–57]. Available online: https://www.hkjpaed.org/details.asp?id=1323&show=1234 (accessed on 10 May 2023).
- Lewis, M.C.; Inman, C.F.; Patel, D.; Schmidt, B.; Mulder, I.; Miller, B.; Gill, B.P.; Pluske, J.; Kelly, D.; Stokes, C.R.; et al. Direct Experimental Evidence That Early-Life Farm Environment Influences Regulation of Immune Responses. Pediatr. Allergy Immunol. 2012, 23, 265–269. [Google Scholar] [CrossRef]
- Okada, H.; Kuhn, C.; Feillet, H.; Bach, J.F. The “hygiene Hypothesis” for Autoimmune and Allergic Diseases: An Update. Clin. Exp. Immunol. 2010, 160, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Grobe, W.; Bieber, T.; Novak, N. Pathophysiology of Atopic Dermatitis. J. Dtsch. Dermatol. Ges. 2019, 17, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Reddel, S.; Del Chierico, F.; Quagliariello, A.; Giancristoforo, S.; Vernocchi, P.; Russo, A.; Fiocchi, A.; Rossi, P.; Putignani, L.; El Hachem, M. Gut Microbiota Profile in Children Affected by Atopic Dermatitis and Evaluation of Intestinal Persistence of a Probiotic Mixture. Sci. Rep. 2019, 9, 4996. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Du, X.; Zhai, S.; Tang, X.; Liu, C.; Li, W. Gut Microbiota and Atopic Dermatitis in Children: A Scoping Review. BMC Pediatr. 2022, 22, 323. [Google Scholar] [CrossRef]
- Petersen, E.B.M.; Skov, L.; Thyssen, J.P.; Jensen, P. Role of the Gut Microbiota in Atopic Dermatitis: A Systematic Review. Acta. Derm. Venereol. 2018, 99, 5–11. [Google Scholar] [CrossRef]
- Candela, M.; Rampelli, S.; Turroni, S.; Severgnini, M.; Consolandi, C.; De Bellis, G.; Masetti, R.; Ricci, G.; Pession, A.; Brigidi, P. Unbalance of Intestinal Microbiota in Atopic Children. BMC Microbiol. 2012, 12, 95. [Google Scholar] [CrossRef]
- Penders, J.; Stobberingh, E.E.; van den Brandt, P.A.; Thijs, C. The Role of the Intestinal Microbiota in the Development of Atopic Disorders. Allergy 2007, 62, 1223–1236. [Google Scholar] [CrossRef]
- Chan, C.W.H.; Chan, J.Y.W.; Leung, T.F.; Choi, K.C.; Tsui, S.K.W.; Wong, C.L.; Chow, K.M. Altered Gut Microbiome and Environmental Factors Associated with Development of Eczema in Hong Kong Infants: A 4-Month Pilot Study. Int. J. Environ. Res. Public Health 2020, 17, 7634. [Google Scholar] [CrossRef]
- Melli, L.C.F.L.; do Carmo-Rodrigues, M.S.; Araújo-Filho, H.B.; Mello, C.S.; Tahan, S.; Pignatari, A.C.C.; Solé, D.; de Morais, M.B. Gut Microbiota of Children with Atopic Dermatitis: Controlled Study in the Metropolitan Region of São Paulo, Brazil. Allergol. Immunopathol. 2020, 48, 107–115. [Google Scholar] [CrossRef]
- Zheng, H.; Liang, H.; Wang, Y.; Miao, M.; Shi, T.; Yang, F.; Liu, E.; Yuan, W.; Ji, Z.S.; Li, D.K. Altered Gut Microbiota Composition Associated with Eczema in Infants. PLoS ONE 2016, 11, e0166026. [Google Scholar] [CrossRef]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Björkstén, B.; Engstrand, L.; Jenmalm, M.C. Low Diversity of the Gut Microbiota in Infants with Atopic Eczema. J. Allergy Clin. Immunol. 2012, 129, 434–440. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Yoo, Y.; Hwang, J.; Na, Y.C.; Kim, H.S. Faecalibacterium prausnitzii Subspecies-Level Dysbiosis in the Human Gut Microbiome Underlying Atopic Dermatitis. J. Allergy Clin. Immunol. 2016, 137, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Pike, M.G.; Heddle, R.J.; Boulton, P.; Turner, M.W.; Atherton, D.J. Increased Intestinal Permeability in Atopic Eczema. J. Invest. Dermatol. 1986, 86, 101–104. [Google Scholar] [CrossRef]
- De Filippis, F.; Paparo, L.; Nocerino, R.; Della Gatta, G.; Carucci, L.; Russo, R.; Pasolli, E.; Ercolini, D.; Berni Canani, R. Specific Gut Microbiome Signatures and the Associated Pro-Inflamatory Functions Are Linked to Pediatric Allergy and Acquisition of Immune Tolerance. Nat. Commun. 2021, 12, 5958. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.S. The Role of Microorganisms in Atopic Dermatitis. Clin. Exp. Immunol. 2006, 144, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.J.; Kang, S.M.; Xie, J.L.; Huang, L.; Wen, Q.; Fan, Y.Y.; Lu, L.J.; Jiang, L. Early-Life Gut Microbial Colonization Shapes Th1/Th2 Balance in Asthma Model in BALB/c Mice. BMC Microbiol. 2017, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Björkstén, B.; Engstrand, L.; Andersson, A.F. Decreased Gut Microbiota Diversity, Delayed Bacteroidetes Colonisation and Reduced Th1 Responses in Infants Delivered by Caesarean Section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Gensollen, T.; Blumberg, R.S. Correlation between Early-Life Regulation of the Immune System by Microbiota and Allergy Development. J. Allergy Clin. Immunol. 2017, 139, 1084–1091. [Google Scholar] [CrossRef]
- Mashiah, J.; Karady, T.; Fliss-Isakov, N.; Sprecher, E.; Slodownik, D.; Artzi, O.; Samuelov, L.; Ellenbogen, E.; Godneva, A.; Segal, E.; et al. Clinical Efficacy of Fecal Microbial Transplantation Treatment in Adults with Moderate-to-Severe Atopic Dermatitis. Immun. Inflamm. Dis. 2022, 10, e570. [Google Scholar] [CrossRef]
- Bath-Hextall, F.; Delamere, F.M.; Williams, H.C. Dietary Exclusions for Improving Established Atopic Eczema in Adults and Children: Systematic Review. Allergy 2009, 64, 258–264. [Google Scholar] [CrossRef]
- Fiocchi, A.; Pawankar, R.; Cuello-Garcia, C.; Ahn, K.; Al-Hammadi, S.; Agarwal, A.; Beyer, K.; Burks, W.; Canonica, G.W.; Ebisawa, M.; et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ. J. 2015, 8, 10. [Google Scholar] [CrossRef]
- Makrgeorgou, A.; Leonardi-Bee, J.; Bath-Hextall, F.J.; Murrell, D.F.; Tang, M.L.K.; Roberts, A.; Boyle, R.J. Probiotics for Treating Eczema. Cochrane Database Syst. Rev. 2018, 11, CD006135. [Google Scholar] [CrossRef] [PubMed]
- Osborn, D.A.; Sinn, J.K. Probiotics in Infants for Prevention of Allergic Disease and Food Hypersensitivity. Cochrane Database Syst. Rev. 2007, 11, CD006475. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Ni, B.; Liu, Z.; Liu, X.; Xie, W.; Wu, I.X.Y.; Li, X. The Role of Probiotics in the Prevention and Treatment of Atopic Dermatitis in Children: An Updated Systematic Review and Meta-Analysis of Randomized Controlled Trials. Paediatr. Drugs 2020, 22, 535–549. [Google Scholar] [CrossRef] [PubMed]
- Tan-Lim, C.S.C.; Esteban-Ipac, N.A.R.; Recto, M.S.T.; Castor, M.A.R.; Casis-Hao, R.J.; Nano, A.L.M. Comparative Effectiveness of Probiotic Strains on the Prevention of Pediatric Atopic Dermatitis: A Systematic Review and Network Meta-Analysis. Pediatr. Allergy Immunol. 2021, 32, 1255–1270. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Su, J. Association of Probiotics with Atopic Dermatitis among Infant: A Meta-Analysis of Randomized Controlled Trials. Oxid. Med. Cell. Longev. 2022, 1–7. [Google Scholar] [CrossRef]
- Chen, L.; Ni, Y.; Wu, X.; Chen, G. Probiotics for the Prevention of Atopic Dermatitis in Infants from Different Geographic Regions: A Systematic Review and Meta-Analysis. J. Dermatolog. Treat. 2022, 33, 2931–2939. [Google Scholar] [CrossRef]
- Zhao, M.; Shen, C.; Ma, L. Treatment Efficacy of Probiotics on Atopic Dermatitis, Zooming in on Infants: A Systematic Review and Meta-Analysis. Int. J. Dermatol. 2018, 57, 635–641. [Google Scholar] [CrossRef]
- Huang, R.; Ning, H.; Shen, M.; Li, J.; Zhang, J.; Chen, X. Probiotics for the Treatment of Atopic Dermatitis in Children: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Front Cell Infect. Microbiol. 2017, 7, 392. [Google Scholar] [CrossRef]
- Nermes, M.; Kantele, J.M.; Atosuo, T.J.; Salminen, S.; Isolauri, E. Interaction of Orally Administered Lactobacillus rhamnosus GG with Skin and Gut Microbiota and Humoral Immunity in Infants with Atopic Dermatitis. Clin. Exp. Allergy 2011, 41, 370–377. [Google Scholar] [CrossRef]
- Fölster-Holst, R.; Müller, F.; Schnopp, N.; Abeck, D.; Kreiselmaier, I.; Lenz, T.; Von Rüden, U.; Schrezenmeir, J.; Christophers, E.; Weichenthal, M. Prospective, Randomized Controlled Trial on Lactobacillus rhamnosus in Infants with Moderate to Severe Atopic Dermatitis. Br. J. Dermatol. 2006, 155, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Pessi, T.; Sütas, Y.; Hurme, M.; Isolauri, E. Interleukin-10 Generation in Atopic Children Following Oral Lactobacillus rhamnosus GG. Clin. Exp. Allergy 2000, 30, 1804–1808. [Google Scholar] [CrossRef] [PubMed]
- Costea, P.I.; Hildebrand, F.; Manimozhiyan, A.; Bäckhed, F.; Blaser, M.J.; Bushman, F.D.; De Vos, W.M.; Ehrlich, S.D.; Fraser, C.M.; Hattori, M.; et al. Enterotypes in the Landscape of Gut Microbial Community Composition. Nat. Microbiol. 2018, 3, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Guo, R.; Zhong, H.; Feng, Q.; Lan, Z.; Qin, B.; Ward, K.J.; Jackson, M.A.; Xia, Y.; Chen, X.; et al. Shotgun Metagenomics of 250 Adult Twins Reveals Genetic and Environmental Impacts on the Gut Microbiome. Cell Syst. 2016, 3, 572–584.e3. [Google Scholar] [CrossRef] [PubMed]
- Climent, E.; Martinez-blanch, J.F.; Llobregat, L.; Ruzafa-costas, B.; Carrión-gutiérrez, M.Á.; Ramírez-boscá, A.; Prieto-merino, D.; Genovés, S.; Codoñer, F.M.; Ramón, D.; et al. Changes in Gut Microbiota Correlates with Response to Treatment with Probiotics in Patients with Atopic Dermatitis. A Post Hoc Analysis of a Clinical Trial. Microorganisms 2021, 9, 854. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Swanson, K.S.; Gibson, G.R.; Hutkins, R.; Reimer, R.A.; Reid, G.; Verbeke, K.; Scott, K.P.; Holscher, H.D.; Azad, M.B.; Delzenne, N.M.; et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Synbiotics. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 687–701. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact Sequence Variants Should Replace Operational Taxonomic Units in Marker-Gene Data Analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2’s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-Species Living Tree Project (LTP)” Taxonomic Frameworks. Nucleic. Acids Res. 2013, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic. Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef: Stat. Ref. Online 2017, 1–15. [Google Scholar] [CrossRef]
- Anderson, M.J. A New Method for Non-Parametric Multivariate Analysis of Variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- Lin, H.; Peddada, S. Das Analysis of Compositions of Microbiomes with Bias Correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Meinshausen, N.; Bühlmann, P. High-Dimensional Graphs and Variable Selection with the Lasso. Ann. Statist. 2006, 34, 1436–1462. [Google Scholar] [CrossRef]
- Stewart, C.J.; Ajami, N.J.; O’Brien, J.L.; Hutchinson, D.S.; Smith, D.P.; Wong, M.C.; Ross, M.C.; Lloyd, R.E.; Doddapaneni, H.V.; Metcalf, G.A.; et al. Temporal Development of the Gut Microbiome in Early Childhood from the TEDDY Study. Nature 2018, 562, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Laman, J.D.; ’t Hart, B.A.; Power, C.; Dziarski, R. Bacterial Peptidoglycan as a Driver of Chronic Brain Inflammation. Trends Mol. Med. 2020, 26, 670–682. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg, A.; Barbarot, S.; Bieber, T.; Christen-Zaech, S.; Deleuran, M.; Fink-Wagner, A.; Gieler, U.; Girolomoni, G.; Lau, S.; Muraro, A.; et al. Consensus-Based European Guidelines for Treatment of Atopic Eczema (Atopic Dermatitis) in Adults and Children: Part I. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 657–682. [Google Scholar] [CrossRef]
- Wollenberg, A.; Barbarot, S.; Bieber, T.; Christen-Zaech, S.; Deleuran, M.; Fink-Wagner, A.; Gieler, U.; Girolomoni, G.; Lau, S.; Muraro, A.; et al. Consensus-Based European Guidelines for Treatment of Atopic Eczema (Atopic Dermatitis) in Adults and Children: Part II. J. Eur. Acad. Dermatol. Venereol. 2018, 32, 850–878. [Google Scholar] [CrossRef] [PubMed]
- Blome, C.; Radtke, M.A.; Eissing, L.; Augustin, M. Quality of Life in Patients with Atopic Dermatitis: Disease Burden, Measurement, and Treatment Benefit. Am. J. Clin. Dermatol. 2016, 17, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Her, Y.; Kim, C.W.; Kim, S.S. Topical Corticosteroid Phobia among Parents of Children with Atopic Eczema in Korea. Ann. Dermatol. 2015, 27, 499. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, Stability and Resilience of the Human Gut Microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The Long-Term Stability of the Human Gut Microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Greenhalgh, K.; Meyer, K.M.; Aagaard, K.M.; Wilmes, P. The Human Gut Microbiome in Health: Establishment and Resilience of Microbiota over a Lifetime. Environ. Microbiol. 2016, 18, 2103–2116. [Google Scholar] [CrossRef]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The Resilience of the Intestinal Microbiota Influences Health and Disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Meng, D.; Weng, M.; Zhu, W.; Wu, W.; Kasper, D.; Walker, W.A. The Symbiotic Bacterial Surface Factor Polysaccharide A on Bacteroides fragilis Inhibits IL-1β-Induced Inflammation in Human Fetal Enterocytes via Toll Receptors 2 and 4. PLoS ONE 2017, 12, e0172738. [Google Scholar] [CrossRef] [PubMed]
- Troy, E.B.; Kasper, D.L. Beneficial Effects of Bacteroides fragilis Polysaccharides on the Immune System. Front. Biosci. 2010, 15, 25. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, C.; Kujawski, M.; Chu, H.; Li, L.; Mazmanian, S.K.; Cantin, E.M. Bacteroides fragilis Polysaccharide A Induces IL-10 Secreting B and T Cells That Prevent Viral Encephalitis. Nat. Commun. 2019, 10, 2153. [Google Scholar] [CrossRef]
- Tang, C.; Tao, J.; Sun, J.; Lv, F.; Lu, Z.; Lu, Y. Regulatory Mechanisms of Energy Metabolism and Inflammation in Oleic Acid-Treated HepG2 Cells from Lactobacillus acidophilus NX2-6 Extract. J. Food Biochem. 2021, 45, e13925. [Google Scholar] [CrossRef]
- Chen, Y.H.; Tsai, W.H.; Wu, H.Y.; Chen, C.Y.; Yeh, W.L.; Chen, Y.H.; Hsu, H.Y.; Chen, W.W.; Chen, Y.W.; Chang, W.W.; et al. Probiotic Lactobacillus Spp. Act Against Helicobacter pylori-Induced Inflammation. J. Clin. Med. 2019, 8, 90. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, S.; Lee, J.H.; Kim, J.H.; Che, X.; Ma, H.W.; Seo, D.H.; Kim, T.I.I.; Kim, W.H.; Kim, S.W.; et al. Lactobacillus acidophilus Suppresses Intestinal Inflammation by Inhibiting Endoplasmic Reticulum Stress. J. Gastroenterol. Hepatol. 2019, 34, 178–185. [Google Scholar] [CrossRef]
- Kim, H.W.; Ju, D.B.; Kye, Y.C.; Ju, Y.J.; Kim, C.G.; Lee, I.K.; Park, S.M.; Choi, I.S.; Cho, K.K.; Lee, S.H.; et al. Galectin-9 Induced by Dietary Probiotic Mixture Regulates Immune Balance to Reduce Atopic Dermatitis Symptoms in Mice. Front. Immunol. 2020, 10, 3063. [Google Scholar] [CrossRef]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.H.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human Gut Microbes Impact Host Serum Metabolome and Insulin Sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A Gut Bacterial Pathway Metabolizes Aromatic Amino Acids into Nine Circulating Metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Zierer, J.; Jackson, M.A.; Kastenmüller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The Fecal Metabolome as a Functional Readout of the Gut Microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef] [PubMed]
- Iikura, Y.; Odajima, Y.; Nagakura, T.; Iinuma, K.; Hayakawa, K.; Oizumi, J. Oral Biotin Treatment Is Effective for Atopic Dermatitis in Children with Low Biotinidase Activity. Acta Paediatr. Scand. 1988, 77, 762–763. [Google Scholar] [CrossRef]
- Kimura, M.; Fukui, T.; Tagami, Y.; Fujiwaki, T.; Yokoyama, M.; Ishioka, C.; Kumasaka, K.; Terada, N.; Yamaguchi, S. Normalization of Low Biotinidase Activity in a Child with Biotin Deficiency after Biotin Supplementation. J. Inherit. Metab. Dis. 2003, 26, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Stücker, M.; Pieck, C.; Stoerb, C.; Niedner, R.; Hartung, J.; Altmeyer, P. Topical Vitamin B12—A New Therapeutic Approach in Atopic Dermatitis-Evaluation of Efficacy and Tolerability in a Randomized Placebo-Controlled Multicentre Clinical Trial. Br. J. Dermatol. 2004, 150, 977–983. [Google Scholar] [CrossRef]
- Chesini, D.; Caminati, M. Vitamin B12 and Atopic Dermatitis: Any Therapeutic Relevance for Oral Supplementation? J. Diet. Suppl. 2022, 19, 238–242. [Google Scholar] [CrossRef]
- Galimberti, F.; Mesinkovska, N.A. Skin Findings Associated with Nutritional Deficiencies. Cleve Clin. J. Med. 2016, 83, 731–739. [Google Scholar] [CrossRef]
- Victoire, A.; Magin, P.; Coughlan, J.; Van Driel, M.L. Interventions for Infantile Seborrhoeic Dermatitis (Including Cradle Cap). Cochrane Database Syst. Rev. 2019, 4, CD011380. [Google Scholar] [CrossRef]
- Mardhiah, M.; Azize, N.A.A.; Yakob, Y.; Affandi, O.; Hock, N.L.; Rowani, M.R.; Habib, A. Clinical, Biochemical and Mutational Findings in Biotinidase Deficiency among Malaysian Population. Mol. Genet. Metab. Rep. 2019, 22, 100548. [Google Scholar] [CrossRef]
- Akgun, A.; Sen, A.; Onal, H. Clinical, Biochemical and Genotypical Characteristics in Biotinidase Deficiency. J. Pediatr. Endocrinol. Metab. 2021, 34, 1425–1433. [Google Scholar] [CrossRef]
- Sakurai-Yageta, M.; Mashimo, Y.; Kuroishi, T.; Ishihara, R.; Shimojo, N.; Kohno, Y.; Okamoto, Y.; Hata, A.; Suzuki, Y. Association between Serum Biotin Levels and Cedar Pollinosis in Japanese Schoolchildren. J. Nutr. Sci. Vitaminol. 2021, 67, 211–216. [Google Scholar] [CrossRef]
- Vieira, B.L.; Lim, N.R.; Lohman, M.E.; Lio, P.A. Complementary and Alternative Medicine for Atopic Dermatitis: An Evidence-Based Review. Am. J. Clin. Dermatol. 2016, 17, 557–581. [Google Scholar] [CrossRef] [PubMed]
- Januchowski, R. Evaluation of Topical Vitamin B12 for the Treatment of Childhood Eczema. J. Altern. Complement. Med. 2009, 15, 387–389. [Google Scholar] [CrossRef] [PubMed]
- Nistico, S.P.; Del Duca, E.; Tamburi, F.; Pignataro, E.; De Carvalho, N.; Farnetani, F.; Pellacani, G. Superiority of a Vitamin B12-Barrier Cream Compared with Standard Glycerol-Petrolatum-Based Emollient Cream in the Treatment of Atopic Dermatitis: A Randomized, Left-to-Right Comparative Trial. Dermatol. Ther. 2017, 30, 12523. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).