Methylprednisolone Promotes Mycobacterium smegmatis Survival in Macrophages through NF-κB/DUSP1 Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture of RAW 264.7 Cells
2.2. Culture of M. smegmatis
2.3. Colony Forming Unit (CFU) Assay
2.4. Western Blot Analysis
2.5. Flow Cytometry
2.6. Transcriptome High-Throughput Sequencing
2.7. Screening of Differentially Expressed Genes (DEGs)
2.8. Construction of a Protein–Protein Interaction (PPI) Network
2.9. Analysis of the Kyoto Encyclopedia of Genes and Genomes (KEGG)
2.10. Analysis of Gene Ontology (GO)
2.11. Construction of the Heat Map
2.12. Eenzyme Linked Immunosorbent Assay (ELISA)
2.13. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
2.14. Data Processing
2.15. Assay of Cell Viability
3. Results
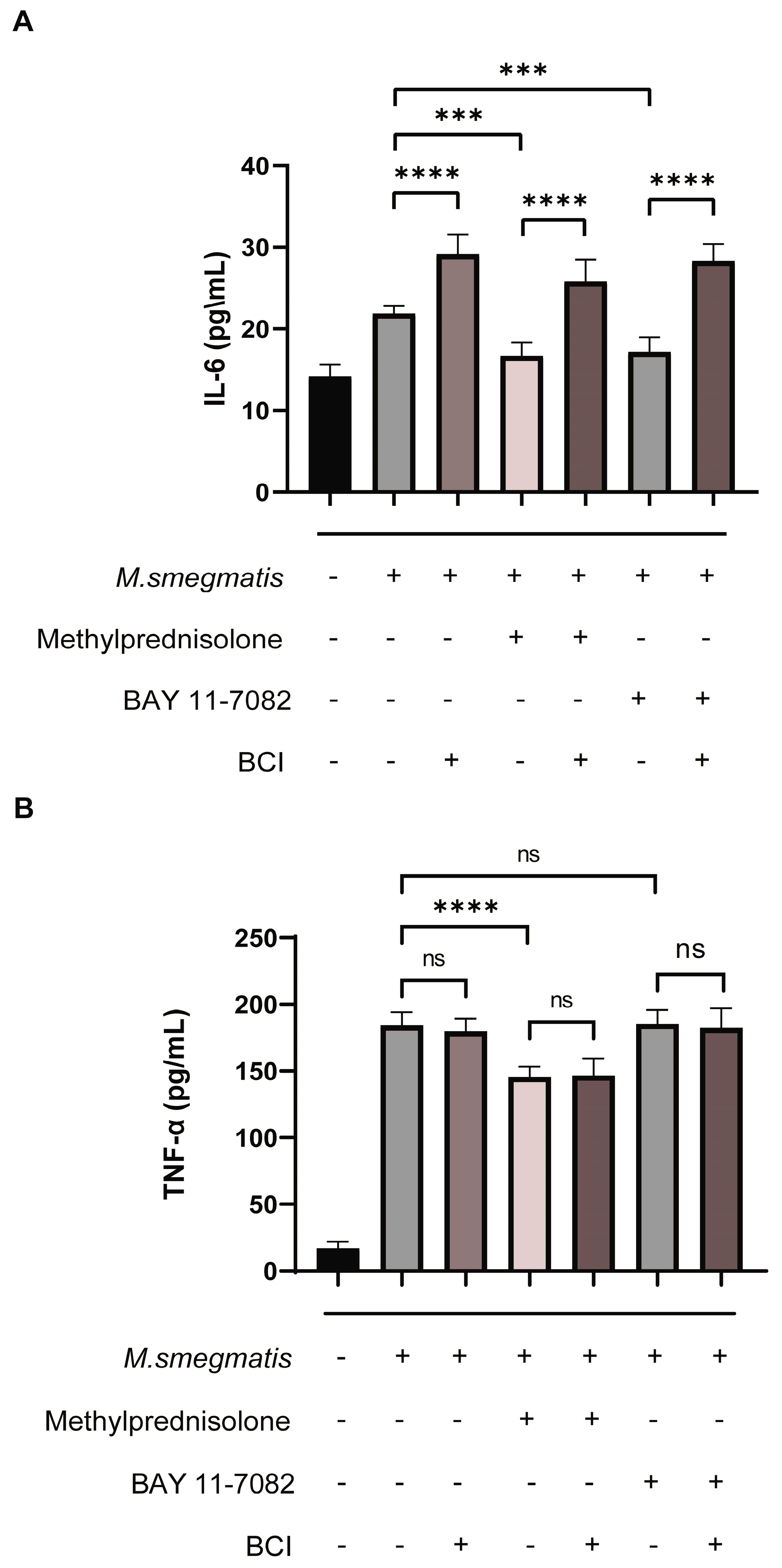
3.1. Methylprednisolone Facilitates M. smegmatis Survival in Macrophages and Suppresses Cellular Secretion of TNF-α and IL-6
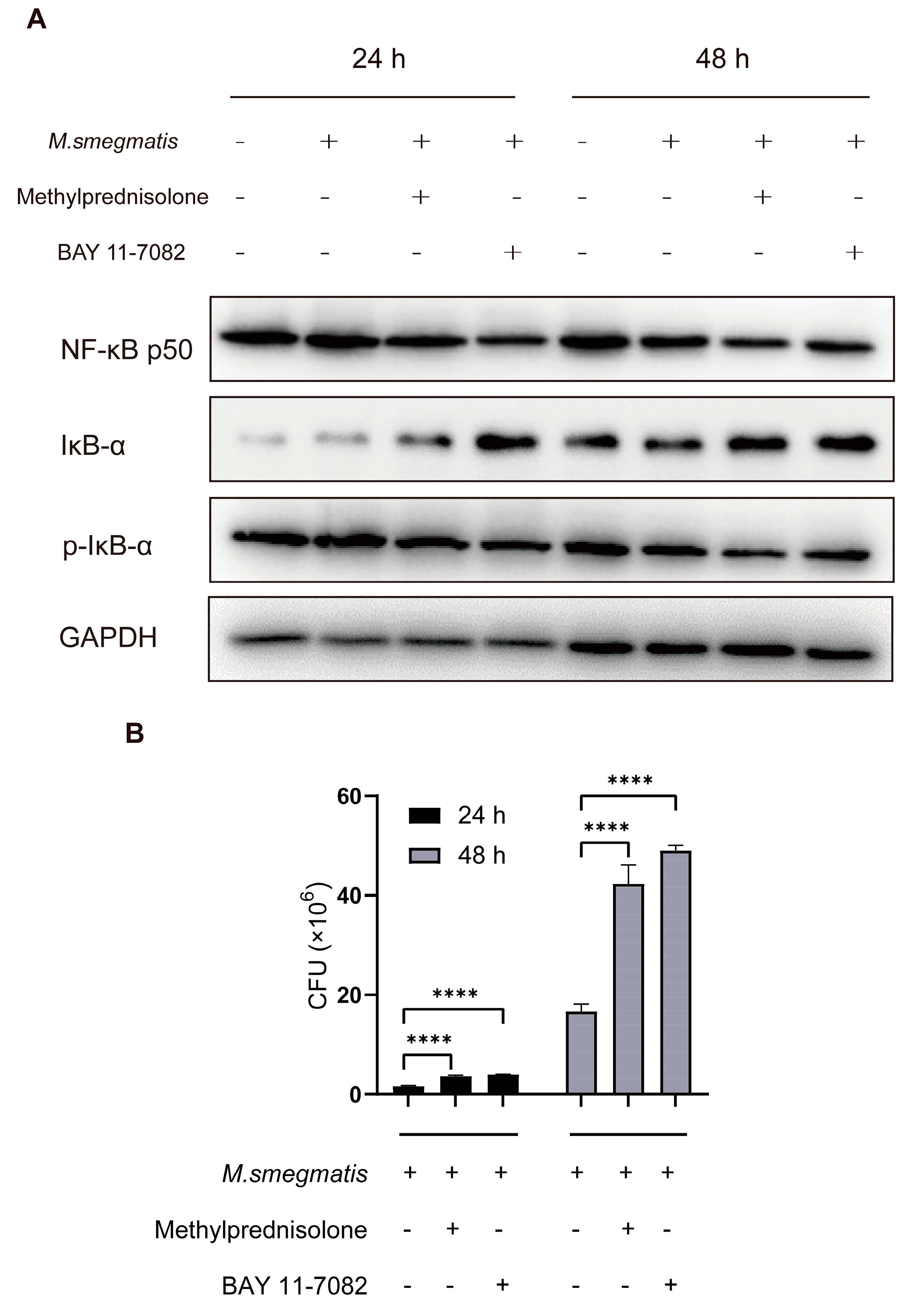
3.2. Methylprednisolone Promotes M. smegmatis Survival in Macrophages by Suppressing the Expression of NF-κB
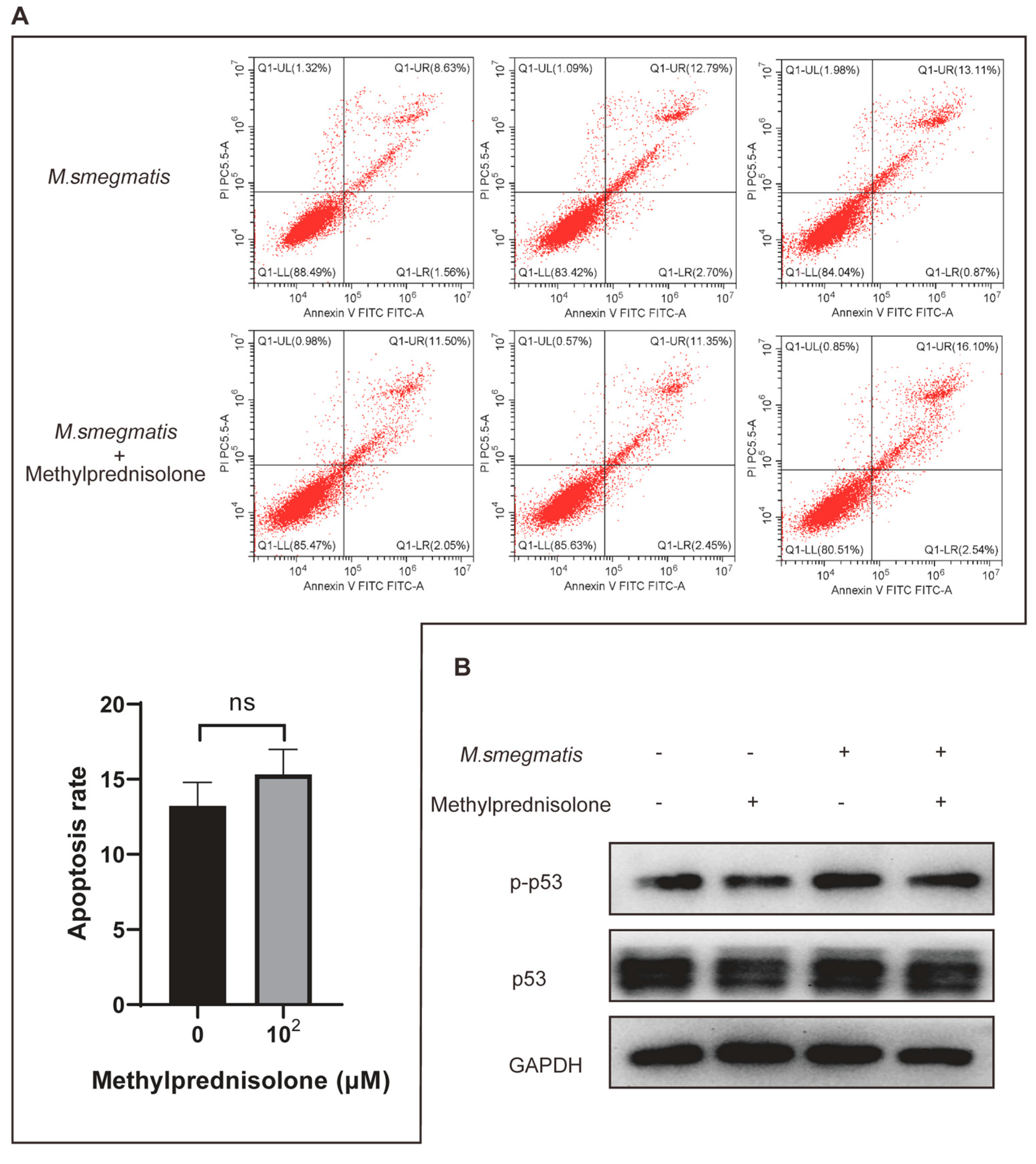
3.3. Methylprednisolone Promotes M. smegmatis Survival in Macrophages, but Not by Inhibiting Infected Cell Apoptosis
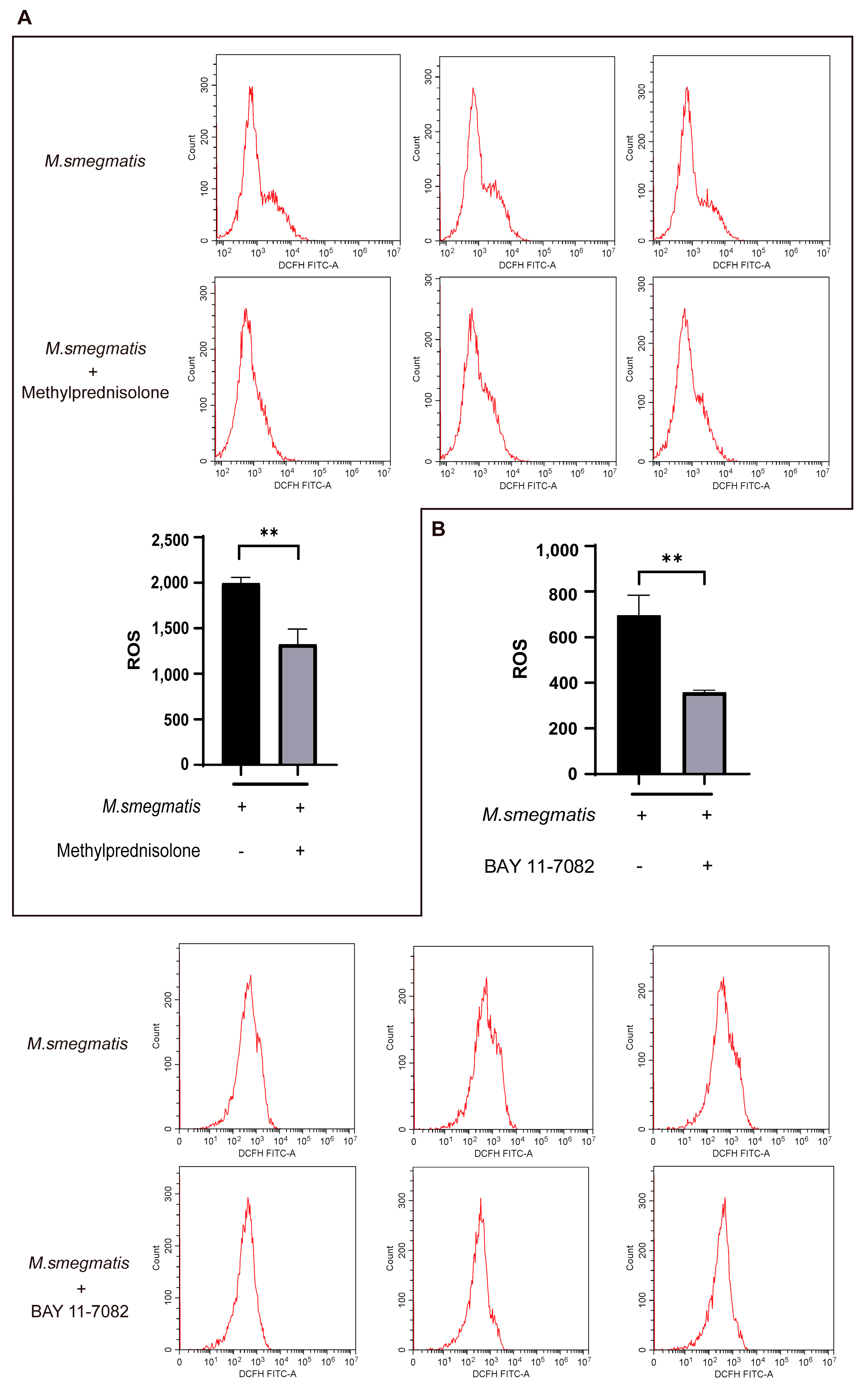
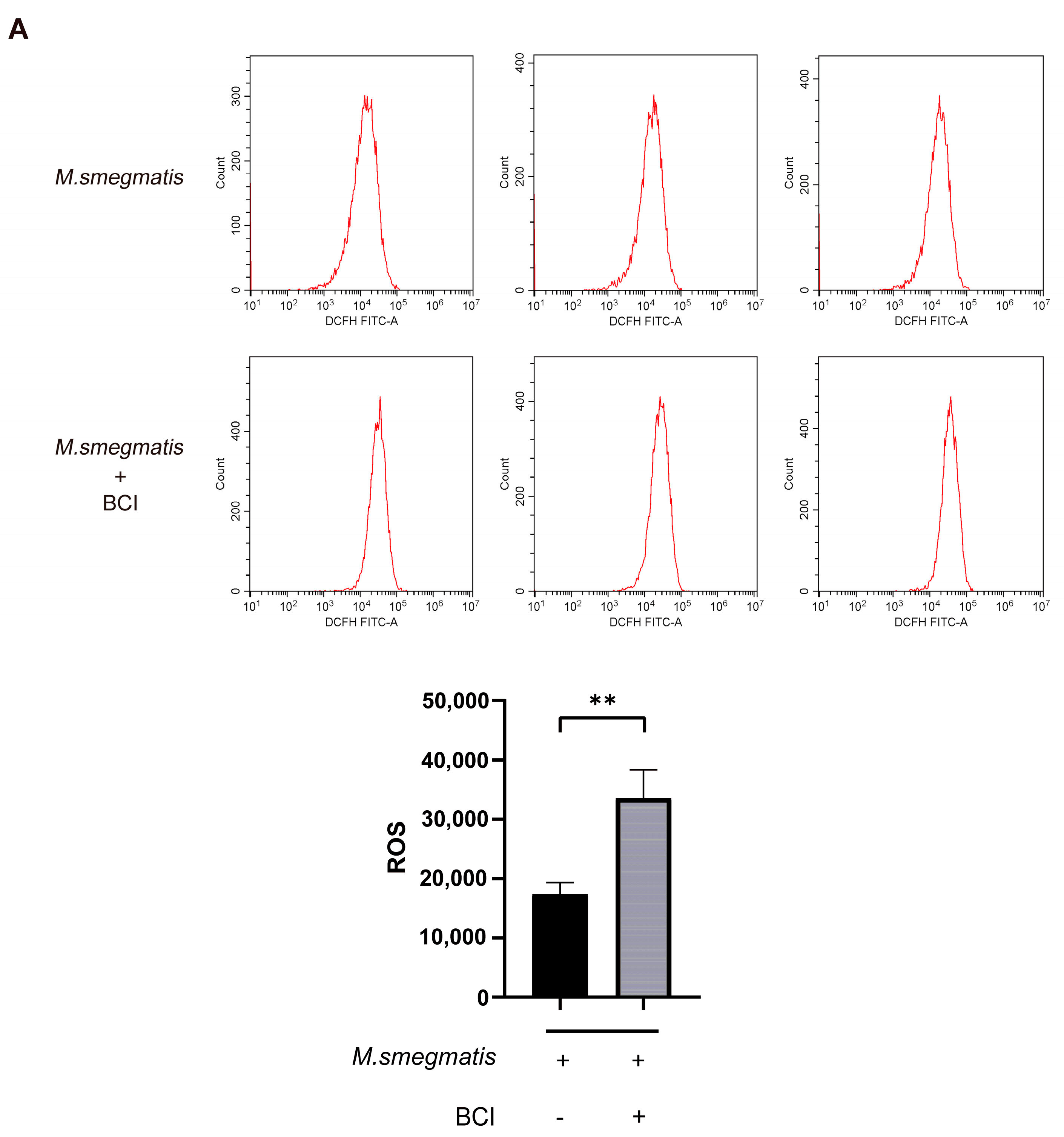
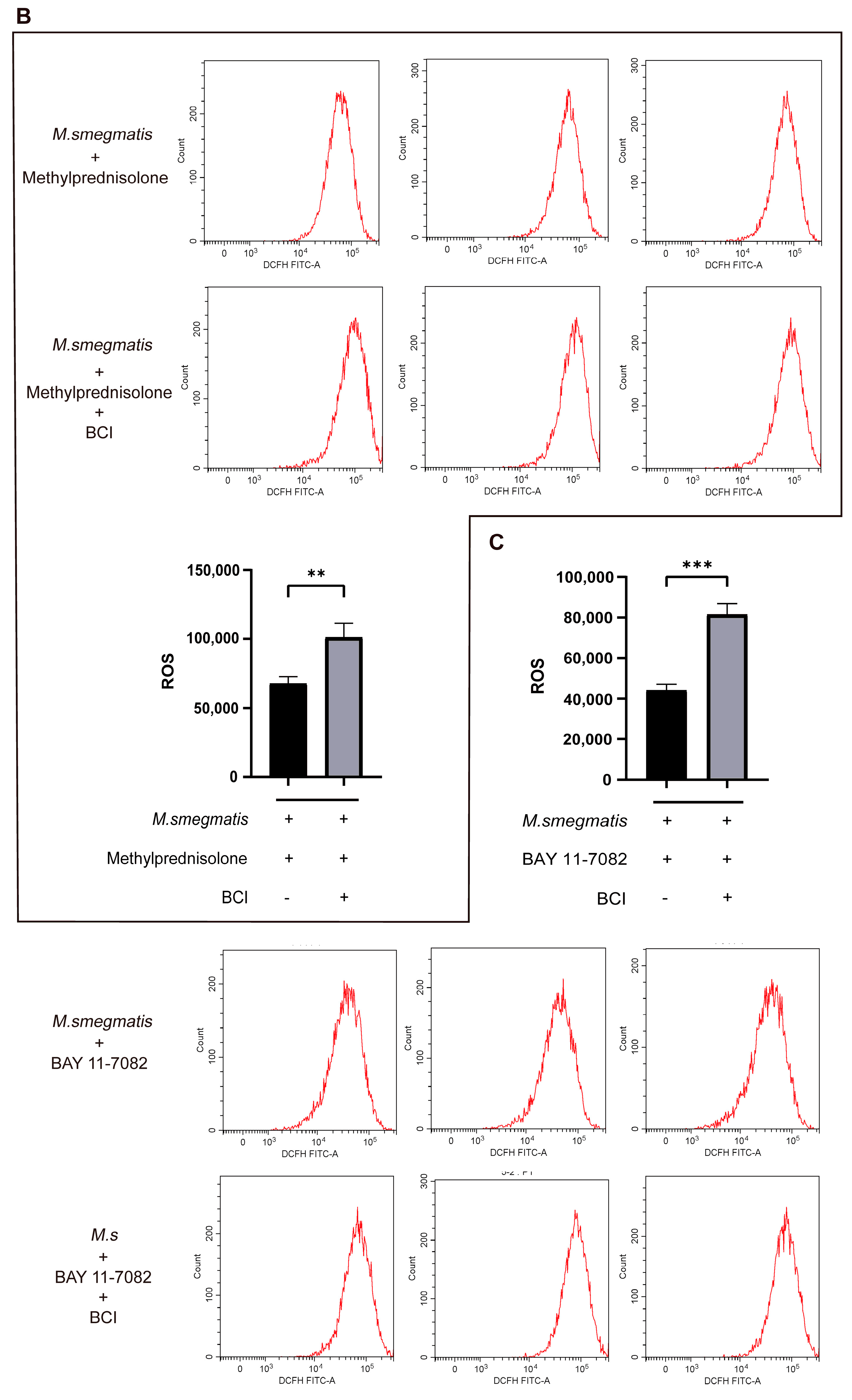
3.4. Methylprednisolone Suppresses Cellular ROS Production through Inhibiting the Expression of NF-κB
3.5. High-Throughput Sequencing Analyzed the Expression of Genes in M. smegmatis Infected Macrophages after BAY 11-7082 Treatment
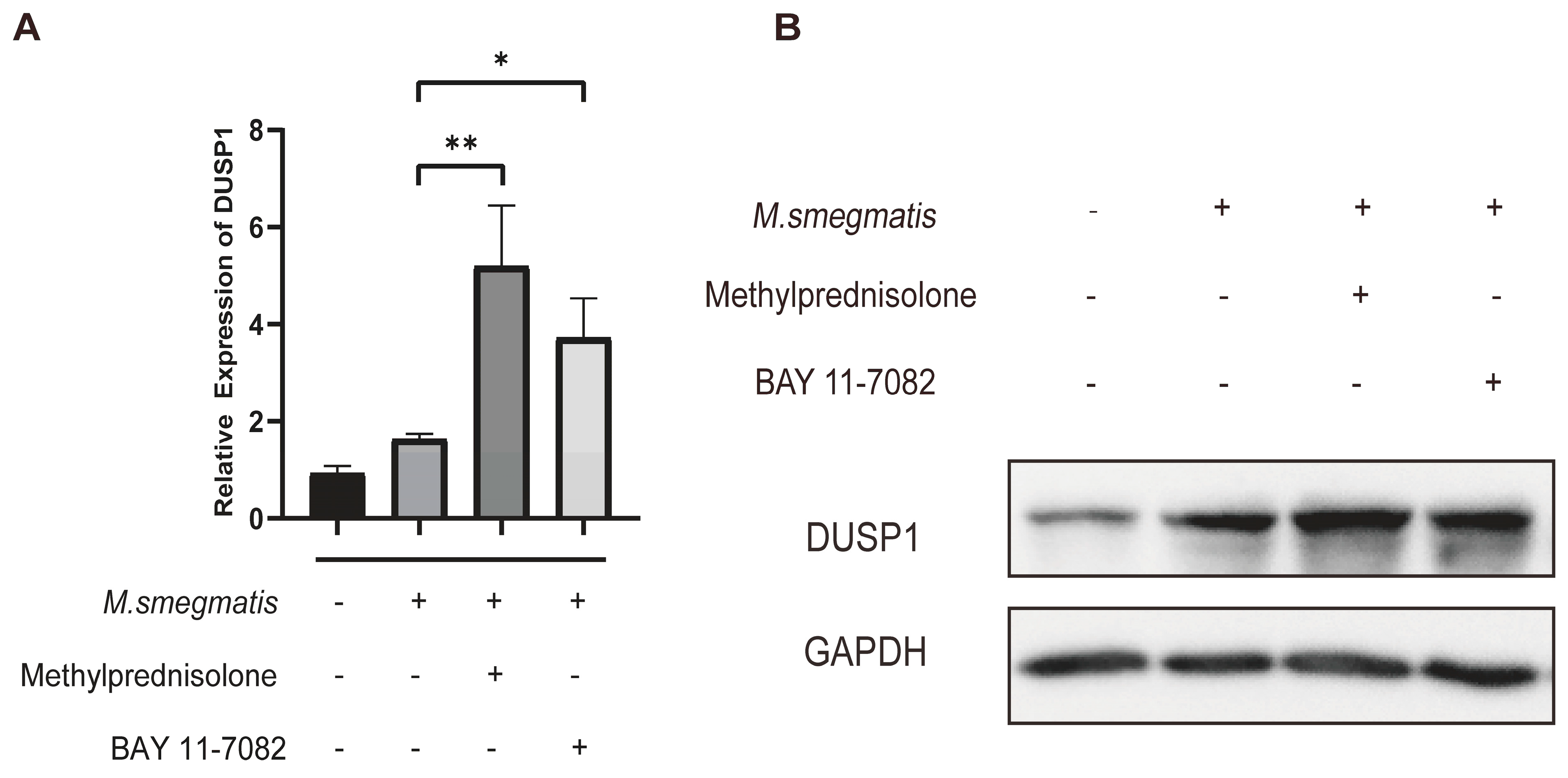
3.6. Methylprednisolone and BAY 11-7082 Up-Regulate the Expression of DUSP1
3.7. Low Level of DUSP1 Inhibited the Survival of M. smegmatis in Macrophages through NF-κB/DUSP1 Pathway
3.8. Inhibiting DUSP1 Prompts Cellular ROS Production and IL-6 Secretion in Macrophages Infected with M. smegmatis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2021. 2021. Available online: https://www.who.int/publications/i/item/9789240037021 (accessed on 1 December 2022).
- World Health Organization. Global Tuberculosis Report 2015. 2015. Available online: https://apps.who.int/iris/bitstream/handle/10665/191102/9789241565059_eng.pdf?sequence=1 (accessed on 1 December 2022).
- Getahun, H.; Matteelli, A.; Abubakar, I.; Aziz, M.; Baddeley, A.; Barreira, D.; Den Boon, S.; Borroto Gutierrez, S.; Bruchfeld, J.; Burhan, E.; et al. Management of latent Mycobacterium tuberculosis infection: WHO guidelines for low tuberculosis burden countries. Eur. Respir. J. 2015, 46, 1563–1576. [Google Scholar] [CrossRef] [PubMed]
- Shea, K.; Kammerer, J.; Winston, C.; Navin, T.; Horsburgh, C. Estimated rate of reactivation of latent tuberculosis infection in the United States, overall and by population subgroup. Am. J. Epidemiol. 2014, 179, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Selmi, C.; Generali, E.; Massarotti, M.; Bianchi, G.; Sciré, C. New treatments for inflammatory rheumatic disease. Immunol. Res. 2014, 60, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Cai, F.; Wang, X.; Zheng, N.; Wu, R. High risk of activation of latent tuberculosis infection in rheumatic disease patients. Infect. Dis. 2020, 52, 80–86. [Google Scholar] [CrossRef]
- Vozoris, N.; Seemangal, J.; Batt, J. Prevalence, screening and treatment of latent tuberculosis among oral corticosteroid recipients. Eur. Respir. J. 2014, 44, 1373–1375. [Google Scholar] [CrossRef]
- Ai, J.; Ruan, Q.; Liu, Q.; Zhang, W. Updates on the risk factors for latent tuberculosis reactivation and their managements. Emerg. Microbes Infect. 2016, 5, e10. [Google Scholar] [CrossRef]
- Jick, S.; Lieberman, E.; Rahman, M.; Choi, H. Glucocorticoid use, other associated factors, and the risk of tuberculosis. Arthritis Rheum. 2006, 55, 19–26. [Google Scholar] [CrossRef]
- Castellana, G.; Castellana, M.; Castellana, C.; Castellana, G.; Resta, E.; Carone, M.; Resta, O. Inhaled Corticosteroids and Risk of Tuberculosis in Patients with Obstructive Lung Diseases: A Systematic Review and Meta-Analysis of Non-randomized Studies. Int. J. Chronic Obstr. Pulm. Dis. 2019, 14, 2219–2227. [Google Scholar] [CrossRef]
- Edwards, A.; Gao, Y.; Allan, R.; Ball, D.; de Graaf, H.; Coelho, T.; Clifford, V.; Curtis, N.; Williams, A.; Faust, S.; et al. Corticosteroids and infliximab impair the performance of interferon-γ release assays used for diagnosis of latent tuberculosis. Thorax 2017, 72, 946–949. [Google Scholar] [CrossRef]
- Cantini, F.; Nannini, C.; Niccoli, L.; Iannone, F.; Delogu, G.; Garlaschi, G.; Sanduzzi, A.; Matucci, A.; Prignano, F.; Conversano, M.; et al. Guidance for the management of patients with latent tuberculosis infection requiring biologic therapy in rheumatology and dermatology clinical practice. Autoimmun. Rev. 2015, 14, 503–509. [Google Scholar] [CrossRef]
- Lai, C.; Lee, M.; Lee, S.; Lee, S.; Chang, S.; Lee, C. Risk of incident active tuberculosis and use of corticosteroids. Int. J. Tuberc. Lung Dis. Off. J. Int. Union Tuberc. Lung Dis. 2015, 19, 936–942. [Google Scholar] [CrossRef] [PubMed]
- Ranjitha, J.; Rajan, A.; Shankar, V. Features of the biochemistry of Mycobacterium smegmatis, as a possible model for Mycobacterium tuberculosis. J. Infect. Public Health 2020, 13, 1255–1264. [Google Scholar]
- Krug, S.; Parveen, S.; Bishai, W.R. Host-Directed Therapies: Modulating Inflammation to Treat Tuberculosis. Front. Immunol. 2021, 12, 660916. [Google Scholar] [CrossRef] [PubMed]
- Barnabei, L.; Laplantine, E.; Mbongo, W.; Rieux-Laucat, F.; Weil, R. NF-κB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021, 12, 716469. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Dixit, V.M. Signaling in innate immunity and inflammation. Cold Spring Harb. Perspect. Biol. 2012, 4, a006049. [Google Scholar] [CrossRef]
- Sun, S.-C.; Chang, J.-H.; Jin, J. Regulation of nuclear factor-κB in autoimmunity. Trends Immunol. 2013, 34, 282–289. [Google Scholar] [CrossRef]
- Kumar, M.; Sahu, S.K.; Kumar, R.; Subuddhi, A.; Maji, R.K.; Jana, K.; Gupta, P.; Raffetseder, J.; Lerm, M.; Ghosh, Z.; et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-κB pathway. Cell Host Microbe 2015, 17, 345–356. [Google Scholar] [CrossRef]
- Niu, W.; Sun, B.; Li, M.; Cui, J.; Huang, J.; Zhang, L. TLR-4/microRNA-125a/NF-κB signaling modulates the immune response to Mycobacterium tuberculosis infection. Cell Cycle 2018, 17, 1931–1945. [Google Scholar] [CrossRef]
- Hoppstädter, J.; Ammit, A.J. Role of Dual-Specificity Phosphatase 1 in Glucocorticoid-Driven Anti-inflammatory Responses. Front. Immunol. 2019, 10, 1446. [Google Scholar] [CrossRef]
- Han, J.; Ulevitch, R.J. Limiting inflammatory responses during activation of innate immunity. Nat. Immunol. 2005, 6, 1198–1205. [Google Scholar] [CrossRef]
- Liew, F.Y.; Xu, D.; Brint, E.K.; O’Neill, L.A.J. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 2005, 5, 446–458. [Google Scholar] [CrossRef]
- Chi, H.; Flavell, R.A. Acetylation of MKP-1 and the control of inflammation. Sci. Signal. 2008, 1, pe44. [Google Scholar] [CrossRef] [PubMed]
- Matta, R.; Barnard, J.A.; Wancket, L.M.; Yan, J.; Xue, J.; Grieves, J.; Frazier, W.J.; Nelin, L.; Cato, A.C.B.; Liu, Y. Knockout of Mkp-1 exacerbates colitis in Il-10-deficient mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1322–G1335. [Google Scholar] [CrossRef] [PubMed]
- Kjellerup, R.B.; Johansen, C.; Kragballe, K.; Iversen, L. The expression of dual-specificity phosphatase 1 mRNA is downregulated in lesional psoriatic skin. Br. J. Dermatol. 2013, 168, 339–345. [Google Scholar] [CrossRef]
- Yang, J.; Sun, L.; Han, J.; Zheng, W.; Peng, W. DUSP1/MKP-1 regulates proliferation and apoptosis in keratinocytes through the ERK/Elk-1/Egr-1 signaling pathway. Life Sci. 2019, 223, 47–53. [Google Scholar] [CrossRef]
- Chang, W.J.; Niu, X.P.; Hou, R.X.; Li, J.Q.; Liu, R.F.; Wang, Q.; Wang, C.F.; Li, X.H.; Yin, G.H.; Zhang, K.M. LITAF, HHEX, and DUSP1 expression in mesenchymal stem cells from patients with psoriasis. Genet. Mol. Res. 2015, 14, 15793–15801. [Google Scholar] [CrossRef]
- Chi, H.; Barry, S.P.; Roth, R.J.; Wu, J.J.; Jones, E.A.; Bennett, A.M.; Flavell, R.A. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 2274–2279. [Google Scholar] [CrossRef] [PubMed]
- Hammer, M.; Mages, J.; Dietrich, H.; Servatius, A.; Howells, N.; Cato, A.C.B.; Lang, R. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J. Exp. Med. 2006, 203, 15–20. [Google Scholar] [CrossRef]
- Salojin, K.V.; Owusu, I.B.; Millerchip, K.A.; Potter, M.; Platt, K.A.; Oravecz, T. Essential role of MAPK phosphatase-1 in the negative control of innate immune responses. J. Immunol. 2006, 176, 1899–1907. [Google Scholar] [CrossRef]
- Zhao, Q.; Wang, X.; Nelin, L.D.; Yao, Y.; Matta, R.; Manson, M.E.; Baliga, R.S.; Meng, X.; Smith, C.V.; Bauer, J.A.; et al. MAP kinase phosphatase 1 controls innate immune responses and suppresses endotoxic shock. J. Exp. Med. 2006, 203, 131–140. [Google Scholar] [CrossRef]
- Gräb, J.; Suárez, I.; van Gumpel, E.; Winter, S.; Schreiber, F.; Esser, A.; Hölscher, C.; Fritsch, M.; Herb, M.; Schramm, M.; et al. Corticosteroids inhibit Mycobacterium tuberculosis-induced necrotic host cell death by abrogating mitochondrial membrane permeability transition. Nat. Commun. 2019, 10, 688. [Google Scholar] [CrossRef] [PubMed]
- Su, L.-J.; Zhang, J.-H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.-Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell. Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef] [PubMed]
- Sakellariou, G.K.; Jackson, M.J.; Vasilaki, A. Redefining the major contributors to superoxide production in contracting skeletal muscle. The role of NAD(P)H oxidases. Free Radic. Res. 2014, 48, 12–29. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, G.; Trocchia, S.; Abdel-Gawad, F.K.; Ciarcia, G. Roles of reactive oxygen species in the spermatogenesis regulation. Front. Endocrinol. 2014, 5, 56. [Google Scholar] [CrossRef] [PubMed]
- Herb, M.; Schramm, M. Functions of ROS in Macrophages and Antimicrobial Immunity. Antioxidants 2021, 10, 313. [Google Scholar] [CrossRef]
- Herb, M.; Gluschko, A.; Wiegmann, K.; Farid, A.; Wolf, A.; Utermöhlen, O.; Krut, O.; Krönke, M.; Schramm, M. Mitochondrial reactive oxygen species enable proinflammatory signaling through disulfide linkage of NEMO. Sci. Signal. 2019, 12, eaar5926. [Google Scholar] [CrossRef]
- Gluschko, A.; Herb, M.; Wiegmann, K.; Krut, O.; Neiss, W.F.; Utermöhlen, O.; Krönke, M.; Schramm, M. The β Integrin Mac-1 Induces Protective LC3-Associated Phagocytosis of Listeria monocytogenes. Cell Host Microbe 2018, 23, 324–337.e5. [Google Scholar] [CrossRef] [PubMed]
- Wink, D.A.; Hines, H.B.; Cheng, R.Y.S.; Switzer, C.H.; Flores-Santana, W.; Vitek, M.P.; Ridnour, L.A.; Colton, C.A. Nitric oxide and redox mechanisms in the immune response. J. Leukoc. Biol. 2011, 89, 873–891. [Google Scholar] [CrossRef] [PubMed]
- Queval, C.J.; Brosch, R.; Simeone, R. The Macrophage: A Disputed Fortress in the Battle against. Front. Microbiol. 2017, 8, 2284. [Google Scholar] [CrossRef]
- Hunter, C.A.; Jones, S.A. IL-6 as a keystone cytokine in health and disease. Nat. Immunol. 2015, 16, 448–457. [Google Scholar] [CrossRef] [PubMed]
- Waage, A.; Brandtzaeg, P.; Halstensen, A.; Kierulf, P.; Espevik, T. The complex pattern of cytokines in serum from patients with meningococcal septic shock. Association between interleukin 6, interleukin 1, and fatal outcome. J. Exp. Med. 1989, 169, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Nagabhushanam, V.; Solache, A.; Ting, L.-M.; Escaron, C.J.; Zhang, J.Y.; Ernst, J.D. Innate inhibition of adaptive immunity: Mycobacterium tuberculosis-induced IL-6 inhibits macrophage responses to IFN-gamma. J. Immunol. 2003, 171, 4750–4757. [Google Scholar] [CrossRef]
- Sebba, A. Tocilizumab: The first interleukin-6-receptor inhibitor. Am. J. Health Syst. Pharm. 2008, 65, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Campbell, C.; Andersson, M.I.; Ansari, M.A.; Moswela, O.; Misbah, S.A.; Klenerman, P.; Matthews, P.C. Risk of Reactivation of Hepatitis B Virus (HBV) and Tuberculosis (TB) and Complications of Hepatitis C Virus (HCV) Following Tocilizumab Therapy: A Systematic Review to Inform Risk Assessment in the COVID-19 Era. Front. Med. 2021, 8, 706482. [Google Scholar] [CrossRef]



| Primer Sets Name | Real-Time Quantitative PCR Primer (5′ to 3′) |
|---|---|
| GAPDH | F:AGGTCGGTGTGAACGGATTTG R:GGGGTCGTTGATGGCAACA |
| DUSP1 | F:TGTTGTTGGATTGTCGCTCCT R:TTGGGCACGATATGCTCCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, A.; He, Y.; Yang, C.; Lu, N.; Bao, J.; Gao, S.; Hosyanto, F.F.; He, X.; Fu, H.; Yan, H.; et al. Methylprednisolone Promotes Mycobacterium smegmatis Survival in Macrophages through NF-κB/DUSP1 Pathway. Microorganisms 2023, 11, 768. https://doi.org/10.3390/microorganisms11030768
Li A, He Y, Yang C, Lu N, Bao J, Gao S, Hosyanto FF, He X, Fu H, Yan H, et al. Methylprednisolone Promotes Mycobacterium smegmatis Survival in Macrophages through NF-κB/DUSP1 Pathway. Microorganisms. 2023; 11(3):768. https://doi.org/10.3390/microorganisms11030768
Chicago/Turabian StyleLi, Anlong, Yonglin He, Chun Yang, Nan Lu, Jiajia Bao, Sijia Gao, Felycia Fernanda Hosyanto, Xintong He, Huichao Fu, Huajian Yan, and et al. 2023. "Methylprednisolone Promotes Mycobacterium smegmatis Survival in Macrophages through NF-κB/DUSP1 Pathway" Microorganisms 11, no. 3: 768. https://doi.org/10.3390/microorganisms11030768
APA StyleLi, A., He, Y., Yang, C., Lu, N., Bao, J., Gao, S., Hosyanto, F. F., He, X., Fu, H., Yan, H., Ding, N., & Xu, L. (2023). Methylprednisolone Promotes Mycobacterium smegmatis Survival in Macrophages through NF-κB/DUSP1 Pathway. Microorganisms, 11(3), 768. https://doi.org/10.3390/microorganisms11030768





