A Bacteriological Comparison of the Hemolymph from Healthy and Moribund Unionid Mussel Populations in the Upper Midwestern U.S.A. Prompts the Development of Diagnostic Assays to Detect Yokenella regensburgei
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Sites
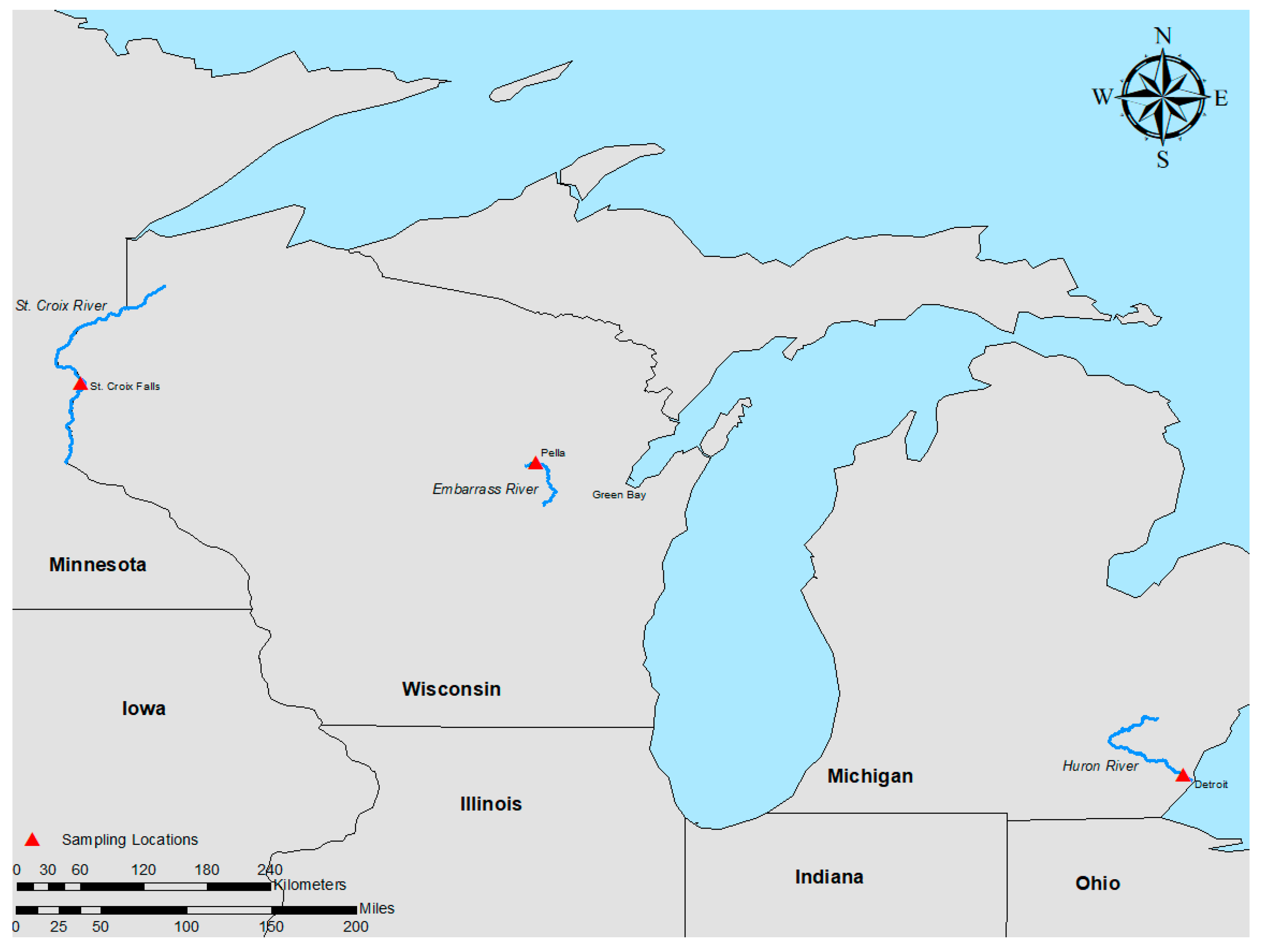
2.1.1. Embarrass River (Wisconsin; 44°44′23.44″ N; 88°47′54.94″ W)
2.1.2. Huron River Sampling (Michigan; 42°5′19.79″ N; 83°16′57.98″ W)
2.1.3. St. Croix River Sampling (Wisconsin; 45°23′56.03″ N; 92°38′59.53″ W)
2.2. Hemolymph Sample Collection
2.3. Development of Yokenella-Specific Diagnostic Assays
2.3.1. qPCR Development and Evaluation
2.3.2. End-Point Conventional PCR (cPCR) Development and Evaluation
2.4. Molecular Evaluation of Bacterial Isolates
3. Results
3.1. Survey Results and Bacterial Isolation
3.2. Assay Development
3.2.1. qPCR Assay
3.2.2. End-Point cPCR Assay
3.2.3. Molecular Evaluation of Bacterial Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vaughn, C.C. Ecosystem Services Provided by Freshwater Mussels. Hydrobiologia 2018, 810, 15–27. [Google Scholar] [CrossRef]
- Williams, J.D.; Warren, M.L.; Cummings, K.S.; Harris, J.L.; Neves, R.J. Conservation Status of Freshwater Mussels of the United States and Canada. Fisheries 1993, 18, 6–22. [Google Scholar] [CrossRef]
- Strayer, D.L.; Downing, J.A.; Haag, W.R.; King, T.L.; Layzer, J.B.; Newton, T.J.; Nichols, S.J. Changing Perspectives on Pearly Mussels, North America’s Most Imperiled Animals. BioScience 2004, 54, 429. [Google Scholar] [CrossRef]
- Haag, W.R. Reassessing Enigmatic Mussel Declines in the United States. Freshw. Mollusk Biol. Conserv. 2019, 22, 43. [Google Scholar] [CrossRef]
- Grizzle, J.M.; Brunner, C.J. Infectious Diseases of Freshwater Mussels and Other Freshwater Bivalve Mollusks. Rev. Fish. Sci. 2009, 17, 425–467. [Google Scholar] [CrossRef]
- Leis, E.; Erickson, S.; Waller, D.; Richard, J.; Goldberg, T. A Comparison of Bacteria Cultured from Unioinid Mussel Hemolymph Between Stable Populations in the Upper Mississippi River Basin and Populations Affected by a Mortality Event in the Clinch River. Freshw. Mollusk Biol. Conserv. 2019, 22, 70–80. [Google Scholar]
- Richard, J.C.; Leis, E.; Dunn, C.D.; Agbalog, R.; Waller, D.; Knowles, S.; Putnam, J.; Goldberg, T.L. Mass Mortality in Freshwater Mussels (Actinonaias Pectorosa) in the Clinch River, USA, Linked to a Novel Densovirus. Sci. Rep. 2020, 10, 14498. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.C.; Campbell, L.J.; Leis, E.M.; Agbalog, R.E.; Dunn, C.D.; Waller, D.L.; Knowles, S.; Putnam, J.G.; Goldberg, T.L. Mussel Mass Mortality and the Microbiome: Evidence for Shifts in the Bacterial Microbiome of a Declining Freshwater Bivalve. Microorganisms 2021, 9, 1976. [Google Scholar] [CrossRef]
- Richard, J.C.; Leis, E.M.; Dunn, C.D.; Harris, C.; Agbalog, R.E.; Campbell, L.J.; Knowles, S.; Waller, D.L.; Putnam, J.G.; Goldberg, T.L. Freshwater Mussels Show Elevated Viral Richness and Intensity during a Mortality Event. Viruses 2022, 14, 2603. [Google Scholar] [CrossRef]
- Leis, E.M.; Dziki, S.; Richard, J.; Agbalog, R.; Waller, D.; Putnam, J.; Knowles, S.; Goldberg, T. Further Bacteriological Analysis of Annual Pheasantshell (Actinonaias Pectorosa) Mussel Mortality Events in the Clinch River (Virginia/Tennessee), USA, Reveals a Consistent Association with Yokenella Regensburgei. Freshw. Mollusk Biol. Conserv. 2023, 26, 1–10. [Google Scholar] [CrossRef]
- Knowles, S.; Leis, E.M.; Richard, J.C.; Cole, R.; Agbalog, R.E.; Putnam, J.G.; Goldberg, T.L.; Waller, D.L. A Novel Gonadotropic Microsporidian Parasite (Microsporidium Clinchi n. Sp.) Infecting a Declining Population of Pheasantshell Mussels (Actinonaias Pectorosa) (Unioinidae) from the Clinch River, USA. Parasitologia 2022, 2, 1–12. [Google Scholar] [CrossRef]
- Starliper, C.E.; Villella, R.; Morrison, P.; Mathias, J. Studies on the Bacterial Flora of Native Freshwater Bivalves from the Ohio River. Biomed. Lett. 1998, 58, 85–95. [Google Scholar]
- Starliper, C.E.; Neves, R.J.; Hanlon, S.; Whittington, P. A Survey of the Indigenous Microbiota (Bacteria) in Three Species of Mussels from the Clinch and Holston Rivers, Virginia. J. Shellfish Res. 2008, 27, 1311–1317. [Google Scholar] [CrossRef]
- Starliper, C.E.; Powell, J.; Garner, J.T.; Schill, W.B. Predominant Bacteria Isolated from Moribund Fusconaia Ebena Ebonyshells Experiencing Die-Offs in Pickwick Reservoir, Tennessee River, Alabama. J. Shellfish Res. 2011, 30, 359–366. [Google Scholar] [CrossRef]
- Balamayooran, G.; Cooper, C.; Paul, N.C.; Ferro, P.J.; Rice, L.; Gomez, G.; Díaz-Delgado, J. Yokenella Regensburgei, a Novel Pathogen in Farmed American Alligators. Vet. Pathol. 2022, 59, 476–481. [Google Scholar] [CrossRef]
- Piette, R. Distribution and Relative Abundance of Snuffbox (Epioblasma Triquetra) in the Wolf River System; Wisconsin Department of Natural Resources: Madison, WI, USA, 2014; p. 51. [Google Scholar]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Standish, I.; Leis, E.; Erickson, S.; McCann, R.; Puzach, C.; Katona, R.; Lark, E.; Bailey, J.; Kleman, E.; Buening, J.; et al. Vagococcus Salmoninarum II—QPCR, Tropism and Egg-associated Transmission. J. Fish Dis. 2020, 43, 317–325. [Google Scholar] [CrossRef]
- Standish, I.; Leis, E.; Schmitz, N.; Credico, J.; Erickson, S.; Bailey, J.; Kerby, J.; Phillips, K.; Lewis, T. Optimizing, Validating, and Field Testing a Multiplex QPCR for the Detection of Amphibian Pathogens. Dis. Aquat. Org. 2018, 129, 1–13. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Gonçalves Pessoa, R.B.; de Oliveira, W.F.; Marques, D.S.C.; dos Santos Correia, M.T.; de Carvalho, E.V.M.M.; Coelho, L.C.B.B. The Genus Aeromonas: A General Approach. Microb. Pathog. 2019, 130, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Schilling, K.; Zessner, M. Foam in the Aquatic Environment. Water Res. 2011, 45, 4355–4366. [Google Scholar] [CrossRef] [PubMed]

| Sampling Date | Location | Mussel Species | Number Sampled | Genus Identification | Prevalence |
|---|---|---|---|---|---|
| 21 August 2018 | St. Croix River | Mucket | 12 | Acinetobacter | 2/12 |
| Actinonaias ligamentina | Aeromonas | 1/12 | |||
| Arthrobacter | 2/12 | ||||
| Bacillus | 5/12 | ||||
| Brevundimonas | 1/12 | ||||
| Ensifer | 1/12 | ||||
| Enterobacter | 1/12 | ||||
| Exiguobacterium | 7/12 | ||||
| Microbacterium | 1/12 | ||||
| Paenibacillus | 1/12 | ||||
| Pseudarthrobacter | 1/12 | ||||
| Pseudomonas | 2/12 | ||||
| Rhodococcus | 1/12 | ||||
| Terracoccus | 1/12 | ||||
| 5 October 2018 | Embarrass River | Mucket | 8 | Aeromonas | 3/8 |
| Actinonaias ligamentina | Chitinibacter | 1/8 | |||
| Flavobacterium | 1/8 | ||||
| Pseudomonas | 1/8 | ||||
| Yokenella | 1/8 | ||||
| Plain Pocketbook | 4 | Aeromonas | 2/4 | ||
| Lampsilis cardium | Yokenella | 1/4 | |||
| Fragile Papershell | 3 | Aeromonas | 1/3 | ||
| Leptodea fragilis | Bacillus | 1/3 | |||
| Flavobacterium | 1/3 | ||||
| Pseudomonas | 1/3 | ||||
| Fat Mucket | 2 | Aeromonas | 2/2 | ||
| Lampsilis siliquoidea | Bacillus | 1/2 | |||
| Chryseobacterium | 1/2 | ||||
| Pseudomonas | 1/2 | ||||
| Serratia | 1/2 | ||||
| Pink Heelsplitter | 4 | Aeromonas | 2/4 | ||
| Potamilus alatus | Yokenella | 2/4 | |||
| Deertoe | 7 | Aeromonas | 3/7 | ||
| Truncilla truncata | Bacillus | 2/7 | |||
| Curtobacterium | 1/7 | ||||
| Pseudomonas | 3/7 | ||||
| Sphingobacterium | 2/7 | ||||
| Yokenella | 2/7 | ||||
| 10 September 2019 | Huron River | Mucket | 20 | Aeromonas | 4/20 |
| Actinonaias ligamentina | Bacillus | 1/20 | |||
| Erwinia | 1/20 | ||||
| Microbacterium | 1/20 | ||||
| Micrococcus | 1/20 | ||||
| Pseudomonas | 1/20 | ||||
| Rhodococcus | 1/20 | ||||
| Staphylococcus | 1/20 | ||||
| Vogesella | 2/20 | ||||
| 23 September 2020 | Huron River | Mucket | 21 | Acidovorax | 1/21 |
| Actinonaias ligamentina | Acinetobacter | 2/21 | |||
| Aeromonas | 1/21 | ||||
| Bacillus | 1/21 | ||||
| Chromobacterium | 1/21 | ||||
| Chryseobacterium | 1/21 | ||||
| Delftia | 1/21 | ||||
| Flavobacterium | 1/21 | ||||
| Novosphingobium | 1/21 | ||||
| Pseudomonas | 1/21 | ||||
| Rheinheimera | 1/21 | ||||
| Staphylococcus | 1/21 |
| gBlock® Copies | Cq Mean | Cq SD | Cq CV(%) |
|---|---|---|---|
| Intra-assay | |||
| 10,000,000 | 16.94 | 0.12 | 0.69 |
| 1,000,000 | 20.30 | 0.09 | 0.43 |
| 100,000 | 23.20 | 0.10 | 0.43 |
| 10,000 | 26.74 | 0.12 | 0.45 |
| 1000 | 29.68 | 0.09 | 0.29 |
| 100 | 33.30 | 0.13 | 0.40 |
| 10 | 36.60 | 0.31 | 0.84 |
| 1 | 39.24 | 0.38 | 0.96 |
| Inter-assay | |||
| 10,000,000 | 17.15 | 0.25 | 1.46 |
| 1,000,000 | 20.34 | 0.19 | 0.91 |
| 100,000 | 23.02 | 0.29 | 1.25 |
| 10,000 | 26.54 | 0.44 | 1.65 |
| 1000 | 29.41 | 0.36 | 1.22 |
| 100 | 33.47 | 0.33 | 1.00 |
| 10 | 36.49 | 0.34 | 0.93 |
| 1 | 39.36 | 0.35 | 0.89 |
| gBlock® Copies | SQ Mean | SQ SD | SQ CV(%) |
|---|---|---|---|
| 10,000,000 | 11,070,000.00 | 594,474.56 | 5.37 |
| 1,000,000 | 988,175.00 | 53,841.36 | 5.45 |
| 100,000 | 123,175.00 | 11,160.73 | 9.06 |
| 10,000 | 10,369.25 | 343.27 | 3.31 |
| 1000 | 1172.50 | 51.65 | 4.40 |
| 100 | 91.14 | 7.07 | 7.76 |
| 10 | 8.31 | 1.55 | 18.68 |
| 1 | 1.50 | NA | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leis, E.M.; Dziki, S.; Standish, I.; Waller, D.; Richard, J.; Weinzinger, J.; Harris, C.; Knowles, S.; Goldberg, T. A Bacteriological Comparison of the Hemolymph from Healthy and Moribund Unionid Mussel Populations in the Upper Midwestern U.S.A. Prompts the Development of Diagnostic Assays to Detect Yokenella regensburgei. Microorganisms 2023, 11, 1068. https://doi.org/10.3390/microorganisms11041068
Leis EM, Dziki S, Standish I, Waller D, Richard J, Weinzinger J, Harris C, Knowles S, Goldberg T. A Bacteriological Comparison of the Hemolymph from Healthy and Moribund Unionid Mussel Populations in the Upper Midwestern U.S.A. Prompts the Development of Diagnostic Assays to Detect Yokenella regensburgei. Microorganisms. 2023; 11(4):1068. https://doi.org/10.3390/microorganisms11041068
Chicago/Turabian StyleLeis, Eric M., Sara Dziki, Isaac Standish, Diane Waller, Jordan Richard, Jesse Weinzinger, Cleyo Harris, Susan Knowles, and Tony Goldberg. 2023. "A Bacteriological Comparison of the Hemolymph from Healthy and Moribund Unionid Mussel Populations in the Upper Midwestern U.S.A. Prompts the Development of Diagnostic Assays to Detect Yokenella regensburgei" Microorganisms 11, no. 4: 1068. https://doi.org/10.3390/microorganisms11041068
APA StyleLeis, E. M., Dziki, S., Standish, I., Waller, D., Richard, J., Weinzinger, J., Harris, C., Knowles, S., & Goldberg, T. (2023). A Bacteriological Comparison of the Hemolymph from Healthy and Moribund Unionid Mussel Populations in the Upper Midwestern U.S.A. Prompts the Development of Diagnostic Assays to Detect Yokenella regensburgei. Microorganisms, 11(4), 1068. https://doi.org/10.3390/microorganisms11041068


_Brussaard.png)



