Capture ELISA for KPC Detection in Gram-Negative Bacilli: Development and Standardisation
Abstract
1. Introduction
2. Materials and Methods
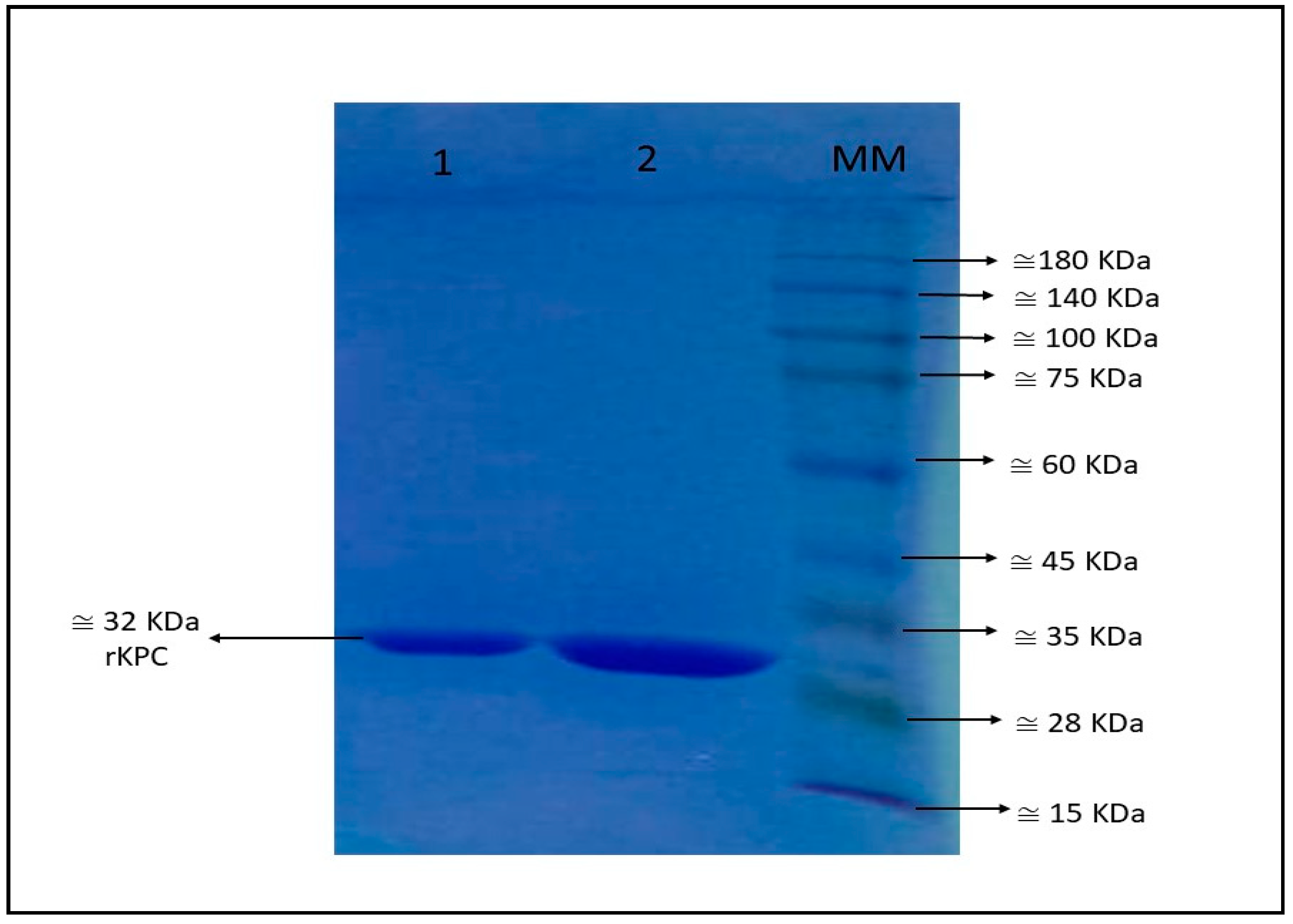
2.1. Expression and Purification of Recombinant KPC (rKPC-2)
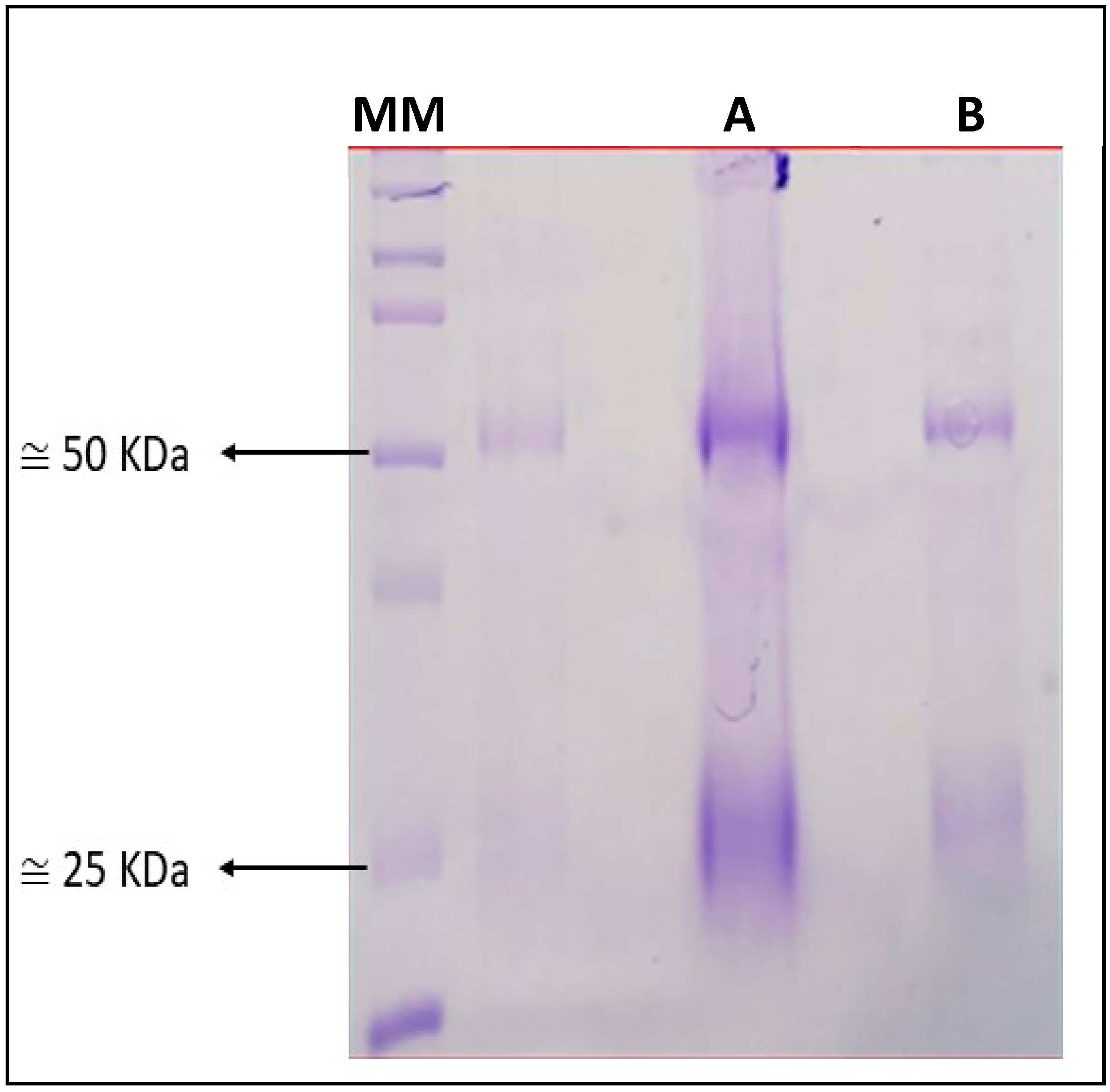
2.2. Production of Antibodies
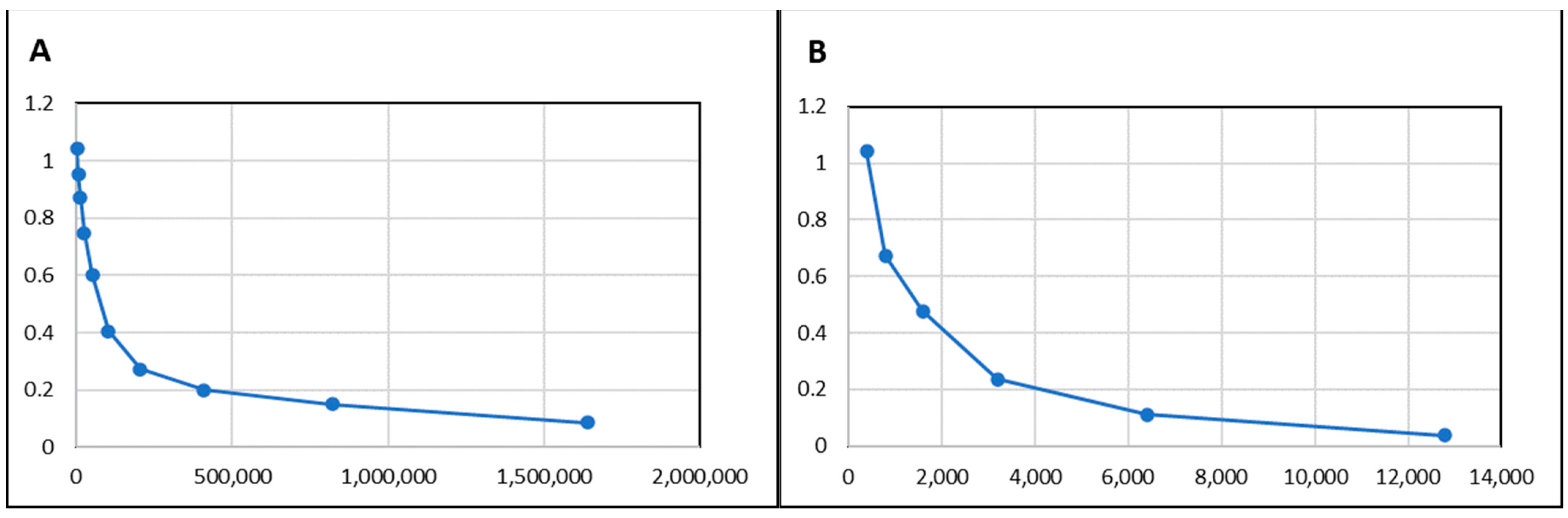
2.3. Checkboard Titration
2.4. Capture ELISA Immunoassay
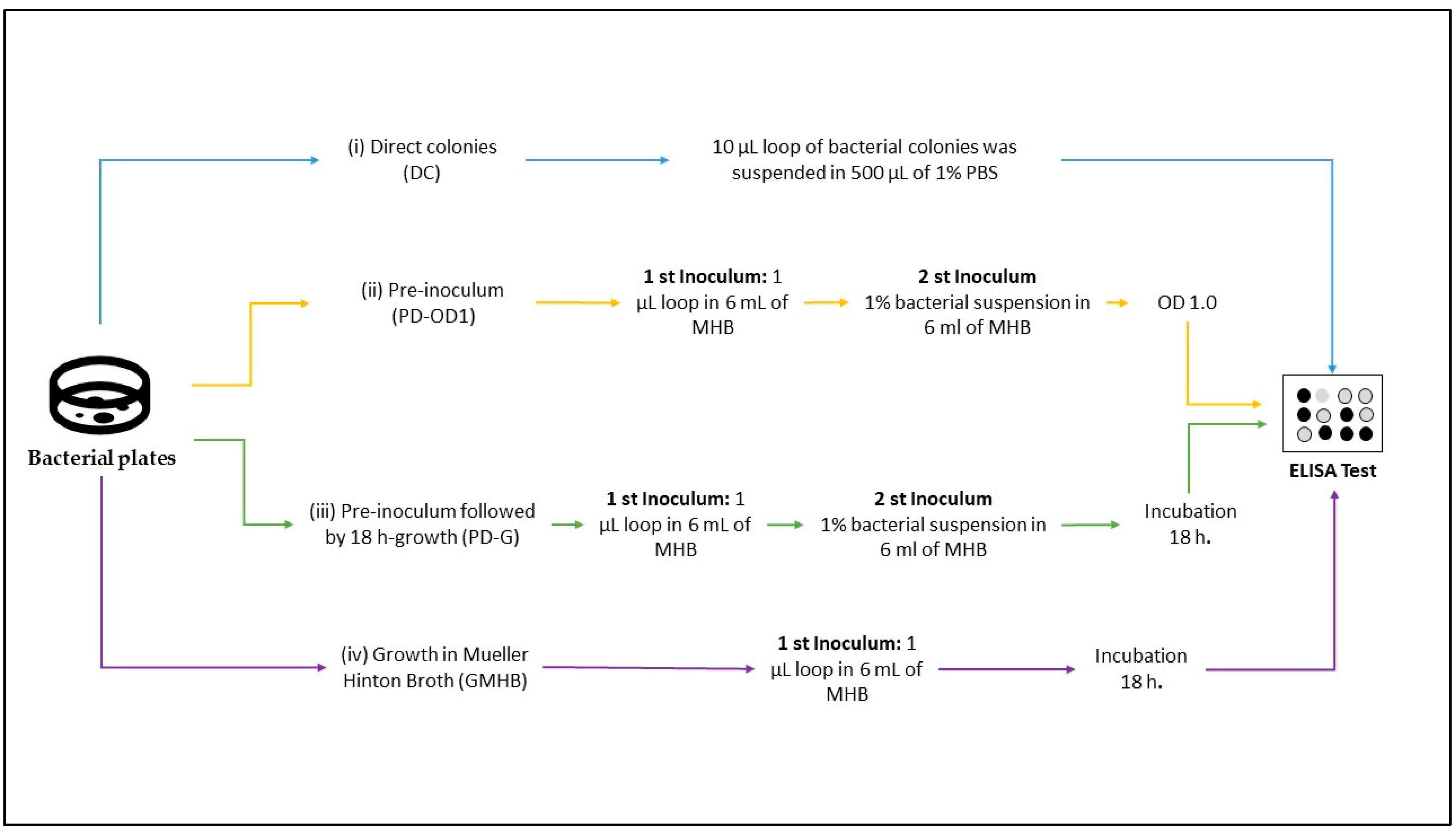
2.5. Bacterial Sample Preparation
2.6. Validation of Capture ELISA
2.7. Data Analysis
3. Results
3.1. Recombinant blaKPC-2
3.2. Immunisation with rKPC-2 Induced Specific Serum Antibody Response in Rabbit and Mice and No Cross-Reactivity
3.3. Different Bacterial Preparation Induced Different Results in ELISA
3.4. Validation of KPC Detection
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization Ten Threats to Global Health in 2019. Available online: https://www.who.int/news-room/spotlight/ten-threats-to-global-health-in-2019 (accessed on 20 February 2023).
- Dadgostar, P. Antimicrobial Resistance: Implications and Costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P. The structure of β-lactamases. Philos. Trans. R Soc. Lond. B Biol. Sci. 1980, 289, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Worthington, R.J.; Melander, C. Overcoming resistance to β-lactam antibiotics. J. Org. Chem. 2013, 78, 4207–4213. [Google Scholar] [CrossRef]
- Lee, C.R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F.; et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among Carbapenem-Resistant Enterobacteriaceae Isolated from Adult and Children Patients in China. Front. Cell Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Reyes, J.A.; Melano, R.; Cárdenas, P.A.; Trueba, G. Mobile genetic elements associated with carbapenemase genes in South American Enterobacterales. Braz. J. Infect. Dis. 2020, 24, 231–238. [Google Scholar] [CrossRef]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef]
- Ducomble, T.; Faucheux, S.; Helbig, U.; Kaisers, U.X.; König, B.; Knaust, A.; Lübbert, C.; Möller, I.; Rodloff, A.C.; Schweickert, B.; et al. Large hospital outbreak of KPC-2-producing Klebsiella pneumoniae: Investigating mortality and the impact of screening for KPC-2 with polymerase chain reaction. J. Hosp. Infect. 2015, 89, 179–185. [Google Scholar] [CrossRef]
- Martin, A.; Fahrbach, K.; Zhao, Q.; Lodise, T. Association between Carbapenem Resistance and Mortality among Adult, Hospitalized Patients with Serious Infections Due to Enterobacteriaceae: Results of a Systematic Literature Review and Meta-analysis. Open Forum. Infect. Dis. 2018, 5, ofy150. [Google Scholar] [CrossRef] [PubMed]
- Karaiskos, I.; Lagou, S.; Pontikis, K.; Rapti, V.; Poulakou, G. The “Old” and the “New” Antibiotics for MDR Gram-Negative Pathogens: For Whom, When, and How. Front. Public Health 2019, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L.; Dortet, L. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 2012, 18, 1503–1507. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. Epidemiology and Diagnostics of Carbapenem Resistance in Gram-negative Bacteria. Clin. Infect. Dis. 2019, 69 (Suppl. S7), S521–S528. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.C.; Gales, A.C.; Monteiro, J.; Silbert, S.; Chagas-Neto, T.; Machado, A.M.O.; Carvalhaes, C.G. Evaluation of a rapid immunochromatographic test for detection of distinct variants of Klebsiella pneumoniae carbapenemase (KPC) in Enterobacteriaceae. J. Microbiol. Methods 2017, 142, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Bianco, G.; Boattini, M.; van Asten, S.A.V.; Iannaccone, M.; Zanotto, E.; Zaccaria, T.; Bernards, A.T.; Cavallo, R.; Costa, C. RESIST-5 O.O.K.N.V. and NG-Test Carba 5 assays for the rapid detection of carbapenemase-producing cultures: A comparative study. J. Hosp. Infect. 2020, 105, 162–166. [Google Scholar] [CrossRef]
- Castanheira, M.; Arends, S.J.R.; Davis, A.P.; Woosley, L.N.; Bhalodi, A.A.; MacVane, S.H. Analyses of a Ceftazidime-Avibactam-Resistant Citrobacter freundii Isolate Carrying blaKPC-2 Reveals a Heterogenous Population and Reversible Genotype. mSphere 2018, 3, e00408-18. [Google Scholar] [CrossRef]
- Nicoletti, A.G.; Marcondes, M.F.; Martins, W.M.; Almeida, L.G.; Nicolas, M.F.; Vasconcelos, A.T.; Oliveira, V.; Gales, A.C. Characterization of BKC-1 class A carbapenemase from Klebsiella pneumoniae clinical isolates in Brazil. Antimicrob. Agents Chemother. 2015, 59, 5159–5164. [Google Scholar] [CrossRef]
- Alhajj, M.; Farhana, A. Enzyme Linked Immunosorbent Assay. [Updated 6 February 2021]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK555922 (accessed on 12 June 2021).
- Kumar, M.; Nandi, S.; Chidri, S. Development of a polyclonal antibody-based AC-ELISA and its comparison with PCR for diagnosis of Canine parvovirus infection. Virol. Sin. 2010, 25, 352–360. [Google Scholar] [CrossRef]
- Sreenivasa Murthy, G.S.; D’Souza, P.E.; Shrikrishna Isloor, K. Evaluation of a polyclonal antibody-based sandwich ELISA for the detection of faecal antigens in Schistosoma spindale infection in bovines. J. Parasit. Dis. 2013, 37, 47–51. [Google Scholar] [CrossRef]
- Chen, R.; Shang, H.; Niu, X.; Huang, J.; Miao, Y.; Sha, Z.; Zhu, R. Establishment and evaluation of an indirect ELISA for detection of antibodies to goat Klebsiella pneumonia. BMC Vet. Res. 2021, 17, 107. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, A.; Plückthun, A. Reproducibility: Standardize antibodies used in research. Nature 2015, 518, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, M.; Mukumoto, M.; Miyatake, K. Production and purification of polyclonal antibodies. Methods Mol. Biol. 2010, 657, 63–74. [Google Scholar] [CrossRef]
- Ascoli, C.A.; Aggeler, B. Overlooked benefits of using polyclonal antibodies. Biotechniques 2018, 65, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Aitken, S.L.; Tarrand, J.J.; Deshpande, L.M.; Tverdek, F.P.; Jones, A.L.; Shelburne, S.A.; Prince, R.A.; Bhatti, M.M.; Rolston, K.V.I.; Jones, R.N.; et al. High Rates of Nonsusceptibility to Ceftazidime-avibactam and Identification of New Delhi Metallo-β-lactamase Production in Enterobacteriaceae Bloodstream Infections at a Major Cancer Center. Clin. Infect. Dis. 2016, 63, 954–958. [Google Scholar] [CrossRef]
- Fujikawa, H.; Morozumi, S. Modeling surface growth of Escherichia coli on agar plates. Appl. Environ. Microbiol. 2005, 71, 7920–7926. [Google Scholar] [CrossRef]
- Fortuin, S.; Nel, A.J.M.; Blackburn, J.M.; Soares, N.C. Comparison between the proteome of Escherichia coli single colony and during liquid culture. J. Proteomics. 2020, 228, 103929. [Google Scholar] [CrossRef]
- Warren, M.R.; Sun, H.; Yan, Y.; Cremer, J.; Li, B.; Hwa, T. Spatiotemporal establishment of dense bacterial colonies growing on hard agar. eLife 2019, 8, e41093. [Google Scholar] [CrossRef]
- Gray, D.A.; Dugar, G.; Gamba, P.; Strahl, H.; Jonker, M.J.; Hamoen, L.W. Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat. Commun. 2019, 10, 890. [Google Scholar] [CrossRef]
- Jaishankar, J.; Srivastava, P. Molecular Basis of Stationary Phase Survival and Applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef]
- Kragh, K.N.; Alhede, M.; Rybtke, M.; Stavnsberg, C.; Jensen, P.Ø.; Tolker-Nielsen, T.; Whiteley, M.; Bjarnsholt, T. The Inoculation Method Could Impact the Outcome of Microbiological Experiments. Appl. Environ. Microbiol. 2018, 84, e02264-17. [Google Scholar] [CrossRef] [PubMed]
- Somerville, G.A.; Proctor, R.A. Cultivation conditions and the diffusion of oxygen into culture media: The rationale for the flask-to-medium ratio in microbiology. BMC Microbiol. 2013, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2021, 72, e169–e183. [Google Scholar] [CrossRef] [PubMed]
- Shields, R.K.; Nguyen, M.H.; Press, E.G.; Chen, L.; Kreiswirth, B.N.; Clancy, C.J. Emergence of Ceftazidime-Avibactam Resistance and Restoration of Carbapenem Susceptibility in Klebsiella pneumoniae Carbapenemase-Producing K pneumoniae: A Case Report and Review of Literature. Open Forum. Infect. Dis. 2017, 4, ofx101. [Google Scholar] [CrossRef] [PubMed]
- Alsenani, T.A.; Viviani, S.L.; Kumar, V.; Taracila, M.A.; Bethel, C.R.; Barnes, M.D.; Papp-Wallace, K.M.; Shields, R.K.; Nguyen, M.H.; Clancy, C.J.; et al. Structural Characterization of the D179N and D179Y Variants of KPC-2 β-Lactamase: Ω-Loop Destabilization as a Mechanism of Resistance to Ceftazidime-Avibactam. Antimicrob. Agents Chemother. 2022, 66, e0241421. [Google Scholar] [CrossRef]
- Tsolaki, V.; Mantzarlis, K.; Mpakalis, A.; Malli, E.; Tsimpoukas, F.; Tsirogianni, A.; Papagiannitsis, C.; Zygoulis, P.; Papadonta, M.E.; Petinaki, E.; et al. Ceftazidime-Avibactam To Treat Life-Threatening Infections by Carbapenem-Resistant Pathogens in Critically Ill Mechanically Ventilated Patients. Antimicrob. Agents Chemother. 2020, 64, e02320-19. [Google Scholar] [CrossRef]
- Giddins, M.J.; Macesic, N.; Annavajhala, M.K.; Stump, S.; Khan, S.; McConville, T.H.; Mehta, M.; Gomez-Simmonds, A.; Uhlemann, A.C. Successive Emergence of Ceftazidime-Avibactam Resistance through Distinct Genomic Adaptations in blaKPC-2-Harboring Klebsiella pneumoniae Sequence Type 307 Isolates. Antimicrob. Agents Chemother. 2018, 62, e02101-17. [Google Scholar] [CrossRef]
- Gaibani, P.; Campoli, C.; Lewis, R.E.; Volpe, S.L.; Scaltriti, E.; Giannella, M.; Pongolini, S.; Berlingeri, A.; Cristini, F.; Bartoletti, M.; et al. In vivo evolution of resistant subpopulations of KPC-producing Klebsiella pneumoniae during ceftazidime/avibactam treatment. J. Antimicrob. Chemother. 2018, 73, 1525–1529. [Google Scholar] [CrossRef]
- Oueslati, S.; Tlili, L.; Exilie, C.; Bernabeu, S.; Iorga, B.; Bonnin, R.A.; Dortet, L.; Naas, T. Different phenotypic expression of KPC β-lactamase variants and challenges in their detection. J. Antimicrob. Chemother. 2020, 75, 769–771. [Google Scholar] [CrossRef]
- Tiseo, G.; Falcone, M.; Leonildi, A.; Giordano, C.; Barnini, S.; Arcari, G.; Carattoli, A.; Menichetti, F. Meropenem-Vaborbactam as Salvage Therapy for Ceftazidime-Avibactam-, Cefiderocol-Resistant ST-512 Klebsiella pneumoniae-Producing KPC-31, a D179Y Variant of KPC-3. Open Forum. Infect. Dis. 2021, 8, ofab141. [Google Scholar] [CrossRef]
- Ding, L.; Shen, S.; Han, R.; Yin, D.; Guo, Y.; Hu, F. Ceftazidime-Avibactam in Combination with Imipenem as Salvage Therapy for ST11 KPC-33-Producing Klebsiella pneumoniae. Antibiotics 2022, 11, 604. [Google Scholar] [CrossRef] [PubMed]
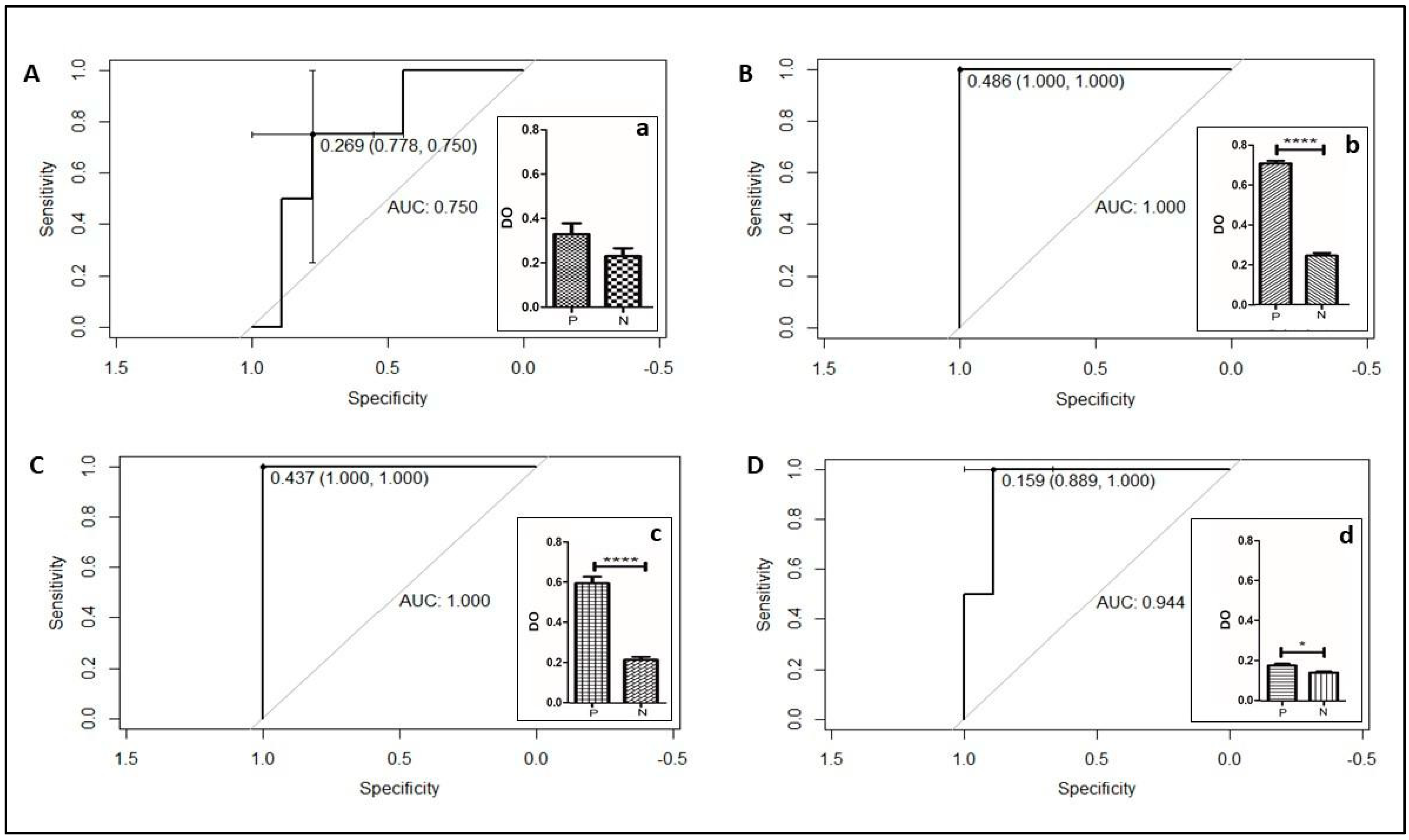
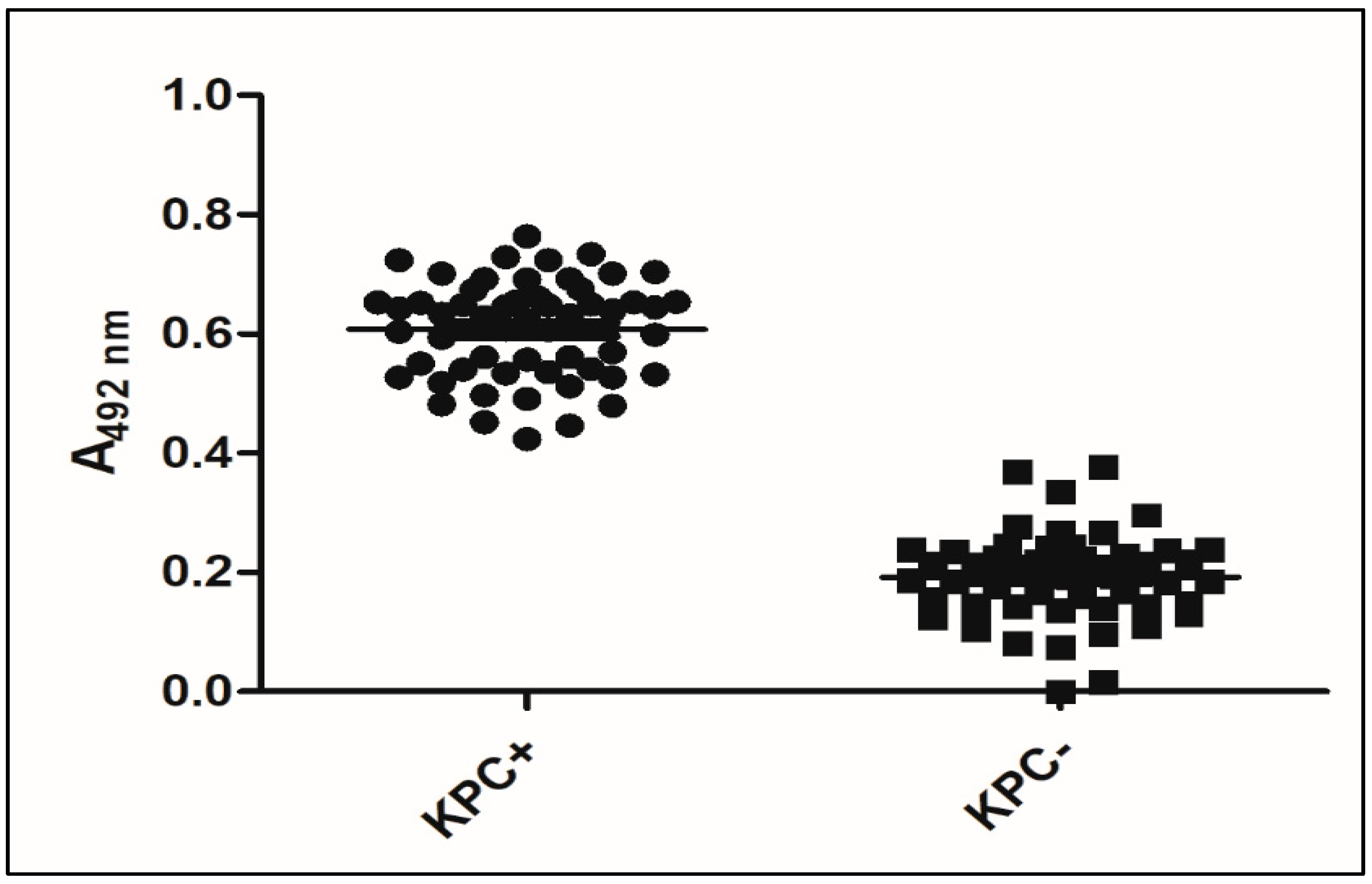
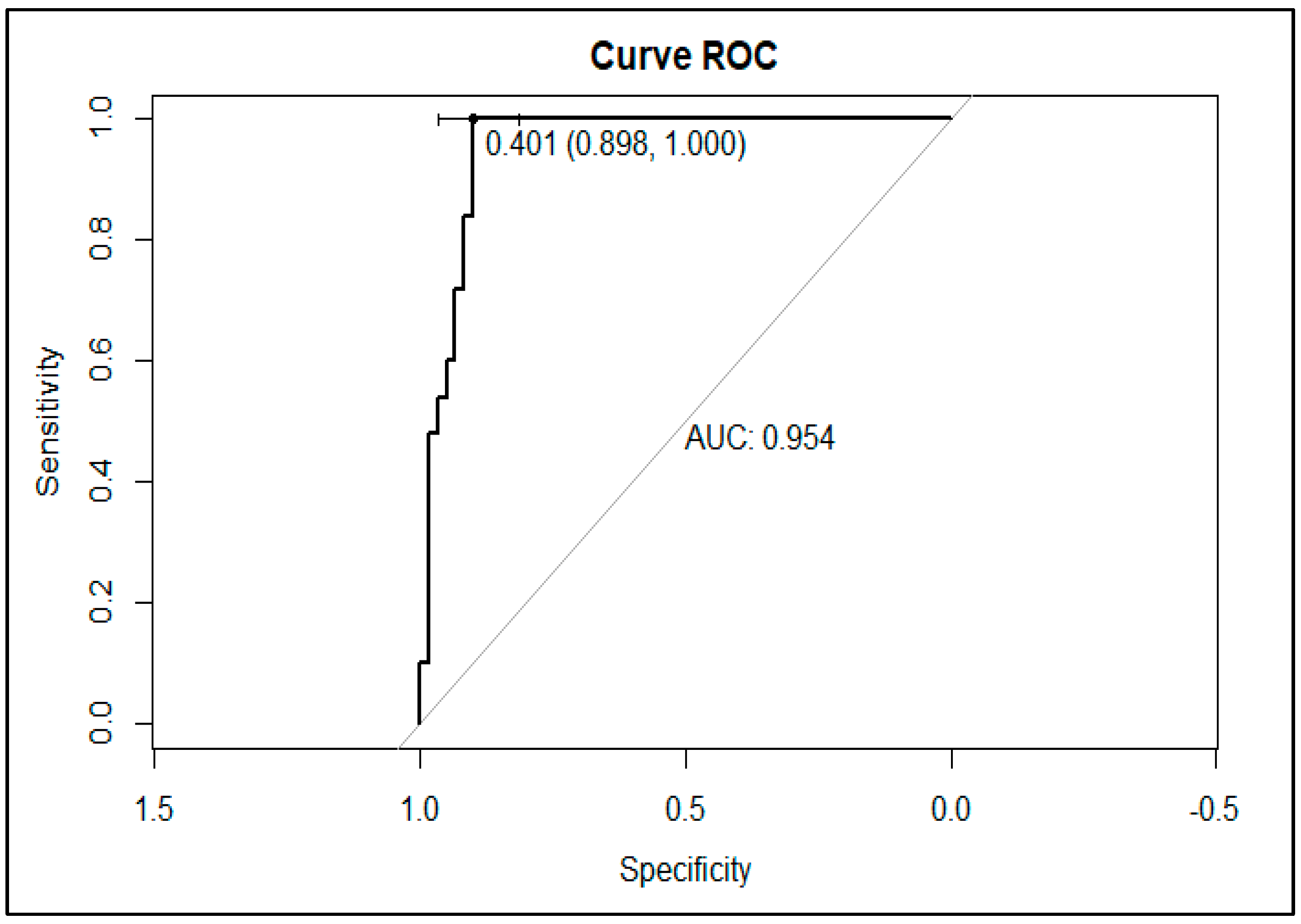
| Bacterial Species | Type of Carbapenemases (Number of Isolates Tested) | |
|---|---|---|
| Carbapenemase-producing isolates | ||
| Klebsiella pneumoniae (7) | KPC-2 (4) | |
| BKC-1 (3) | ||
| Pseudomonas aeruginosa (3) | VIM-1 (1) | |
| GES-5 (1) | ||
| SPM-1 (1) | ||
| Escherichia coli (1) | NDM-1 (1) | |
| Non-Carbapenemase-producing isolates | ||
| Klebsiella pneumoniae (1) | SHV-18 (1) | |
| Non-β-lactamase producing isolates | ||
| Escherichia coli (1) | None | |
| Total: | 13 isolates | |
| Carbapenemase-Producing Isolates/Species | Type of Carbapenemase (Number of Isolates Tested) | |
|---|---|---|
| KPC-producing isolates positive (N = 50) | ||
| Klebsiella pneumoniae (47) | ||
| KPC-2 (35) | ||
| KPC-3 (7) | ||
| KPC-7 (1) | ||
| KPC-11 (1) | ||
| KPC-33 (1) | ||
| KPC-66 (1) | ||
| KPC-144 (1) | ||
| Klebsiella oxytoca (1) | ||
| KPC-2 (1) | ||
| Pseudomonas aeruginosa (2) | ||
| KPC-2 (2) | ||
| Double-carbapenemase producer (N = 3) | ||
| Klebsiella pneumoniae (3) | ||
| KPC-2/BKC-1 (3) | ||
| Other carbapenemases (N = 24) | ||
| Klebsiella pneumoniae (12) | ||
| BKC-1 (6) | ||
| BKC-2 (1) | ||
| NDM-1 (1) | ||
| OXA-48 (1) | ||
| GES-5 (2) | ||
| IMP-1 (1) | ||
| Pseudomonas aeruginosa (8) | ||
| NDM-1 (1) | ||
| IMP-1 (2) | ||
| GIM-1 (2) | ||
| SPM-1 (3) | ||
| Pseudomonas monteilli (1) | ||
| VIM-1 (1) | ||
| Enterobacter spp. (3) | ||
| NDM-1 (3) | ||
| Non-carbapenemase-producing isolates (N = 19) | ||
| Klebsiella pneumoniae (2) | ||
| OmpK-36 (1) | ||
| OmpK (35/36) (1) | ||
| Pseudomonas aeruginosa (16) | ||
| Non carbapenemase producer | ||
| Serratia marcescens (1) | ||
| GES-16 (1) | ||
| ESBL-producing isolates (N = 3) | ||
| Klebsiella pneumoniae (3) | ||
| CTX-M8 (2) | ||
| SHV-18 (1) | ||
| Isolates susceptible to carbapenems (N = 10) | ||
| Klebsiella pneumoniae (4) | ||
| Escherichia coli (2) | ||
| Acinetobacter baumannii (1) | ||
| Proteus mirabilis (1) | ||
| Shigella flexneri (1) | ||
| Salmonella enterica subsp. Enterica (1) | ||
| Total N = 109 isolates | ||
| A | Rabbit (µg/mL) | |||
| Mice (µg/mL) | 25 | 12.5 | 6.25 | 3.125 |
| 12.5 | 0.195 | 0.146 | 0.11 | 0.104 |
| 6.25 | 0.181 | 0.116 | 0.104 (*) | 0.073 |
| 3.125 | 0.158 | 0.109 | 0.078 | 0.063 |
| 1.5625 | 0.154 | 0.095 | 0.07 | 0.057 |
| 0.78125 | 0.151 | 0.098 | 0.076 | 0.059 |
| 0.390625 | 0.146 | 0.091 | 0.066 | 0.057 |
| 0.1953125 | 0.147 | 0.1 | 0.073 | 0.058 |
| 0.09765625 | 0.149 | 0.096 | 0.078 | 0.06 |
| B | Rabbit (µg/mL) | |||
| Mice (µg/mL) | 25 | 12.5 | 6.25 | 3.125 |
| 12.5 | 1.165 | 1.091 | 1.062 | 0.948 |
| 6.25 | 1.055 | 1.097 | 1.026 (*) | 0.925 |
| 3.125 | 0.855 | 0.825 | 0.835 | 0.708 |
| 1.5625 | 0.833 | 0.714 | 0.666 | 0.571 |
| 0.78125 | 0.923 | 0.754 | 0.695 | 0.518 |
| 0.390625 | 0.936 | 0.695 | 0.722 | 0.57 |
| 0.1953125 | 0.87 | 0.829 | 0.682 | 0.609 |
| 0.09765625 | 0.988 | 0.871 | 0.737 | 0.576 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valencio, A.; da Silva, M.A.; Santos, F.F.; Polatto, J.M.; Machado, M.M.F.; Piazza, R.M.F.; Gales, A.C. Capture ELISA for KPC Detection in Gram-Negative Bacilli: Development and Standardisation. Microorganisms 2023, 11, 1052. https://doi.org/10.3390/microorganisms11041052
Valencio A, da Silva MA, Santos FF, Polatto JM, Machado MMF, Piazza RMF, Gales AC. Capture ELISA for KPC Detection in Gram-Negative Bacilli: Development and Standardisation. Microorganisms. 2023; 11(4):1052. https://doi.org/10.3390/microorganisms11041052
Chicago/Turabian StyleValencio, André, Miriam Aparecida da Silva, Fernanda Fernandes Santos, Juliana Moutinho Polatto, Marcelo Marcondes Ferreira Machado, Roxane Maria Fontes Piazza, and Ana Cristina Gales. 2023. "Capture ELISA for KPC Detection in Gram-Negative Bacilli: Development and Standardisation" Microorganisms 11, no. 4: 1052. https://doi.org/10.3390/microorganisms11041052
APA StyleValencio, A., da Silva, M. A., Santos, F. F., Polatto, J. M., Machado, M. M. F., Piazza, R. M. F., & Gales, A. C. (2023). Capture ELISA for KPC Detection in Gram-Negative Bacilli: Development and Standardisation. Microorganisms, 11(4), 1052. https://doi.org/10.3390/microorganisms11041052







