Abstract
Rhizobium is a Gram-negative bacterium, which dissolves minerals, produces growth hormones, promotes root growth, and protects plants from different soil-borne pathogens. In the present study, roots, stalks, and fresh weight of maize (Zea mays L.) were significantly increased after soaking in Bradyrhizobium japonicum compared with the control. Subsequently, transcriptome sequencing results of the whole maize plant soaked in B. japonicum showed that multiple growth and development-related genes were up-regulated more than 100-fold compared to the control. Furthermore, the abundance of plant growth promoting bacteria, such as Acidobacteria Subgroup_6 and Chloroflexi KD4-96, were increased significantly. On the contrary, the abundance of multiple pathogens, such as Curvularia, Fusarium and Mycocentrospora, were significantly decreased. Moreover, inoculation with B. japonicum could inhibit the infection of the pathogen Fusarium graminearum in maize. These results suggest that soaking seeds in B. japonicum may affect the expression of maize growth and development-related genes as the bacteria changes the soil microorganism community structure. These findings may help to expand the application of B. japonicum in crop production and provide new opportunities for food security.
1. Introduction
Maize (Zea mays L.) is a cereal crop that is planted extensively worldwide [1,2]; it originated in Mexico or South America about 9000 years ago from teosinte (Zea mays ssp. parviglumis) [3,4]. Subsequently, maize was domesticated in southwestern Amazon [5]. As a staple food, animal feed, and industrial raw material, today maize has become an important part of national food security [1].
Rhizobia are Gram-negative bacteria widely distributed in soil, which infect the roots of legumes to form root nodules, fix molecular nitrogen in the air to form ammonia, provide nitrogen nutrition for plants, and have the ability to improve soil effects. Rhizobia can be divided into the α- proteobacteria class and β- proteobacteria class, of which the former mainly includes Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, Sinorhizobium, Methylobacterium, Ochrobactrum, Aminobacter, Burkholderia, Microvirga, and Phyllobacterium [6,7,8]. Rhizobia can be extracted from a variety of crops, which is important for soil environmental improvement and agricultural production [9,10,11]. For example, rhizobia PEPV16 could produce siderophores and indole acetic acid and solubilize phosphate, as well as promote the growth of Lactuca sativa and Daucus carota [12]. Rhizobia is able to promote the growth and vitality of rice seedlings, and the benefits of early seedling development can be extended to significantly increased grain yield at maturity [13]. In addition, co-inoculation of Rhizobia with endophytes can increase the nodulation ability of α- and β-rhizobia, thus improving the growth of legumes [14]. The inoculation of Rhizobium could promote nitrogen efficiency and increase wheat yield in the Nile delta [15]. Meanwhile, Rhizobia inoculated with Pseudomonas putida, P. fluorescens, or Bacillus cereus can significantly increase the growth and nodulation rate of peas [16].
Soil microorganisms include microscopic organisms present in large numbers in the soil, such as bacteria, fungi, oomycetes, nematodes, protozoa, algae, archaea, and arthropods [17]. The rhizosphere soil of plants contains many microorganisms that are involved in important processes, mainly regulating plant physiology and morphology, promoting plant growth through the production of plant hormones, and acting as protective agents against plant pathogens [18,19]. For example, inoculation with plant growth promoting bacteria can ameliorate the adverse effects of salt stress on wheat yield [20]. Inoculation of Achromobacter piechaudii ARV8 promotes the production of ACC (1-aminocyclopropane-1-carboxylate) deaminase to alleviate drought stress in tomato and pepper [21]. In a different study, two plant growth-promoting bacteria (B. subtilis BS87 and B. megaterium BM89) isolated from the sugarcane rhizosphere increased crop yield potential [22].
The aim of this study was to investigate the relationship between the growth and development of maize soaked in Rhizobium and the microorganism changes in maize rhizosphere soil. We found that root length, stem length, and fresh weight of maize increased significantly after rhizobia treatment. In addition, transcriptome sequencing results showed that maize growth and development-related genes were significantly up-regulated. Furthermore, rhizosphere soil microbial diversity changed significantly, and while the abundance of plant growth promoting bacteria increased significantly, the abundance of pathogenic fungi (Fusarium spp.) decreased significantly. Moreover, B. japonicum soaking could significantly reduce Fusarium damage to maize growth. Our results preliminarily explained the mechanism by which Rhizobium promoted maize growth, and provide new insights for the application of rhizobia to maize.
2. Materials and Methods
2.1. Maize, Rhizobium, and Soil
Maize seeds were provided by the Anhui Agricultural University. The soybean Rhizobium (B. japonicum) was purchased from Ruichu Biotechnology Co., Ltd. (Shanghai, China), while soil was collected from the maize fields at the Anhui Academy of Agricultural Sciences (31°53′ N, 117°14′ E).
2.2. Seed Treatment and Germination Test
Maize seeds were surface sterilized with 2% sodium hypochlorite for 5 min, and then washed thoroughly with autoclaved distilled water. The seeds were soaked in a B. japonicum solution at 25 °C for 3 h. After dressing, the seeds were air dried for 30 min at room temperature. Then, 10 seeds were sown per germination box (with soil) and were watered daily with distilled water. The amount of distilled water per treatment remained the same. Seedling length was recorded after 11 days, and root length and fresh weight were recorded after 20 days. All treatments were placed in a light incubator under the following conditions: 25 ± 1 °C, photoperiod of 16:8 h (L:D), and relative humidity of 70~80%. The B. japonicum soaking group was named ND-372 and the control group was named UT. Each experiment was performed with three replicates.
2.3. RNA Isolation, cDNA Synthesis, and Next-Generation Sequencing
We extracted RNA from the whole maize plant after 20 days. The whole samples were carried out in clean bench. Total RNA was isolated using Trizol Reagent (Invitrogen Life Technologies, Carlsbad, CA, USA), after which the concentration, quality, and integrity were determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA). Three micrograms of RNA were used as input material for RNA sample preparations. Sequencing libraries were generated according to the following steps. First, mRNA was purified from total RNA using poly-T oligomeric magnetic beads. Fragmentation was performed using divalent cations in Illumina proprietary fragmentation buffer at an elevated temperature. First-strand cDNA was synthesized using random oligonucleotides and Super Script II. This was followed by second-strand cDNA synthesis using DNA polymerase I and ribonuclease H. The remaining overhang was converted to blunt ends by exonuclease/polymerase activity, and the enzyme was removed. After adenylation of the 3′ ends of the DNA fragments, Illumina PE adapter oligonucleotides were ligated in preparation for hybridization. Library fragments were purified using the AMPure XP system to screen for cDNA fragments with an optimal length of 400–500 bp (Beckman Coulter, Beverly, CA, USA). DNA fragments connecting adaptor molecules at both ends were selectively enriched in 15 cycles of PCR reactions using an Illumina PCR primer cocktail. Products were purified (AMPure XP system, Beckman, Shanghai, China) and quantified using an Agilent high sensitivity DNA assay on a Bioanalyzer 2100 system (Agilent). The sequencing libraries were sequenced by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China) on a NovaSeq 6000 platform (Illumina).
2.4. DNA Extraction and 16S rDNA Gene Amplicon Sequencing
The rhizosphere soil samples which tightly attached to plant roots (<5 mm) from ND-372 and UT were gently collected. OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, USA) was used to extract the total genomic DNA samples according to the manufacturer’s instructions and stored at −20 °C until further analysis. The quantity and quality of extracted DNAs were determined using a NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis, respectively.
PCR amplification for the bacteria (338F 5′-ACTCCTACGGGAGGCAGCA-3′, 806R 5′-GGACTACHVGGGTWTCTAAT-3′) and fungi (ITS5F GGAAGTAAAAGTCGTAACAAGG, ITS2R GCTGCGTTCTTCATCGATGC) were performed. Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. The PCR components included buffer (5×) 5 μL, fast pfu DNA polymerase (5 U/μL) 0.25 μL, dNTPs (2.5 mM) 2 μL, forward and reverse primer each (10 uM) 1 μL, DNA template 1 μL, and ddH2O 14.75 μL. Thermal cycling consisted of an initial denaturation at 98 °C for 5 min, followed by 25 cycles consisting of denaturation at 98 °C for 30 s, annealing at 53 °C for 30 s, and extension at 72 °C for 45 s, with a final extension at 72 °C for 5 min. PCR amplicons were purified using Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China). Quantification was performed via the Quant-iT PicoGreen double-stranded DNA Detection kit (Invitrogen, Carlsbad, CA, USA). Paired-end 2 × 250 bp sequencing was performed using the Illumina MiSeq platform and MiSeq Kit v3 at Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China).
2.5. Statistical Analysis
Data were expressed as the mean ± standard deviation (SD). The data were statistically analyzed separately for each experiment using one-way ANOVA in SPSS software (v.22.0; SPSS Company, Chicago, IL, USA). Statistical significance was defined at a p-value of 0.05. GraphPad Prism 7 was used to plot the data in the study (GraphPad Software, Inc., San Diego, CA, USA).
HTSeq (0.9.1) statistic was used to compare the read count values of each gene as the original expression of that gene, and then FPKM was used to standardize the expression. Then, DESeq (1.30.0) was used to analyze gene differential expression; the screening conditions were as follows: expression difference multiple |log2FoldChange| > 1, and significant p-value < 0.05. At the same time, the R language Pheatmap (v.1.0.8) software package was used to perform bi-directional clustering analysis of all the different genes of samples. We calculated the distance using the Euclidian method based on the expression levels of the same gene in different samples and the expression patterns of different genes in the same sample, and cluster them using the complete linkage method to generate a heatmap.
Microbiome bioinformatics were conducted using QIIME2 2019.4 with slight modifications based on the official tutorials (https://docs.qiime2.org/2019.4/tutorials/, accessed on 16 February 2022). Sequence data analyses were mainly conducted using QIIME2 and R packages (v.3.2.0). Simply put, QIIME2 and R package (v3.2.0) were used to analyze the sequence data. The ASV table in QIME2 was used to calculate the Chao1 richness estimator, observed species, Shannon diversity index, Simpson index, Faith’s PD, Pielou’s uniformity and Good’s coverage; it was visualized in the form of block diagram. ASV sorting abundance curves were generated and the abundance and evenness of ASVs between samples were compared. Using the Jaccard metric, Bray–Curtis metric, and UniFrac distance metric, beta diversity analysis and visualized group method (UPGMA) hierarchical clustering by principal coordinate analysis (PCoA), non-metric multidimensional scale (NMDS), and unweighted arithmetic means were performed. Principal Component Analysis (PCA) was also based on general level component spectra. Multivariate analysis of variance (PERMANOVA) and similarity analysis (ANOSIM) were used to evaluate the significance of microbial community structure in the inter group differentiation. MEGAN and GraPhlAn were used to visualize their taxonomy composition and abundance. The R package “VennDiagram” was used to generate Venn diagrams to visualize samples or groups based on the occurrence of shared and unique ASVs (regardless of their relative abundance). We used MetagenomeSeq to statistically compare the taxonomic abundance of ASV levels between samples or groups and visualize it as a Manhattan map. Linear discriminant analysis effect size (LEfSe) with default parameters was used to screen out the classification groups with large differences between groups. At the same time, orthogonal partial least squares discriminant analysis (OPLS-DA) was introduced as a supervised model, and the R package “muma” was used to reveal the changes in microbiota between groups. Random forest analysis was performed using QIIME2 default settings to distinguish samples from different groups.
3. Results
3.1. B. japonicum Treatment Promoted Maize Growth
To investigate the role of B. japonicum in maize growth, maize seeds were soaked in a B. japonicum solution (Figure 1A). Root length was significantly increased (1.45 cm on average) in treatment groups (ND-372s) compared with control groups (UTs) (Figure 1B). In addition, seedling length was also significantly increased (4.30 cm on average) in ND-372s compared with UTs (Figure 1C). Moreover, the fresh weight of ND-372 groups were significantly increased (2.00 g on average) compared with UT groups (Figure 1D). These results indicate that soaking seed in B. japonicum can promote maize growth.
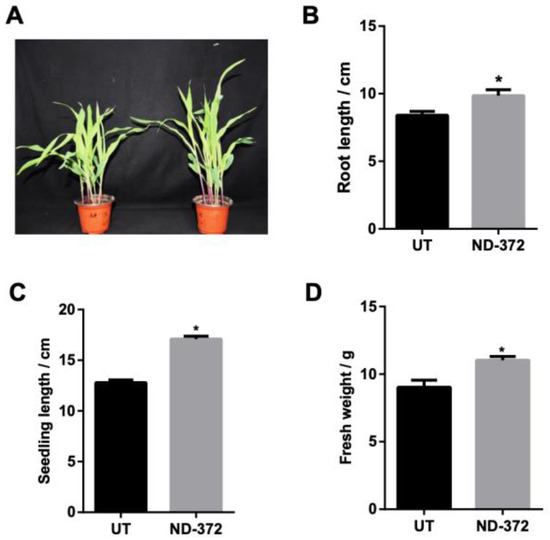
Figure 1.
B. japonicum treatment promotes maize growth. (A) The growth situation of maize. (B) Root length; (C) Seedling length; (D) Fresh weight. Note: ND-372, B. japonicum soaking groups; UT, without B. japonicum soaking groups; the significant differences are marked by stars (Student’s t-test, p < 0.05).
3.2. The Expression of Maize Growth-Related Genes Was Induced by B. japonicum
In order to explore the role of B. japonicum in promoting maize growth, transcriptome sequencing was performed. First, the sample correlation test and PCA analysis showed that the correlation between the treatments was appropriate in both the UT (CK, untreated) and ND-372 (T, treatment) groups, respectively (Figure S1A,B). Second, gene differential expression analysis indicated that 8782 genes were differentially expressed: 5123 were up-regulated and 3659 were down-regulated (Table 1, Figure 2A). In addition, GO enrichment analysis showed that differentially expressed genes were distributed in molecular functions, biological processes, and cell components (Figure 2B). Furthermore, a total of 16 genes related to growth and development were significantly up-regulated more than 20-fold (Table 2). It was reasonable to speculate that B. japonicum treatment was related to maize growth and development.

Table 1.
Number of differentially expressed genes.

Figure 2.
Analysis of differentially expressed genes. (A) Volcanic map of differentially expressed genes. The abscissa is log2FoldChange, and the ordinate is −log10 (p-value). The two vertical dashed lines in the figure are the 2-fold differential expression threshold. The dotted line indicates the threshold of p-value = 0.05. The red dots are the up-regulated genes, the blue dots are the down-regulated genes, and gray dots indicate non-significantly differentially expressed genes. (B) GO enrichment analysis of differentially expressed genes. The abscissa is the term of Go level2, and the ordinate is the enriched −log10 (p-value) of each term.

Table 2.
Significantly up-regulated genes related to growth and development of seed soaked in B. japonicum.
3.3. B. japonicum Treatment Increased the Abundance of Maize Rhizosphere Bacteria
To reveal the role of soil bacteria on maize growth, an Ilumina HiSeq PE250 platform was used to investigate bacteria changes in soil. In total, 527,312 valid sequences and 25,071 operational taxonomic units (OTUs) were detected in eight groups (Table 3). All UT and ND_372 samples contained 4966 OTUs, and conventional OTUs accounted for nearly 19.81%, while the remaining 37.23% and 42.97% were found in UT and ND_372, respectively (Figure S2A).

Table 3.
General statistics of bacteria via the 16S rDNA sequencing.
In order to explore the relationship between soil bacteria and maize growth, we conducted bacteria alpha diversity analysis. Compared with the UT, the observed species values and Chao1 diversity index were higher in the ND_372 groups (Figure 3A,B), indicating that ND_372 had a greater richness. The Shannon and Simpson’s diversity indexes were increased significantly, indicating that bacterial community diversity and evenness in the ND_372 groups were higher than that in UT groups (Figure 3C,D). In addition, Pielou’s evenness and Good’s coverage index were also increased significantly, which indicated a higher level of diversity evolution and evenness (Figure 3E,F). To evaluate the contribution of bacteria, genus composition analysis was performed. The results showed that the abundance of many plant growth promotion-related bacteria, such as subgroups_6 and KD4-96, increased significantly (Figure 3G). The results suggested that soaking seed in B. japonicum may promote the growth of maize by altering the abundance of bacteria.
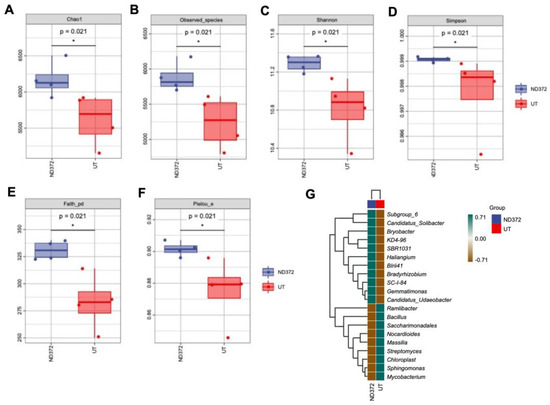
Figure 3.
Alpha diversity index, species differences and marker species of rhizosphere soil bacteria (A) Chao1; (B) Observed_species; (C) Shannon; (D) Simpson; (E) Faith_pd; (F) Pielou_e. Each panel corresponds to an alpha diversity index, identified in the gray area at the top. In each panel, the abscissa is the group label, and the ordinate is the value of the corresponding alpha diversity index. (G) Species differences and marker species of rhizosphere soil bacteria. The samples were clustered according to the Euclidean distance of species composition data (the default clustering algorithm) via UPGMA, and arranged according to the clustering results by default. The significant differences are marked by stars (Student’s t-test, p < 0.05).
3.4. B. japonicum Treatment Affected the Variation of Maize Rhizosphere Fungi
To explore the effects of soil fungi on maize growth, 705,404 valid sequences and 1324 operational taxonomic units (OTUs) were detected in eight groups (Table 4). All UT and ND_372 samples contained 310 OTUs, and conventional OTUs accounted for nearly 23.42%, while the remaining 20.24% and 54.98% were found in UT and ND_372, respectively (Figure S2B).

Table 4.
General statistics of fungi via the 16S rDNA sequencing.
In order to explore the relationship between soil microorganisms and maize growth, we conducted a fungal alpha diversity analysis. Compared with the UT, the observed species values and Chao1 diversity index were higher in ND_372 (Figure 4A,B), indicating that ND_372 had greater richness. There were no significant differences between UT and ND_372 in Shannon, Simpson’s diversity, or Pielou’s evenness indexes, indicating that the microbial community diversity and evenness were similar (Figure 4C–E). However, the Good’s coverage index of the UT group was significantly higher than that of the ND_372 treatment group, indicating that the UT group had better coverage (Figure 4F). To evaluate the contribution of fungi, species composition analysis was performed. The results showed that the abundance of many kinds of microorganisms changed, among which the pathogenic fungi Curvularia, Mycocentrospora and Fusarium decreased significantly (Figure 4G). Moreover, inoculation of rhizobia could significantly reduce the infection of Fusarium graminearum on maize root (Figure 5A,B). Meanwhile, the root length was significantly increased (3.48 cm on average) in treatment groups (ND-372) compared with the control groups (UT) (Figure 5C). The results suggest that B. japonicum treatment changed the soil fungal microbial environment, which may inhibit the pathogenic fungi (e.g., Fusarium species).
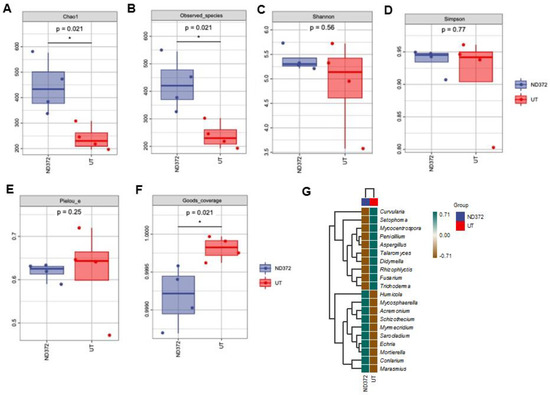
Figure 4.
Alpha diversity index, species differences and marker species of rhizosphere soil fungi. (A) Chao1; (B) Observed_species; (C) Shannon; (D) Simpson; (E) Faith_pd; (F) Pielou_e. Each panel corresponds to an alpha diversity index, identified in the gray area at the top. In each panel, the abscissa is the group label, and the ordinate is the value of the corresponding alpha diversity index. (G) Species differences and marker genera of rhizosphere soil fungi. The samples were clustered according to the Euclidean distance of genera composition data (the default clustering algorithm) via UPGMA, and arranged according to the clustering results by default. The significant differences are marked by stars (Student’s t-test, p < 0.05).
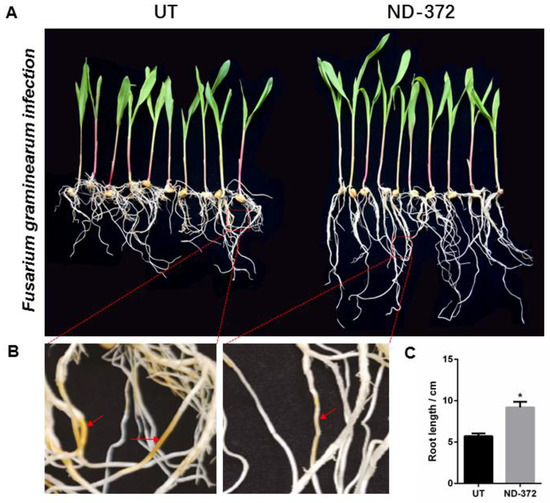
Figure 5.
B. japonicum treatment inhibit the infection of Fusarium on maize roots. (A,B) The left is Fusarium infestation of maize roots; and the right is Fusarium infestation of maize roots with B. japonicum infiltration. (C) Root length. The significant differences are marked by stars (Student’s t-test, p < 0.05).
4. Discussion
Rhizobia are Gram-negative bacilli that exist widely in soil and play an important role in agricultural production [23]. Previous studies have shown that rhizobia are associated with promoting plant growth and improving plant resistance to stress. For example, Rhizobium leguminosarum, isolated from non-leguminous plants, is reported to promote rice growth [11]. Furthermore, dual-inoculation of Rhizobium and mycorrhizal fungi reduced drought stress and increased the relative water content, fat content, and yield of soybean plants [24]. But the effects of B. japonicum on maize growth and rhizosphere soil microorganisms have rarely been reported. In the present study, B. japonicum infiltration was shown to alter the structure of rhizosphere soil microorganisms and to have played a role in regulating the growth and development of maize. In this study, we found that the typical growth indexes, including the root length, seedling length, and fresh weight of maize were significantly increased following B. japonicum treatment. Similarly, co-inoculation with MesoRhizobium ciceris, Pseudomonas sp., and Bacillus sp. significantly increased seed germination and shoot and root length compared with an uninoculated control in Cicer arietinum [25]. It is therefore reasonable to speculate that B. japonicum is involved in promoting maize growth.
Genes contain complete sequences of nucleotides required to produce a polypeptide chain or functional RNA, which notably affect living processes in organisms. In crops, a large number of genes are known to be involved in regulating crop growth and morphological characters. For instance, functions of the TaYUCCA gene family are diverse, and may play an important role in the growth and development of wheat [26]. Gmpskγ1 is a novel PSK encoding gene in soybean, which promotes cell expansion in transgenic plants, thereby increasing seed size and yield [27]. OsGASR9 promotes germination, width, and thickness of rice, while Ghgasa10-1 promotes seedling germination and root elongation of transgenic Arabidopsis thaliana, indicating that GhGASA10-1 promotes cell elongation [28]. Recent studies have shown that knockout of KRN2 in maize or OsKRN2 in rice increases grain yield by ~10% and ~8%, respectively [29]. The Tacol-b5 gene regulates spikelet number, tiller number, and yield per plant in wheat, and field yield tests show that Tacol-b5 significantly promotes wheat yield [30]. Here, we found that several growth and development-related genes of maize, including Zm00001eb397820, Zm00001eb125420, Zm00001eb244140, Zm00001eb204390, and Zm00001eb147260 were significantly up-regulated by >100-fold under B. japonicum treatment, suggesting that B. japonicum infiltration promotes the growth and development of maize. Similar to our results, RNA-seq results from Zhang et al. show that the expression of Zm00001d005892, Zm00001d027925, Zm00001d047017, and Zm00001d039245 are related to root architecture or development in arabidopsis, rice, and maize [31].
The rhizosphere is a narrow zone that is around and affected by plant roots; a large number of microorganisms exist in this region and it is considered as one of the most complex ecosystems [32]. The rhizosphere soil microbial community is considered an indicator of soil quality and yield, which greatly affects the formation of soil fertility and the transformation of plant nutrition. A stable soil microbial community is very important to agricultural ecosystems and is conducive to maintaining the stability of soil systems [33]. Plant rhizosphere bacteria (PGPR) are bacteria that promote plant growth through a variety of mechanisms, including phosphate dissolution, siderophore production, biological nitrogen fixation, and rhizosphere engineering [34]. A number of PGPR have been applied, for example, Pseudomonas, Bacillus, Enterobacter, Klebsiella, Azobacter, Variovorax Azosprillum, and Serratia [35]. In our study, the abundance of multiple plant growth-promoting bacteria, such as Acidobacteria subgroup_6 and Chloroflexi_KD4-96, increased significantly. Consistent with our results, inoculation with SinoRhizobium sp. A15, Bacillus sp. A28, and Sphingomonas sp. A55 increased the abundance and species richness of maize rhizosphere bacteria; moreover, the relative abundances of Acidobacteria subgroup_6, Chloroflexi_KD4-96, Verrucomicrobiae, and Mucilaginibacter at the genus level were positively correlated with maize biomass and yield [36]. In addition, the abundance of many types of microorganisms changed, among which the pathogenic fungi Curvularia and Fusarium decreased significantly. Previous studies have demonstrated that Fusarium head blight (FHB), caused by the fungal pathogen F. graminearum, is an important economic disease of wheat and also causes panicle rot and stem rot verticillium wilt that seriously affect maize yield and quality [37,38]. Our study showed that soaking maize seeds in B. japonicum could increase the abundance of growth-promoting bacteria and reduce the abundance of pathogenic fungi in soil, which may greatly improve maize production.
5. Conclusions
In conclusion, this work reveals that B. japonicum treatment is able to promote maize growth by inducing significant expressions of growth-associated genes in maize. We also found that B. japonicum treatment remodels the rhizosphere microbiomes of maize. Thus, our work provides a novel and efficient method to expand the application of B. japonicum in crop production and provide new opportunities for food security in an environmentally friendly manner.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11071654/s1, Figure S1: Correlation test; Figure S2: Summary of different species and markers in rhizosphere soil bacteria.
Author Contributions
Z.L.: Conceptualization; Formal analysis; Methodology; Visualization; Writing—original draft, review and editing. Y.C.: Investigation; Formal analysis; Validation. X.S.: Investigation; Formal analysis; Validation. Z.Y.: Investigation; Formal analysis. X.R.: Project administration; Data curation; Resources Supervision; Validation; Writing review and editing; Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Project of Anhui Academy of Agricultural Sciences (XJBS-202115).
Data Availability Statement
Data will be made available on request.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Myresiotis, C.K.; Vryzas, Z.; Papadopoulou-Mourkidou, E. Effect of specific plant-growth-promoting rhizobacteria (PGPR) on growth and uptake of neonicotinoid insecticide thiamethoxam in corn (Zea mays L.) Seedlings. Pest Manag. Sci. 2015, 71, 1258–1266. [Google Scholar] [CrossRef]
- Wang, B.; Lin, Z.; Li, X.; Zhao, Y.; Zhao, B.; Wu, G.; Ma, X.; Wang, H.; Xie, Y.; Li, Q.; et al. Genome-wide selection and genetic improvement during modern maize breeding. Nat. Genet. 2020, 52, 565–571. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, X.; Chen, W.; Zhang, R.; Li, Z.; Wen, W.; Warburton, M.L.; Li, J.; Li, H.; Yang, X. Population genomics of Zea species identifies selection signatures during maize domestication and adaptation. BMC Plant Biol. 2022, 22, 72. [Google Scholar] [CrossRef]
- Eakin, H.; Perales, H.; Appendini, K.; Sweeney, S. Selling Maize in Mexico: The persistence of peasant farming in an era of global markets. Dev. Chang. 2014, 45, 133–155. [Google Scholar] [CrossRef]
- Kistler, L.; Maezumi, S.Y.; Gregorio de Souza, J.; Przelomska, N.A.S.; Malaquias Costa, F.; Smith, O.; Loiselle, H.; Ramos-Madrigal, J.; Wales, N.; Ribeiro, E.R.; et al. Multiproxy evidence highlights a complex evolutionary legacy of maize in South America. Science 2018, 362, 1309–1313. [Google Scholar] [CrossRef]
- Gyaneshwar, P.; Hirsch, A.M.; Moulin, L.; Chen, W.-M.; Elliott, G.N.; Bontemps, C.; Estrada-de los Santos, P.; Gross, E.; dos Reis, F.B.; Sprent, J.I.; et al. Legume-nodulating betaproteobacteria: Diversity, host range, and future prospects. Mol. Plant-Microbe Interact. 2011, 24, 1276–1288. [Google Scholar] [CrossRef]
- Rogel, M.A.; Ormeño-Orrillo, E.; Martinez Romero, E. Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst. Appl. Microbiol. 2011, 34, 96–104. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial associations with legumes. CRC Crit. Rev. Plant Sci. 2015, 34, 17–42. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Gao, J.-S.; Cao, Y.-H.; Sheirdil, R.A.; Wang, X.-C.; Zhang, L. Rhizobium oryzicola sp. nov., potential plant-growth-promoting endophytic bacteria isolated from rice roots. Int. J. Syst. Evol. Microbiol. 2015, 65, 2931–2936. [Google Scholar] [CrossRef]
- Celador-Lera, L.; Menéndez, E.; Peix, A.; Igual, J.M.; Velázquez, E.; Rivas, R. Rhizobium zeae sp. nov., isolated from maize (Zea mays L.) Roots. Int. J. Syst. Evol. Microbiol. 2017, 67, 2306–2311. [Google Scholar] [CrossRef]
- Mano, H.; Morisaki, H. Endophytic bacteria in the rice plant. Microbes Environ. 2008, 23, 109–117. [Google Scholar] [CrossRef]
- Flores-Félix, J.D.; Menéndez, E.; Rivera, L.P.; Marcos-García, M.; Martínez-Hidalgo, P.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, M.D.l.E.; García-Fraile, P.; Rivas, R. Use of Rhizobium leguminosarum as a potential biofertilizer for Lactuca sativa and Daucus carota crops. J. Plant Nutr. Soil Sci. 2013, 176, 876–882. [Google Scholar] [CrossRef]
- Biswas, J.C.; Ladha, J.K.; Dazzo, F.B.; Yanni, Y.G.; Rolfe, B.G. Rhizobial inoculation influences seedling vigor and yield of rice. Agron. J. 2000, 92, 880–886. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Tavares, M.J.; Franck, J.; Ali, S.; Glick, B.R.; Rossi, M.J. ACC deaminase plays a major role in Pseudomonas fluorescens yss6 ability to promote the nodulation of Alpha- and Betaproteobacteria rhizobial strains. Arch. Microbiol. 2019, 201, 817–822. [Google Scholar] [CrossRef]
- Yanni, Y.G.; Dazzo, F.B.; Squartini, A.; Zanardo, M.; Zidan, M.I.; Elsadany, A.E.Y. Assessment of the natural endophytic association between Rhizobium and wheat and its ability to increase wheat production in the Nile delta. Plant Soil 2016, 407, 367–383. [Google Scholar] [CrossRef]
- Tilak, K.V.B.R.; Ranganayaki, N.; Manoharachari, C. Synergistic effects of plant-growth promoting rhizobacteria and Rhizobium on nodulation and nitrogen fixation by pigeonpea (Cajanus cajan). Eur. J. Soil Sci. 2006, 57, 67–71. [Google Scholar] [CrossRef]
- Hemkemeyer, M.; Schwalb, S.A.; Heinze, S.; Joergensen, R.G.; Wichern, F. Functions of elements in soil microorganisms. Microbiol. Res. 2021, 252, 126832. [Google Scholar] [CrossRef]
- Ali, S.; Xie, L. Plant growth promoting and stress mitigating abilities of soil born microorganisms. Recent Pat. Food. Nutr. Agric. 2020, 11, 96–104. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Orhan, F. Alleviation of salt stress by halotolerant and halophilic plant growth-promoting bacteria in wheat (Triticum aestivum). Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2016, 47, 621–627. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 2004, 166, 525–530. [Google Scholar] [CrossRef]
- Chandra, A.; Chandra, P.; Tripathi, P. Whole genome sequence insight of two plant growth-promoting bacteria (B. subtilis BS87 and B. megaterium BM89) isolated and characterized from sugarcane rhizosphere depicting better crop yield potentiality. Microbiol. Res. 2021, 247, 126733. [Google Scholar] [CrossRef]
- Ouyabe, M.; Tanaka, N.; Shiwa, Y.; Fujita, N.; Kikuno, H.; Babil, P.; Shiwachi, H. Rhizobium dioscoreae sp. nov., a plant growth-promoting bacterium isolated from yam (Dioscorea species). Int. J. Syst. Evol. Microbiol. 2020, 70, 5054–5062. [Google Scholar] [CrossRef]
- Igiehon, N.O.; Babalola, O.O.; Cheseto, X.; Torto, B. Effects of rhizobia and arbuscular mycorrhizal fungi on yield, size distribution and fatty acid of soybean seeds grown under drought stress. Microbiol. Res. 2021, 242, 126640. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, T.; Wang, H.; Feng, D. Genome-wide identification and expression analysis of the tayucca gene family in wheat. Mol. Biol. Rep. 2021, 48, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Liu, Y.; Zeng, S.; Yan, J.; Wang, E.; Luo, L. Expression of a novel PSK-encoding gene from soybean improves seed growth and yield in transgenic plants. Planta 2019, 249, 1239–1250. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Sun, Y.; Tian, Z.; Fu, G.; Pei, X.; Pan, Z.; Nazir, M.F.; Song, S.; Li, H.; Wang, X.; et al. Ghgasa10-1 promotes the cell elongation in fiber development through the phytohormones IAA-induced. BMC Plant Biol. 2021, 21, 448. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 2022, 375, eabg7985. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, H.; Li, T.; Wu, J.; Nagarajan, R.; Lei, L.; Powers, C.; Kan, C.-C.; Hua, W.; Liu, Z.; et al. Tacol-B5 modifies spike architecture and enhances grain yield in wheat. Science 2022, 376, 180–183. [Google Scholar] [CrossRef]
- Zhang, C.; Li, X.; Wang, Z.; Zhang, Z.; Wu, Z. Identifying key regulatory genes of maize root growth and development by RNA sequencing. Genomics 2020, 112, 5157–5169. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottova, D.; Kristufek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef]
- Vejan, P.; Abdullah, R.; Khadiran, T.; Ismail, S.; Nasrulhaq Boyce, A. Role of Plant Growth Promoting Rhizobacteria in Agricultural Sustainability—A Review. Molecules 2016, 21, 573. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Chen, L.; Hao, Z.; Li, K.; Sha, Y.; Wang, E.; Sui, X.; Mi, G.; Tian, C.; Chen, W. Effectsof growth-promoting rhizobacteria on maize growth and rhizosphere microbial community under conservation tillage in Northeast China. Microbiol. Biotechnol. 2021, 14, 535–550. [Google Scholar] [CrossRef]
- Ge, C.; Tang, C.; Zhu, Y.-X.; Wang, G.-F. Genome-wide identification of the maize 2OGD superfamily genes and their response to Fusarium verticillioides and Fusarium graminearum. Gene 2021, 764, 145078. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Zhang, L.; Yin, Z.; Liang, Y.; Chen, L.; Zou, S.; Dong, H. The golgin protein RUD3 regulates Fusarium graminearum growth and virulence. Appl. Environ. Microbiol. 2021, 87, e02522-20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).