The Recent Advances in the Utility of Microbial Lipases: A Review
Abstract
1. Introduction
| Microbial Source | Isolated from | Applications | Reference |
|---|---|---|---|
| Fungal Species | |||
| Penicillium simplicissimum | Food samples | Olive oil as an inductor to increase the production of lipase; can be used in various food industries | [20] |
| Yarrowia lipolytica IMUFRJ50682 | Isolated from an estuary in Guanabara Bay | Potential application in the hydrolysis of fish oil to further produce polyunsaturated fatty acids in a suitable process | [21] |
| Penicillium roqueforti | Organic debris | Uses in ethyl oleate synthesis | [22] |
| Aspergillus niger | Soil samples | Various industries where low-temperature enzymatic reactions are required | [8] |
| Arbuscular mycorrhizal fungi (AMF) | Soil samples | AMF can boost plant nutrient absorption and resilience to a variety of abiotic stresses | [23] |
| Agaricus bisporus | Wood-rotting samples | The Laccase enzyme is responsible for the color change of the Boletus genus mushrooms when they come into contact with air. | [24] |
| Acremonium implicatum | Buds of Panax notoginseng | Detects antimicrobial activity against phytopathogens | [25] |
| Chaetomium spp. | Marine green alga | Has the ability to synthesize a variety of bioactive chemicals that are lethal to leukemia cell lines | [26] |
| Eupenicillium spp. | Soft coral-derived fungus | An important resource for finding active natural products. | [27] |
| Fusarium moniliforme, Paecilomyces sp. | Root tip of plantain | This is the organism most used for the production of gibberellic acid through submerged fermentation | [28] |
| Pestalotiopsis spp. | Chinese mangrove plant Rhizophora stylosa | Used to find chromones, coumarins, isocoumarin derivatives, cytosporones, lactones, alkaloids, and terpenoids, among other bioactive natural compounds | [29] |
| Daedalea quercina | Wood-rotting samples | Capable of producing antioxidative and anti-inflammatory compounds | [30] |
| Penicillium spp. | Soil and water samples | Well recognized for its use in the manufacture and ripening of blue-veined cheese | [31] |
| Talaromyces spp. | Soil samples | Used as food preservatives and coloring for several hundred years and still used in Asia for the production of certain items | [32] |
| Beauveria bassiana | Air samples | Used for the control of insect pests | [33] |
| Chaetomium globosum | Marine green alga | Used to produce a wide range of bioactive substances; these compounds showed considerable cytotoxicity against leukemia cell lines | [26] |
| Alternaria spp. | Catharanthus roseus | Produces alternariol 51, alternariol 5-O-sulfate 52, and alternariol 5-O-methyl, among other polyketides; these chemicals have an anticancer effect on a variety of cancer cells, including oral human epidermal carcinoma (KB) cells | [34] |
| Leptosphaeria maculans | Leaf of canola/rapeseed seedlings | Causes blackleg disease in canola/rapeseed | [35] |
| Colletotrichum falcatum | Sugarcane | Causes red rot disease in sugarcane | [36] |
| Phoma spp. | Crop plants | Commonly occurs on crop plants that are economically important and causes devastating plant diseases | [37] |
| Penicillium roqueforti | Organic debris | Uses in ethyl oleate synthesis | [22] |
| Yeast Species | |||
| Meyerozyma guilliermondii | Cheese whey | Capable source of feed lipase used in cheese whey | [38] |
| Wickerhamomyces anomalus | Grapes and wines | Can increase certain characteristics of wine | [39] |
| Kluyveromyces marxianus | Traditional dairy products | Acts as an efficient cell factory to produce various metabolites including lipase | [40] |
| Saccharomyces cerevisiae | Organic materials | Has the ability to produce wine with the best combination of chemical and aromatic characteristics | [41] |
| Pichia pastoris | Exudates of a chestnut tree | Has the ability to produce recombinant protein | [42] |
| Rhodotorula glutinis | Isolated in Antarctica | Has the capability of producing lipids and growing quickly; can be used in various industrial processes | [43] |
| Trichosporon spp. | Lignocellulosic biomass | Has the potential to produce ethanol under anaerobic conditions in synthetic media and sweet sorghum | [44] |
| Brettanomyces spp. | Brewing products | Has a huge potential to add new flavors to the craft beer repertoire | [45] |
| Cryptococcus neoformans | Bird samples | Is a leading cause of morbidity and mortality in immunocompromised individuals, such as patients suffering from HIV/AIDS | [46] |
| Geotrichum candidum | Tibet kefir milk | Can remove more than 99% of Pb2+ ions in water at low concentrations and a maximum of 325.68 mg lead/g from dry biomass | [47] |
| Hansenula anomala | Fermented soybean | Has the capability of excellent lipase production | [48] |
| Bacterial Species | |||
| Enterobacter cloacae | Acidic conditions | Enantioselective esterification potential for pharmaceutical applications | [49] |
| Serratia nematodiphilia | Paper and pulp effluent | Paper and pulp effluent in deinking process in making of paper and increasing brightness, intensity, and the pulping rate | [50] |
| Exiguobacterium sp. strain (AMBL-20) | Glacial water samples | Exhibits potential in bio-detergent formulation | [51] |
| Escherichia coli | Acidic conditions | Has the ability to grow in moderate acidic conditions | [52] |
| Acidithiobacillus ferrooxidans | Acidophilic and chemolithotrophic sulfur- and iron-oxidizing bacterium | Widely used in the bioleaching process for extracting metals | [53] |
| Leptospirillum spp. | Acidic environments | Has the ability to survive at low pH and can be used in different industrial processes | [54] |
| Roseobacter spp. | Marine environment | Alphaproteo bacteria with diverse metabolic and regulatory capabilities | [55] |
| Bacillus spp. | Abattoir soil | Bacterial isolate showed positive lipase activity at the end of 24 h incubation | [56] |
| Bacillus amyloliquefaciens | Environmental samples | Media was optimized with cheap agro-industrial wastes and investigated for enhanced production of lipases in solid-state fermentation | [57] |
| Anoxybacillus gonensis UF7 | Isolated from a hot spring | Anoxybacillus lipase was used for the first time as catalyst for biodiesel production | [58] |
2. Sources of Microbial Lipases
2.1. Lipases from Fungi
2.2. Lipases from Yeast
2.3. Lipases from Bacteria
3. Purification Strategies of Microbial Lipases
3.1. Classical Purification Techniques
Precipitation and Chromatographic Separation
3.2. Modern Purification Techniques
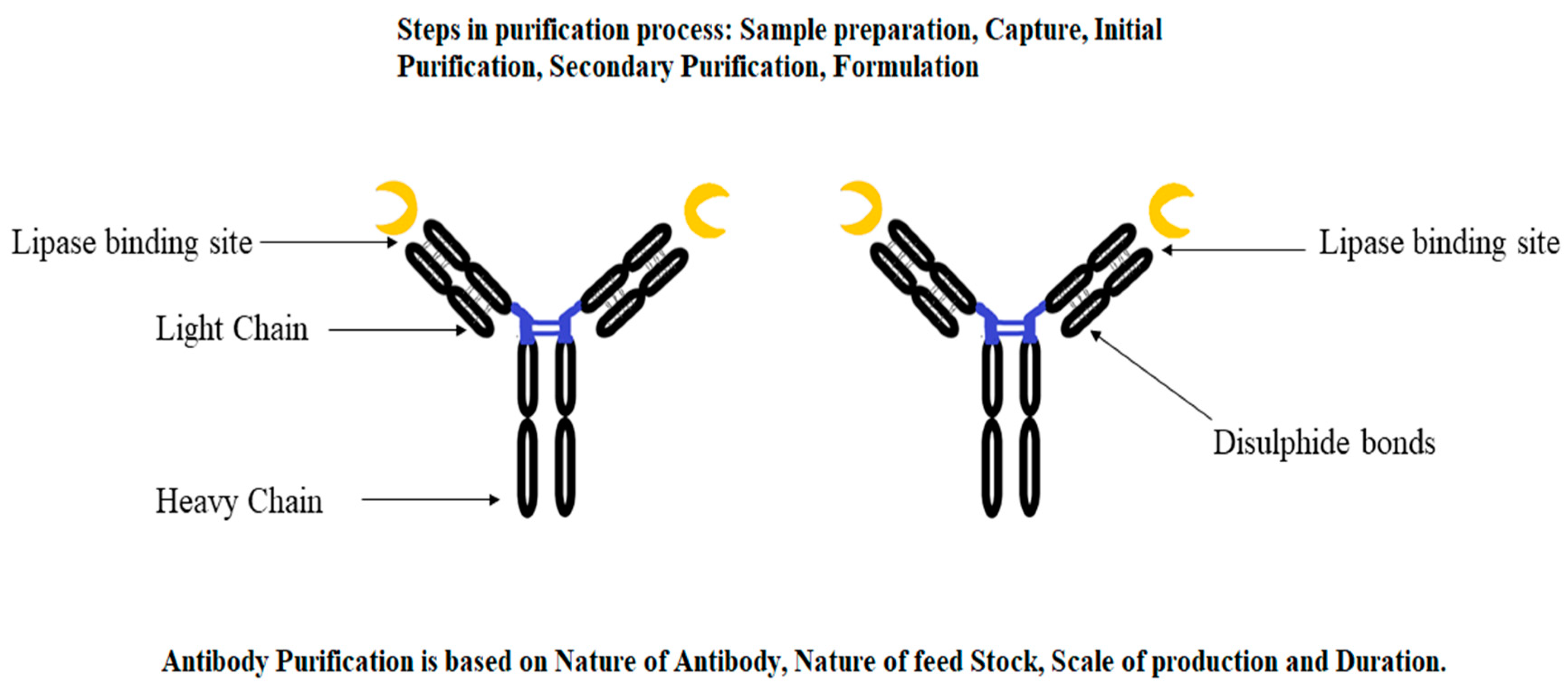
3.2.1. Immunopurification
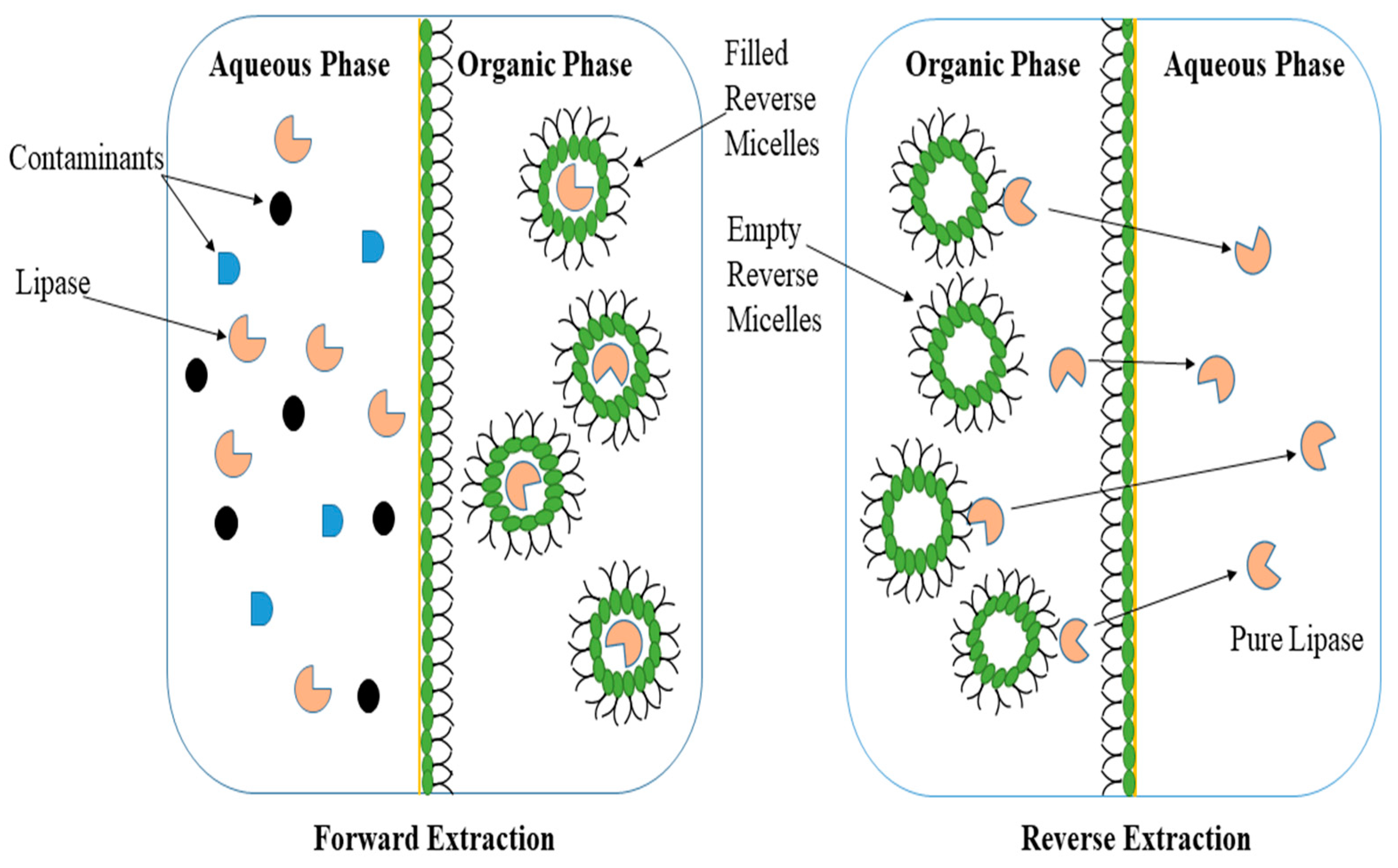
3.2.2. Reverse Micellar Systems (RMS)
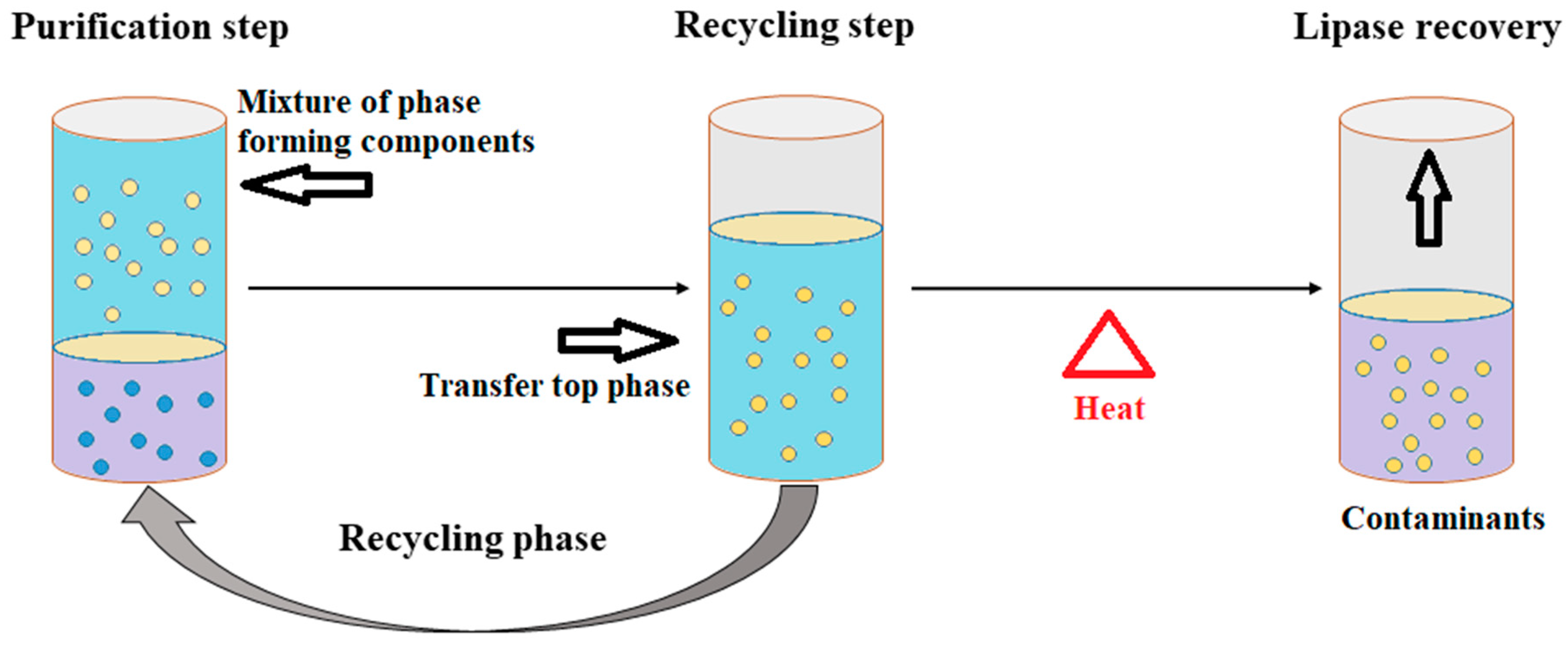
3.2.3. Aqueous Two-Phase System (ATPS)
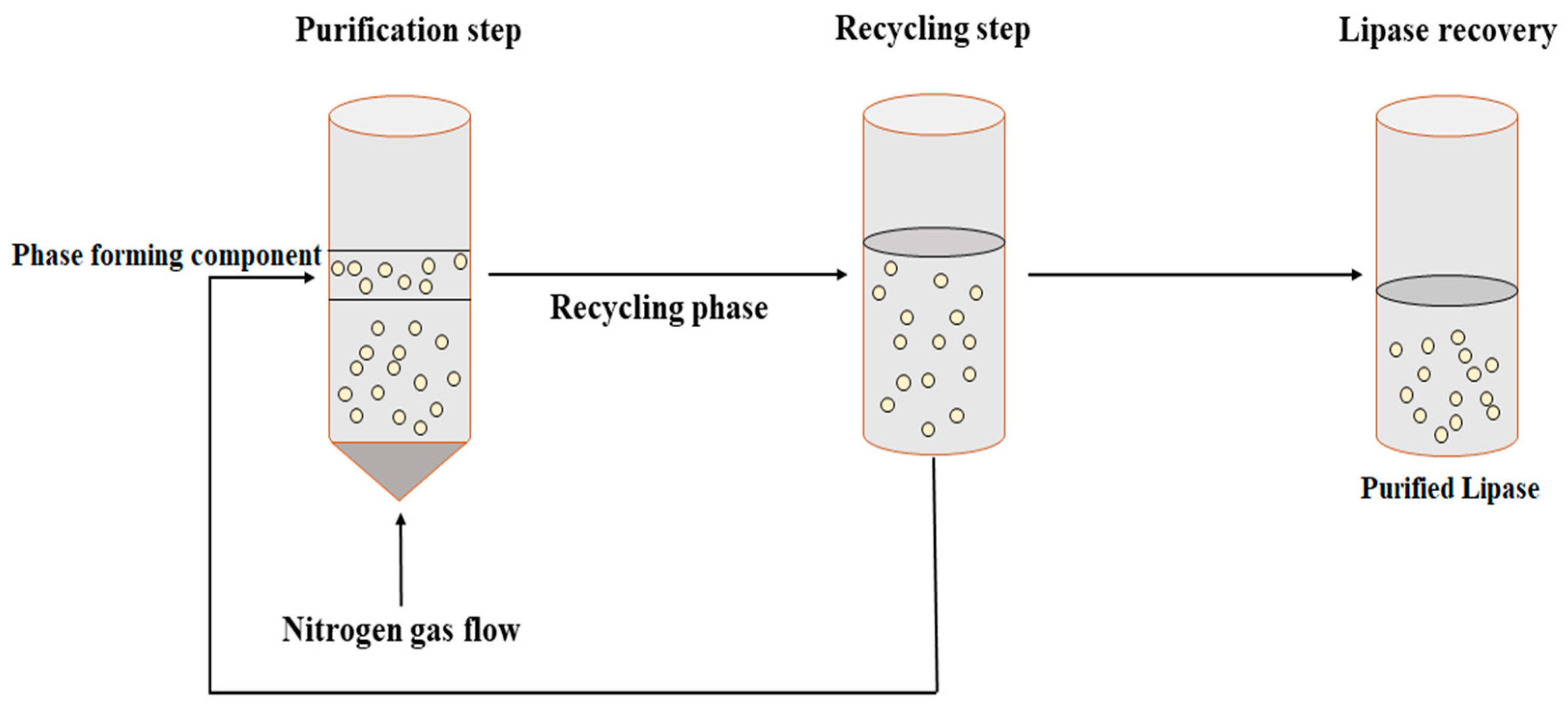
3.2.4. Aqueous Two-Phase Flotation (ATPF)
4. Uses of Microbial Lipases in Various Industrial Processes
4.1. Food Industry
4.2. Textile Industry
4.3. Leather Industry
4.4. Cosmetics Industry
4.5. Paper Industry
4.6. Detergent Industry
5. Role of Lipases in Biosensors
6. Role of Lipases in Biodiesel Production
7. Role of Lipases in the Separation of Racemic Mixtures
8. Role of Lipases in Bioremediation
9. Recent Trends and Targets in Lipase Engineering
9.1. Thermo-Stability
9.2. Catalytic Activity
9.3. Solvent Tolerance
10. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jaeger, K.-E.; Reetz, M.T. Microbial lipases form versatile tools for biotechnology. Trends Biotechnol. 1998, 16, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, P.; Kanwar, S.S. Lipase catalyzed esters syntheses in organic media: A review. Int. J. Inst. Pharm. Life Sci. 2012, 2, 91–119. [Google Scholar]
- Verma, N.; Thakur, S.; Bhatt, A.K. Microbial lipases: Industrial applications and properties (a review). Int. Res. J. Biol. Sci. 2012, 1, 88–92. [Google Scholar]
- Rozi, M.F.A.M.; Rahman, R.N.Z.R.A.; Leow, A.T.C.; Ali, M.S.M. Ancestral sequence reconstruction of ancient lipase from family I. 3 bacterial lipolytic enzymes. Mol. Phylogenet. Evol. 2022, 168, 107381. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, K.-E.; Ransac, S.; Dijkstra, B.W.; Colson, C.; van Heuvel, M.; Misset, O. Bacterial lipases. FEMS Microbiol. Rev. 1994, 15, 29–63. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, L.B.; Audipudi, A.V. Biochemical Characterization of Extracellular Lipase from an Improved Strain of Penicillium Citrinum KU613360. Ann. Rom. Soc. Cell Biol. 2021, 25, 7758–7770. [Google Scholar]
- Rodríguez-Salarichs, J.; Garcia de Lacoba, M.; Prieto, A.; Martínez, M.J.; Barriuso, J. Versatile lipases from the candida rugosa-like family: A mechanistic insight using computational approaches. J. Chem. Inf. Model. 2021, 61, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Kexin, Z.; Yonggang, W.; Ebadi, A.G.; Toughani, M. Optimization of low-temperature lipase production conditions and study on enzymatic properties of Aspergillus Niger. Iran. J. Chem. Chem. Eng. 2021, 40, 1364–1374. [Google Scholar]
- Adio, O.Q.; Kareem, S.O.; Osho, M.B.; Omemu, A.M. Production of lipases in solid-state fermentation by Aspergillus niger F7-02 With Agricultural Residues. J. Microbiol. Biotechnol. Food Sci. 2021, 2021, 509–512. [Google Scholar]
- Helal, S.E.; Abdelhady, H.M.; Abou-Taleb, K.A.; Hassan, M.G.; Amer, M.M. Lipase from Rhizopus oryzae R1: In-depth characterization, immobilization, and evaluation in biodiesel production. J. Genet. Eng. Biotechnol. 2021, 19, 1–13. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Pang, H.; Yuan, S.; Wang, X.; Hu, Z.; Zhou, Q.; He, Y.; Yan, Y.; Xu, L. Codisplay of Rhizopus oryzae and Candida rugosa lipases for biodiesel production. Catalysts 2021, 11, 421. [Google Scholar] [CrossRef]
- Phukon, L.C.; Chourasia, R.; Kumari, M.; Godan, T.K.; Sahoo, D.; Parameswaran, B.; Rai, A.K. Production and characterisation of lipase for application in detergent industry from a novel Pseudomonas helmanticensis HS6. Bioresour. Technol. 2020, 309, 123352. [Google Scholar] [CrossRef] [PubMed]
- Yeon-Sik, C.; In-Jung, L.; Jong-Myeong, K.; Seon-Kap, H.; Seok-Jong, S.; Ho-Youn, K.; Hyeokjun, Y.; Muhammad, H.; Sumera, K.; Ung-Han, Y. Plant growth promotion and Penicillium citrinum. BMC Microbiol. 2008, 8, 231. [Google Scholar]
- Kriegera, N.; Taipa, M.A.; Melo, E.H.M. Kinetic characterization of PC Lipase in AOT/isooctane reverse micelles. Appl. Biochem. Biotechnol. 1997, 67, 87–95. [Google Scholar] [CrossRef]
- Goto, M.; Noda, S.; Kamiya, N.; Nakashio, F. Enzymatic resolution of racemic ibuprofen by surfactant-coated lipases in organic media. Biotechnol. Lett. 1996, 18, 839–844. [Google Scholar] [CrossRef]
- Ou, J.; Yuan, X.; Liu, Y.; Zhang, P.; Xu, W.; Tang, K. Lipase from pseudomonas cepacia immobilized into ZIF-8 as bio-catalyst for enantioselective hydrolysis and transesterification. Process Biochem. 2021, 102, 132–140. [Google Scholar] [CrossRef]
- Pang, Y.; Zhang, Y.; Chen, M.; Lu, W.; Chen, M.; Yan, Y.; Lin, M.; Zhang, W.; Zhou, Z. Pseudomonas nanhaiensis sp. nov., a lipase-producing bacterium isolated from deep-sea sediment of the South China Sea. Antonie Van Leeuwenhoek 2021, 114, 1791–1804. [Google Scholar] [CrossRef]
- Pandey, A.; Benjamin, S.; Soccol, C.R.; Nigam, P.; Krieger, N.; Soccol, V.T. The realm of microbial lipases in biotechnology. Biotechnol. Appl. Biochem. 1999, 29, 119–131. [Google Scholar]
- Fatima, S.; Faryad, A.; Ataa, A.; Joyia, F.A.; Parvaiz, A. Microbial lipase production: A deep insight into the recent advances of lipase production and purification techniques. Biotechnol. Appl. Biochem. 2021, 68, 445–458. [Google Scholar] [CrossRef]
- Greco-Duarte, J.; de Almeida, F.P.; de Godoy, M.G.; Lins, U.; Freire, D.M.G.; Gutarra, M.L.E. Simultaneous lipase production and immobilization: Morphology and physiology study of Penicillium simplicissimum in submerged and solid-state fermentation with polypropylene as an inert support. Enzym. Microb. Technol. 2023, 164, 110173. [Google Scholar] [CrossRef]
- Carvalho, C.M.L.; Cabral, J.M.S. Reverse micelles as reaction media for lipases. Biochimie 2000, 82, 1063–1085. [Google Scholar] [CrossRef] [PubMed]
- Soares, G.A.; Alnoch, R.C.; Silva Dias, G.; Santos Reis, N.d.; Tavares, I.M.d.C.; Ruiz, H.A.; Bilal, M.; de Oliveira, J.R.; Krieger, N.; Franco, M. Production of a fermented solid containing lipases from Penicillium roqueforti ATCC 10110 and its direct employment in organic medium in ethyl oleate synthesis. Biotechnol. Appl. Biochem. 2022, 69, 1284–1299. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.; Qin, C.; Ahanger, M.A.; Raza, S.; Khan, M.I.; Ashraf, M.; Ahmed, N.; Zhang, L. Role of arbuscular mycorrhizal fungi in plant growth regulation: Implications in abiotic stress tolerance. Front. Plant Sci. 2019, 10, 1068. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; Herrera de Los Santos, M.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.N. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Factories 2019, 18, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhou, X.; Zhao, Y.; Zhu, S.; Wu, L.; He, Y.; Ping, X.; Lu, X.; Huang, W.; Qian, J. Colonization of endophyte Acremonium sp. D212 in Panax notoginseng and rice mediated by auxin and jasmonic acid. J. Integr. Plant Biol. 2020, 62, 1433–1451. [Google Scholar] [CrossRef]
- Kamat, S.; Kumari, M.; Sajna, K.V.; Jayabaskaran, C. Endophytic fungus, Chaetomium globosum, associated with marine green alga, a new source of Chrysin. Sci. Rep. 2020, 10, 18726. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tan, Y.; Liu, J.; Zhou, X.; Zeng, S.; Dong, J.; Liu, Y.; Yang, B. A new griseofulvin derivative from a soft coral-derived fungus Eupenicillium sp. SCSIO41208. Nat. Prod. Res. 2020, 34, 2971–2975. [Google Scholar] [CrossRef]
- El-Sheikh, M.A.; Rajaselvam, J.; Abdel-Salam, E.M.; Vijayaraghavan, P.; Alatar, A.A.; Biji, G.D. Paecilomyces sp. ZB is a cell factory for the production of gibberellic acid using a cheap substrate in solid state fermentation. Saudi J. Biol. Sci. 2020, 27, 2431–2438. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, X.; Li, G.; Feng, Z.; Xu, J. Pestalotiopisorin B, a new isocoumarin derivative from the mangrove endophytic fungus Pestalotiopsis sp. HHL101. Nat. Prod. Res. 2020, 34, 1002–1007. [Google Scholar] [CrossRef]
- Jensen, B.; Coolen, B.F.; Smit, T.H. Hymenophore configuration of the oak mazegill (Daedalea quercina). Mycologia 2020, 112, 895–907. [Google Scholar] [CrossRef]
- Coton, E.; Coton, M.; Hymery, N.; Mounier, J.; Jany, J.-L. Penicillium roqueforti: An overview of its genetics, physiology, metabolism and biotechnological applications. Fungal Biol. Rev. 2020, 34, 59–73. [Google Scholar] [CrossRef]
- Isbrandt, T.; Tolborg, G.; Ødum, A.; Workman, M.; Larsen, T.O. Atrorosins: A new subgroup of Monascus pigments from Talaromyces atroroseus. Appl. Microbiol. Biotechnol. 2020, 104, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Dannon, H.F.; Dannon, A.E.; Douro-Kpindou, O.K.; Zinsou, A.V.; Houndete, A.T.; Toffa-Mehinto, J.; Elegbede, I.A.T.; Olou, B.D.; TamÒ, M. Toward the efficient use of Beauveria bassiana in integrated cotton insect pest management. J. Cotton Res. 2020, 3, 1–21. [Google Scholar] [CrossRef]
- Rana, K.L.; Kour, D.; Kaur, T.; Devi, R.; Negi, C.; Yadav, A.N.; Yadav, N.; Singh, K.; Saxena, A.K. Endophytic fungi from medicinal plants: Biodiversity and biotechnological applications. In Microbial Endophytes; Elsevier: Amsterdam, The Netherlands, 2020; pp. 273–305. [Google Scholar]
- Fu, F.; Zhang, X.; Liu, F.; Peng, G.; Yu, F.; Fernando, D. Identification of resistance loci in Chinese and Canadian canola/rapeseed varieties against Leptosphaeria maculans based on genome-wide association studies. BMC Genom. 2020, 21, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, M.; Malathi, P.; Sundar, A.R.; Viswanathan, R. Use of green fluorescent protein expressing Colletotrichum falcatum, the red rot pathogen for precise host–pathogen interaction studies in sugarcane. Sugar Tech 2020, 22, 112–121. [Google Scholar] [CrossRef]
- Deb, D.; Khan, A.; Dey, N. Phoma diseases: Epidemiology and control. Plant Pathol. 2020, 69, 1203–1217. [Google Scholar] [CrossRef]
- Knob, A.; Izidoro, S.C.; Lacerda, L.T.; Rodrigues, A.; de Lima, V.A. A novel lipolytic yeast Meyerozyma guilliermondii: Efficient and low-cost production of acid and promising feed lipase using cheese whey. Biocatal. Agric. Biotechnol. 2020, 24, 101565. [Google Scholar] [CrossRef]
- Padilla, B.; Gil, J.V.; Manzanares, P. Challenges of the non-conventional yeast Wickerhamomyces anomalus in winemaking. Fermentation 2018, 4, 68. [Google Scholar] [CrossRef]
- Karim, A.; Gerliani, N.; Aïder, M. Kluyveromyces marxianus: An emerging yeast cell factory for applications in food and biotechnology. Int. J. Food Microbiol. 2020, 333, 108818. [Google Scholar] [CrossRef]
- Capece, A.; Pietrafesa, R.; Siesto, G.; Romano, P. Biotechnological approach based on selected Saccharomyces cerevisiae starters for reducing the use of sulfur dioxide in wine. Microorganisms 2020, 8, 738. [Google Scholar] [CrossRef]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Maza, D.D.; Viñarta, S.C.; Su, Y.; Guillamón, J.M.; Aybar, M.J. Growth and lipid production of Rhodotorula glutinis R4, in comparison to other oleaginous yeasts. J. Biotechnol. 2020, 310, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Xu, Z.; Chen, S.; Jin, M. Microbial lipid production from dilute acid and dilute alkali pretreated corn stover via Trichosporon dermatis. Bioresour. Technol. 2020, 295, 122253. [Google Scholar] [CrossRef]
- Colomer, M.S.; Funch, B.; Forster, J. The raise of Brettanomyces yeast species for beer production. Curr. Opin. Biotechnol. 2019, 56, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Bermas, A.; Geddes-McAlister, J. Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 2020, 114, 721–734. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Li, Z.; Liu, L.; Chen, X.; Li, W.; Zhang, X.; Dong, M. Lead removal from water by a newly isolated Geotrichum candidum LG-8 from Tibet kefir milk and its mechanism. Chemosphere 2020, 259, 127507. [Google Scholar] [CrossRef]
- Cui, J.; Xia, P.; Zhang, L.; Hu, Y.; Xie, Q.; Xiang, H. A novel fermented soybean, inoculated with selected Bacillus, Lactobacillus and Hansenula strains, showed strong antioxidant and anti-fatigue potential activity. Food Chem. 2020, 333, 127527. [Google Scholar] [CrossRef]
- Asitok, A.; Ekpenyong, M.; Ogarekpe, N.; Antigha, R.; Takon, I.; Rao, A.; Iheanacho, J.; Antai, S. Intracellular-to-extracellular localization switch of acidic lipase in Enterobacter cloacae: Evaluation of production kinetics and enantioselective esterification potential for pharmaceutical applications. Prep. Biochem. Biotechnol. 2022, 1–15. [Google Scholar] [CrossRef]
- Intwala, S.; Barot, J. Characterization & Partial purification of Microbial Lipase enzyme produce by Serratia nematodiphilia isolated from Paper & pulp effluent. South Asian J. Exp. Biol. 2022, 12, 414–421. [Google Scholar]
- Yasin, M.T.; Ali, Y.; Ahmad, K.; Ghani, A.; Amanat, K.; Basheir, M.M.; Faheem, M.; Hussain, S.; Ahmad, B.; Hussain, A. Alkaline lipase production by novel meso-tolerant psychrophilic Exiguobacterium sp. strain (AMBL-20) isolated from glacier of northeastern Pakistan. Arch. Microbiol. 2021, 203, 1309–1320. [Google Scholar] [CrossRef]
- Xu, C.; Suo, H.; Xue, Y.; Qin, J.; Chen, H.; Hu, Y. Experimental and theoretical evidence of enhanced catalytic performance of lipase B from Candida antarctica acquired by the chemical modification with amino acid ionic liquids. Mol. Catal. 2021, 501, 111355. [Google Scholar] [CrossRef]
- Gao, X.-Y.; Liu, X.-J.; Fu, C.-A.; Gu, X.-F.; Lin, J.-Q.; Liu, X.-M.; Pang, X.; Lin, J.-Q.; Chen, L.-X. Novel strategy for improvement of the bioleaching efficiency of Acidithiobacillus ferrooxidans based on the AfeI/R quorum sensing system. Minerals 2020, 10, 222. [Google Scholar] [CrossRef]
- Vergara, E.; Neira, G.; González, C.; Cortez, D.; Dopson, M.; Holmes, D.S. Evolution of predicted acid resistance mechanisms in the extremely acidophilic Leptospirillum genus. Genes 2020, 11, 389. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, G.C.; Gifford, S.M.; Septer, A.N. A model roseobacter, Ruegeria pomeroyi DSS-3, employs a diffusible killing mechanism to eliminate competitors. Msystems 2020, 5, e00443-20. [Google Scholar] [CrossRef]
- Mathew, C.E.; Izomor, R.N. Screening for Lipase Producing Bacteria from Abattoir Soil. Biosci. J. 2023, 11, 37–42. [Google Scholar]
- Mazhar, H.; Ullah, I.; Ali, U.; Abbas, N.; Hussain, Z.; Ali, S.S.; Zhu, H. Optimization of low-cost solid-state fermentation media for the production of thermostable lipases using agro-industrial residues as substrate in culture of Bacillus amyloliquefaciens. Biocatal. Agric. Biotechnol. 2023, 47, 102559. [Google Scholar] [CrossRef]
- Altinok, F.; Albayrak, S.; Arslan, N.P.; Taskin, M.; Aygun, E.; Sisecioglu, M.; Adiguzel, A. Application of Anoxybacillus gonensins UF7 lipase as a catalyst for biodiesel production from waste frying oils. Fuel 2023, 334, 126672. [Google Scholar] [CrossRef]
- Qiu, J.; Han, R.; Wang, C. Microbial halophilic lipases: A review. J. Basic Microbiol. 2021, 61, 594–602. [Google Scholar] [CrossRef]
- Ismail, A.R.; Kashtoh, H.; Baek, K.-H. Temperature-resistant and solvent-tolerant lipases as industrial biocatalysts: Biotechnological approaches and applications. Int. J. Biol. Macromol. 2021, 187, 127–142. [Google Scholar] [CrossRef]
- Elend, C.; Schmeisser, C.; Hoebenreich, H.; Steele, H.L.; Streit, W.R. Isolation and characterization of a metagenome-derived and cold-active lipase with high stereospecificity for (R)-ibuprofen esters. J. Biotechnol. 2007, 130, 370–377. [Google Scholar] [CrossRef]
- Borrelli, G.M.; Trono, D. Recombinant lipases and phospholipases and their use as biocatalysts for industrial applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef]
- Chandra, P.; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 169. [Google Scholar] [CrossRef] [PubMed]
- Hamayun, M.; Afzal Khan, S.; Ahmad, N.; Tang, D.-S.; Kang, S.-M.; Na, C.-I.; Sohn, E.-Y.; Hwang, Y.-H.; Shin, D.-H.; Lee, B.-H. Cladosporium sphaerospermum as a new plant growth-promoting endophyte from the roots of Glycine max (L.) Merr. World J. Microbiol. Biotechnol. 2009, 25, 627–632. [Google Scholar] [CrossRef]
- Hamayun, M.; Hussain, A.; Khan, S.A.; Kim, H.-Y.; Khan, A.L.; Waqas, M.; Irshad, M.; Iqbal, A.; Rehman, G.; Jan, S. Gibberellins producing endophytic fungus Porostereum spadiceum AGH786 rescues growth of salt affected soybean. Front. Microbiol. 2017, 8, 686. [Google Scholar] [CrossRef]
- Ikram, M.; Ali, N.; Jan, G.; Jan, F.G.; Rahman, I.U.; Iqbal, A.; Hamayun, M. IAA producing fungal endophyte Penicillium roqueforti Thom., enhances stress tolerance and nutrients uptake in wheat plants grown on heavy metal contaminated soils. PLoS ONE 2018, 13, e0208150. [Google Scholar] [CrossRef] [PubMed]
- Godoy, C.A.; Klett, J.; Di Geronimo, B.; Hermoso, J.A.; Guisán, J.M.; Carrasco-López, C. Disulfide engineered lipase to enhance the catalytic activity: A structure-based approach on btl2. Int. J. Mol. Sci. 2019, 20, 5245. [Google Scholar] [CrossRef]
- Patel, N.; Rai, D.; Shahane, S.; Mishra, U. Lipases: Sources, production, purification, and applications. Recent Pat. Biotechnol. 2019, 13, 45–56. [Google Scholar] [CrossRef]
- Benjamin, S.; Pandey, A. Candida rugosa lipases: Molecular biology and versatility in biotechnology. Yeast 1998, 14, 1069–1087. [Google Scholar] [CrossRef]
- Triyaswati, D.; Ilmi, M. Lipase-producing filamentous fungi from non-dairy creamer industrial waste. Microbiol. Biotechnol. Lett. 2020, 48, 167–178. [Google Scholar] [CrossRef]
- Lima, A.C.P.; Cammarota, M.C.; Gutarra, M.L.E. Obtaining filamentous fungi and lipases from sewage treatment plant residue for fat degradation in anaerobic reactors. PeerJ 2018, 6, e5368. [Google Scholar] [CrossRef]
- Araujo, S.C.; Ramos, M.R.M.F.; do Espírito Santo, E.L.; de Menezes, L.H.S.; de Carvalho, M.S.; Tavares, I.M.d.C.; Franco, M.; de Oliveira, J.R. Optimization of lipase production by Penicillium roqueforti ATCC 10110 through solid-state fermentation using agro-industrial residue based on a univariate analysis. Prep. Biochem. Biotechnol. 2022, 52, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Vakhlu, J. Yeast lipases: Enzyme purification, biochemical properties and gene cloning. Electron. J. Biotechnol. 2006, 9. [Google Scholar] [CrossRef]
- Kademi, A.; Lee, B.; Houde, A. Production of heterologous microbial lipases by yeasts. Indian J. Biotechnol. 2003, 2, 346–355. [Google Scholar]
- Kasana, R.C.; Pandey, C.B. Exiguobacterium: An overview of a versatile genus with potential in industry and agriculture. Crit. Rev. Biotechnol. 2018, 38, 141–156. [Google Scholar] [CrossRef]
- Hwang, H.T.; Qi, F.; Yuan, C.; Zhao, X.; Ramkrishna, D.; Liu, D.; Varma, A. Lipase-catalyzed process for biodiesel production: Protein engineering and lipase production. Biotechnol. Bioeng. 2014, 111, 639–653. [Google Scholar] [CrossRef]
- Sahoo, R.K.; Kumari, K.S.; Sahoo, S.; Das, A.; Gaur, M.; Dey, S.; Mohanty, S.; Subudhi, E. Bio-statistical optimization of lipase production by thermophilic Pseudomonas formosensis and its application on oral biofilm degradation. Biocatal. Agric. Biotechnol. 2021, 33, 101969. [Google Scholar] [CrossRef]
- Moon, Y.-S.; Ali, S. Isolation and identification of multi-trait plant growth–promoting rhizobacteria from coastal sand dune plant species of Pohang beach. Folia Microbiol. 2022, 67, 523–533. [Google Scholar] [CrossRef]
- Zhao, J.; Ma, M.; Zeng, Z.; Yu, P.; Gong, D.; Deng, S. Production, purification and biochemical characterisation of a novel lipase from a newly identified lipolytic bacterium Staphylococcus caprae NCU S6. J. Enzym. Inhib. Med. Chem. 2021, 36, 249–257. [Google Scholar] [CrossRef]
- Saxena, R.K.; Sheoran, A.; Giri, B.; Davidson, W.S. Purification strategies for microbial lipases. J. Microbiol. Methods 2003, 52, 1–18. [Google Scholar] [CrossRef]
- Bharathi, D.; Rajalakshmi, G.; Komathi, S. Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil. J. King Saud Univ.-Sci. 2019, 31, 898–901. [Google Scholar] [CrossRef]
- Ameri, A.; Shakibaie, M.; Faramarzi, M.A.; Ameri, A.; Amirpour-Rostami, S.; Rahimi, H.R.; Forootanfar, H. Thermoalkalophilic lipase from an extremely halophilic bacterial strain Bacillus atrophaeus FSHM2: Purification, biochemical characterization and application. Biocatal. Biotransform. 2017, 35, 151–160. [Google Scholar] [CrossRef]
- Iftikhar, T.; Niaz, M.; Jabeen, R.; Haq, I.U. Purification and characterization of extracellular lipases. Pak. J. Bot 2011, 43, 1541–1545. [Google Scholar]
- Rahman, M.B.A.; Azaman, R.N.; Omar, E.M.; Latif, M.A.M.; Abdulmalek, E. Candida rugosa lipase immobilized on diethylaminoethyl-cellulose (deae) for esterification of butyl oleate. Malays. J. Anal. Sci. 2019, 23, 376–382. [Google Scholar]
- Hecht, E.S.; Mehta, S.; Wecksler, A.T.; Aguilar, B.; Swanson, N.; Phung, W.; Dubey Kelsoe, A.; Benner, W.H.; Tesar, D.; Kelley, R.F. Insights into ultra-low affinity lipase-antibody noncovalent complex binding mechanisms. mAbs 2022, 14, 2135183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Riccardi, C.; Kamen, D.; Reilly, J.; Mattila, J.; Bak, H.; Xiao, H.; Li, N. Identification of the specific causes of polysorbate 20 degradation in monoclonal antibody formulations containing multiple lipases. Pharm. Res. 2022, 39, 75–87. [Google Scholar] [CrossRef]
- Rahimi, H.; Soro, S.; Rughetti, A.; Palocci, C.; Biffoni, M.; Barachini, S.; Taurino, F.; Cernia, E.; Frati, L.; Nuti, M. Monoclonal antibodies against Candida rugosa lipase. J. Mol. Catal. B Enzym. 2004, 28, 71–74. [Google Scholar] [CrossRef]
- Behera, S.; Das, S.; Balasubramanian, S. An atomistic view of solvent-free protein liquids: The case of Lipase A. Phys. Chem. Chem. Phys. 2021, 23, 7302–7312. [Google Scholar] [CrossRef]
- Gaikaiwari, R.P.; Wagh, S.A.; Kulkarni, B.D. Efficient lipase purification using reverse micellar extraction. Bioresour. Technol. 2012, 108, 224–230. [Google Scholar] [CrossRef]
- Goswami, D. Lipase Catalysis in Mixed Micelles. ChemBioEng Rev. 2022, 9, 409–418. [Google Scholar] [CrossRef]
- Shehata, M.; Ünlü, A.; Iglesias-Fernández, J.; Osuna, S.; Sezerman, O.U.; Timucin, E. Brave new surfactant world revisited by thermoalkalophilic lipases: Computational insights into the role of SDS as a substrate analog. Phys. Chem. Chem. Phys. 2023, 25, 2234–2247. [Google Scholar] [CrossRef]
- Fernandes, M.L.M.; Krieger, N.; Baron, A.M.; Zamora, P.P.; Ramos, L.P.; Mitchell, D.A. Hydrolysis and synthesis reactions catalysed by Thermomyces lanuginosa lipase in the AOT/Isooctane reversed micellar system. J. Mol. Catal. B Enzym. 2004, 30, 43–49. [Google Scholar] [CrossRef]
- Tan, C.H.; Show, P.L.; Ooi, C.W.; Ng, E.P.; Lan, J.C.W.; Ling, T.C. Novel lipase purification methods–a review of the latest developments. Biotechnol. J. 2015, 10, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Souza, R.L.; Lima, R.A.; Coutinho, J.A.P.; Soares, C.M.F.; Lima, Á.S. Novel aqueous two-phase systems based on tetrahydrofuran and potassium phosphate buffer for purification of lipase. Process Biochem. 2015, 50, 1459–1467. [Google Scholar] [CrossRef]
- Souza, R.L.; Lima, R.A.; Coutinho, J.A.P.; Soares, C.M.F.; Lima, Á.S. Aqueous two-phase systems based on cholinium salts and tetrahydrofuran and their use for lipase purification. Sep. Purif. Technol. 2015, 155, 118–126. [Google Scholar] [CrossRef]
- Bi, P.-y.; Li, D.-q.; Dong, H.-r. A novel technique for the separation and concentration of penicillin G from fermentation broth: Aqueous two-phase flotation. Sep. Purif. Technol. 2009, 69, 205–209. [Google Scholar] [CrossRef]
- Show, P.L.; Tan, C.P.; Anuar, M.S.; Ariff, A.; Yusof, Y.A.; Chen, S.K.; Ling, T.C. Direct recovery of lipase derived from Burkholderia cepacia in recycling aqueous two-phase flotation. Sep. Purif. Technol. 2011, 80, 577–584. [Google Scholar] [CrossRef]
- Aravindan, R.; Anbumathi, P.; Viruthagiri, T. Lipase applications in food industry. Indian J. Biotechnol. 2007, 6, 141–158. [Google Scholar]
- Sharma, R.; Chisti, Y.; Banerjee, U.C. Production, purification, characterization, and applications of lipases. Biotechnol. Adv. 2001, 19, 627–662. [Google Scholar] [CrossRef]
- Seyed-Moslemi, S.A.; Hesari, J.; Peighambardoust, S.H.; Peighambardoust, S.J. Effect of microbial lipase and transglutaminase on the textural, physicochemical, and microbial parameters of fresh quark cheese. J. Dairy Sci. 2021, 104, 7489–7499. [Google Scholar] [CrossRef]
- Akram, F.; Mir, A.S.; Haq, I.u.; Roohi, A. An appraisal on prominent industrial and biotechnological applications of bacterial lipases. Mol. Biotechnol. 2022, 1–23. [Google Scholar] [CrossRef]
- Mahboob, S.; Tahir, K.; Ali, S. a systematic overview on the upstreaming, downstreaming and industrial applications of microbial lipases. Int. J. Biol. Biotechnol. 2022, 19, 171–182. [Google Scholar]
- Adetunji, A.I.; Olaniran, A.O. Production strategies and biotechnological relevance of microbial lipases: A review. Braz. J. Microbiol. 2021, 52, 1257–1269. [Google Scholar] [CrossRef]
- Medeiros, A.B.P.; Rossi, S.C.; Bier, M.C.J.; Vandenberghe, L.P.S.; Soccol, C.R. Enzymes for flavor, dairy, and baking industries. In Food Composition and Analysis; Apple Academic Pres: Abingdon, UK, 2014; pp. 37–48. [Google Scholar]
- de Alencar Figueira Angelotti, J.; Battaglia Hirata, D.; Rodrigues dos Santos, M. Lipases: Sources of acquisition, ways of production, and recent applications. Catal. Res. 2022, 2, 1–43. [Google Scholar]
- Ramarethinam, S.; Latha, K.; Rajalakshmi, N. Use of a fungal lipase for enhancement of aroma in black tea. Food Sci. Technol. Res. 2002, 8, 328–332. [Google Scholar] [CrossRef]
- Yao, C.; Yuan, A.; Zhang, H.; Li, B.; Liu, J.; Xi, F.; Dong, X. Facile surface modification of textiles with photocatalytic carbon nitride nanosheets and the excellent performance for self-cleaning and degradation of gaseous formaldehyde. J. Colloid Interface Sci. 2019, 533, 144–153. [Google Scholar] [CrossRef] [PubMed]
- El-Fiky, A.F.; Khalil, E.M.; Mowafi, S.; Zaki, R.A.; El-Sayed, H. A novel approach towards removal of lipid barrier from wool fibers’ surface using thermophilic lipase. J. Nat. Fibers 2022, 19, 9471–9485. [Google Scholar] [CrossRef]
- McKelvey, S.M.; Murphy, R.A. Biotechnological use of fungal enzymes. Fungi Biol. Appl. 2017, 201–225. [Google Scholar] [CrossRef]
- Kalantzi, S.; Kekos, D.; Mamma, D. Bioscouring of cotton fabrics by multienzyme combinations: Application of Box–Behnken design and desirability function. Cellulose 2019, 26, 2771–2790. [Google Scholar] [CrossRef]
- Kumar, J.A.; Kumar, M.S. A study on improving dyeability of polyester fabric using lipase enzyme. Autex Res. J. 2019, 20, 243–249. [Google Scholar] [CrossRef]
- Selvam, K.; Vishnupriya, B.; Maanvizhi, M. Enzymatic synthesis of fragrance ester by lipase from marine Actinomycetes for textile industry. Int. J. Eng. Adv. Technol 2013, 3, 91–96. [Google Scholar]
- Abou Taleb, M.; Gomaa, S.K.; Wahba, M.I.; Zaki, R.A.; El-Fiky, A.F.; El-Refai, H.A.; El-Sayed, H. Bioscouring of wool fibres using immobilized thermophilic lipase. Int. J. Biol. Macromol. 2022, 194, 800–810. [Google Scholar] [CrossRef] [PubMed]
- El-Gendi, H.; Saleh, A.K.; Badierah, R.; Redwan, E.M.; El-Maradny, Y.A.; El-Fakharany, E.M. A comprehensive insight into fungal enzymes: Structure, classification, and their role in mankind’s challenges. J. Fungi 2022, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Moujehed, E.; Zarai, Z.; Khemir, H.; Miled, N.; Bchir, M.S.; Gablin, C.; Bessueille, F.; Bonhommé, A.; Leonard, D.; Carrière, F. Cleaner degreasing of sheepskins by the Yarrowia lipolytica LIP2 lipase as a chemical-free alternative in the leather industry. Colloids Surf. B Biointerfaces 2022, 211, 112292. [Google Scholar] [CrossRef] [PubMed]
- Aloulou, A.; Rodriguez, J.A.; Puccinelli, D.; Mouz, N.; Leclaire, J.; Leblond, Y.; Carrière, F. Purification and biochemical characterization of the LIP2 lipase from Yarrowia lipolytica. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2007, 1771, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Aloulou, A.; Puccinelli, D.; De Caro, A.; Leblond, Y.; Carrière, F. A comparative study on two fungal lipases from Thermomyces lanuginosus and Yarrowia lipolytica shows the combined effects of detergents and pH on lipase adsorption and activity. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2007, 1771, 1446–1456. [Google Scholar] [CrossRef]
- Ben Rejeb, I.; Khemir, H.; Messaoudi, Y.; Miled, N.; Gargouri, M. Optimization of Enzymatic Degreasing of Sheep Leather for an Efficient Approach and Leather Quality Improvement Using Fractional Experimental Design. Appl. Biochem. Biotechnol. 2022, 194, 2251–2268. [Google Scholar] [CrossRef]
- Ansorge-Schumacher, M.B.; Thum, O. Immobilised lipases in the cosmetics industry. Chem. Soc. Rev. 2013, 42, 6475–6490. [Google Scholar] [CrossRef]
- Hube, S.; Wu, B. Mitigation of emerging pollutants and pathogens in decentralized wastewater treatment processes: A review. Sci. Total Environ. 2021, 779, 146545. [Google Scholar] [CrossRef]
- Casanova, F.; Santos, L. Encapsulation of cosmetic active ingredients for topical application–a review. J. Microencapsul. 2016, 33, 1–17. [Google Scholar] [CrossRef]
- Fujiwara, M.; Nakahara, Y. Preparation of inorganic microcapsules and their applications to cosmetics. Fragr. J. 2007, 35, 33. [Google Scholar]
- Miguez, J.P.; Gama, R.S.; Bolina, I.C.A.; de Melo, C.C.; Cordeiro, M.R.; Hirata, D.B.; Mendes, A.A. Enzymatic synthesis optimization of a cosmetic ester catalyzed by a homemade biocatalyst prepared via physical adsorption of lipase on amino-functionalized rice husk silica. Chem. Eng. Res. Des. 2018, 139, 296–308. [Google Scholar] [CrossRef]
- Houde, A.; Kademi, A.; Leblanc, D. Lipases and their industrial applications: An overview. Appl. Biochem. Biotechnol. 2004, 118, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Kuila, A. Lipase and their different industrial applications: A review. Braz. J. Biol. Sci. 2018, 5, 237–247. [Google Scholar] [CrossRef]
- Farrell, R.L.; Hata, K.; Wall, M.B. Solving pitch problems in pulp and paper processes by the use of enzymes or fungi. Biotechnol. Pulp Pap. Ind. 2006, 197–212. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Javed, S.; Hameed, A. Enzymes used in detergents: Lipases. Afr. J. Biotechnol. 2010, 9, 4836–4844. [Google Scholar]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Hot topics in cold adaptation. Nat. Rev. Microbiol. 2003, 1, 200–208. [Google Scholar] [CrossRef]
- Bajpai, D. Laundry detergents: An overview. J. Oleo Sci. 2007, 56, 327–340. [Google Scholar] [CrossRef]
- Maharana, A.K.; Singh, S.M. A cold and organic solvent tolerant lipase produced by Antarctic strain Rhodotorula sp. Y-23. J. Basic Microbiol. 2018, 58, 331–342. [Google Scholar] [CrossRef]
- Rios, N.S.; Pinheiro, B.B.; Pinheiro, M.P.; Bezerra, R.M.; dos Santos, J.C.S.; Gonçalves, L.R.B. Biotechnological potential of lipases from Pseudomonas: Sources, properties and applications. Process Biochem. 2018, 75, 99–120. [Google Scholar] [CrossRef]
- Luo, Y.; Zheng, Y.; Jiang, Z.; Ma, Y.; Wei, D. A novel psychrophilic lipase from Pseudomonas fluorescens with unique property in chiral resolution and biodiesel production via transesterification. Appl. Microbiol. Biotechnol. 2006, 73, 349–355. [Google Scholar] [CrossRef]
- Liu, W.; Li, M.; Yan, Y. Heterologous expression and characterization of a new lipase from Pseudomonas fluorescens Pf0–1 and used for biodiesel production. Sci. Rep. 2017, 7, 15711. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Gupta, N.; Rathi, P. Bacterial lipases: An overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004, 64, 763–781. [Google Scholar] [CrossRef] [PubMed]
- Sandoval, G.; Herrera-López, E.J. Lipase, phospholipase, and esterase biosensors. Lipases Phospholipases Methods Protoc. 2018, 1832, 391–425. [Google Scholar]
- Herrera-López, E.J. Lipase and phospholipase biosensors: A review. Lipases Phospholipases Methods Protoc. 2012, 861, 525–543. [Google Scholar]
- Dutta, S.; Ray, L. Production and characterization of an alkaline thermostable crude lipase from an isolated strain of Bacillus cereus C 7. Appl. Biochem. Biotechnol. 2009, 159, 142–154. [Google Scholar] [CrossRef]
- Kumari, A.; Gupta, R. Extracellular expression and characterization of thermostable lipases, LIP8, LIP14 and LIP18, from Yarrowia lipolytica. Biotechnol. Lett. 2012, 34, 1733–1739. [Google Scholar] [CrossRef]
- Deb, C.; Daniel, J.; Sirakova, T.D.; Abomoelak, B.; Dubey, V.S.; Kolattukudy, P.E. A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 3866–3875. [Google Scholar] [CrossRef]
- Pohanka, M. Biosensors and bioassays based on lipases, principles and applications, a review. Molecules 2019, 24, 616. [Google Scholar] [CrossRef]
- Cavalcante, F.T.T.; Neto, F.S.; de Aguiar Falcão, I.R.; da Silva Souza, J.E.; de Moura Junior, L.S.; da Silva Sousa, P.; Rocha, T.G.; de Sousa, I.G.; de Lima Gomes, P.H.; de Souza, M.C.M. Opportunities for improving biodiesel production via lipase catalysis. Fuel 2021, 288, 119577. [Google Scholar] [CrossRef]
- Quayson, E.; Amoah, J.; Hama, S.; Kondo, A.; Ogino, C. Immobilized lipases for biodiesel production: Current and future greening opportunities. Renew. Sustain. Energy Rev. 2020, 134, 110355. [Google Scholar] [CrossRef]
- Mendes, A.A.; Soares, C.M.F.; Tardioli, P.W. Recent advances and future prospects for biolubricant base stocks production using lipases as environmentally friendly catalysts: A mini-review. World J. Microbiol. Biotechnol. 2023, 39, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, J.Y.N.H.; Cavalcante, F.T.T.; Freitas, L.M.; Castro, A.P.; Borges, P.T.; de Sousa Junior, P.G.; Filho, M.N.R.; Lopes, A.A.S.; da Fonseca, A.M.; Lomonaco, D. A Theoretical and Experimental Study for Enzymatic Biodiesel Production from Babassu Oil (Orbignya sp.) Using Eversa Lipase. Catalysts 2022, 12, 1322. [Google Scholar] [CrossRef]
- Abdulmalek, S.A.; Yan, Y. Recent developments of lipase immobilization technology and application of immobilized lipase mixtures for biodiesel production. Biofuels Bioprod. Biorefining 2022, 16, 1062–1094. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, T.; Kumar, R.; Meena, K.; Kanwar, S.S. Biodiesel and the potential role of microbial lipases in its production. Microb. Technol. Welf. Soc. 2019, 83–99. [Google Scholar] [CrossRef]
- Lee, H.-s.; Lee, D.; Kim, S.; Kim, J. Effect of supercritical carbon dioxide on the enzymatic production of biodiesel from waste animal fat using immobilized Candida antarctica lipase B variant. BMC Biotechnol. 2017, 17, 1–6. [Google Scholar]
- Sharma, R.K.; Saxena, M.; O’Neill, C.A.; Ramos, H.A.R.; Griebenow, K. Synthesis of Rhizopus arrhizus lipase nanoparticles for biodiesel production. ACS Omega 2018, 3, 18203–18213. [Google Scholar] [CrossRef]
- Narwal, S.K.; Saun, N.K.; Dogra, P.; Chauhan, G.; Gupta, R. Production and characterization of biodiesel using nonedible castor oil by immobilized lipase from Bacillus aerius. BioMed Res. Int. 2015, 2015, 281934. [Google Scholar] [CrossRef]
- Hasan, F.; Shah, A.A.; Hameed, A. Industrial applications of microbial lipases. Enzym. Microb. Technol. 2006, 39, 235–251. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Wang, X.; Ching, C.B.; Wu, J.C. Experimental optimization of enzymic kinetic resolution of racemic flurbiprofen. Biotechnol. Appl. Biochem. 2005, 42, 67–71. [Google Scholar]
- Muralidhar, R.V.; Chirumamilla, R.R.; Ramachandran, V.N.; Marchant, R.; Nigam, P. Racemic resolution of RS-baclofen using lipase from Candida cylindracea. Meded. Fak. Land Bouwkun Digeen Toegep. Biol. Wet 2001, 66, 227–232. [Google Scholar]
- Lesny, J.E.; Mohan, A.P.; Vijaya, J.A. Bioremediation of petroleum-polluted soil using biosurfactant producing bacteria, Pseudomonas sp. J. Sci. Res. 2022, 66, 1. [Google Scholar]
- Blandón, L.M.; Marín, M.A.; Quintero, M.; Jutinico-Shubach, L.M.; Montoya-Giraldo, M.; Santos-Acevedo, M.; Gómez-León, J. Diversity of cultivable bacteria from deep-sea sediments of the Colombian Caribbean and their potential in bioremediation. Antonie Van Leeuwenhoek 2022, 115, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Welz, P.; Swanepoel, G.; Weels, S.; Le Roes-Hill, M. Wastewater from the edible oil industry as a potential source of lipase-and surfactant-producing actinobacteria. Microorganisms 2021, 9, 1987. [Google Scholar] [CrossRef] [PubMed]
- Pathak, V.M. Review on the current status of polymer degradation: A microbial approach. Bioresour. Bioprocess. 2017, 4, 1–31. [Google Scholar] [CrossRef]
- Iram, D.; Riaz, R.; Iqbal, R.K. Usage of potential micro-organisms for degradation of plastics. Open J. Environ. Biol 2019, 4, 7–15. [Google Scholar]
- Wang, H.-T.; Ding, J.; Xiong, C.; Zhu, D.; Li, G.; Jia, X.-Y.; Zhu, Y.-G.; Xue, X.-M. Exposure to microplastics lowers arsenic accumulation and alters gut bacterial communities of earthworm Metaphire californica. Environ. Pollut. 2019, 251, 110–116. [Google Scholar] [CrossRef]
- Illanes, A.; Cauerhff, A.; Wilson, L.; Castro, G.R. Recent trends in biocatalysis engineering. Bioresour. Technol. 2012, 115, 48–57. [Google Scholar] [CrossRef]
- Soni, S. Trends in lipase engineering for enhanced biocatalysis. Biotechnol. Appl. Biochem. 2022, 69, 265–272. [Google Scholar] [CrossRef]
- de Miguel Bouzas, T.; Barros-Velázquez, J.; Gonzalez Villa, T. Industrial applications of hyperthermophilic enzymes: A review. Protein Pept. Lett. 2006, 13, 645–651. [Google Scholar] [CrossRef]
- Yasar, G.; Gulhan, U.G.; Guduk, E.; Aktas, F. Screening, partial purification and characterization of the hyper-thermophilic lipase produced by a new isolate of Bacillus subtilis LP2. Biocatal. Biotransform. 2020, 38, 367–375. [Google Scholar] [CrossRef]
- Castilla, A.; Giordano, S.R.; Irazoqui, G. Extremophilic lipases and esterases: Characteristics and industrial applications. In Microbial Extremozymes; Elsevier: Amsterdam, The Netherlands, 2022; pp. 207–222. [Google Scholar]
- Wang, R.; Wang, S.; Xu, Y.; Yu, X. Enhancing the thermostability of Rhizopus chinensis lipase by rational design and MD simulations. Int. J. Biol. Macromol. 2020, 160, 1189–1200. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.-W.; Wang, R.; Zhang, M.; Xu, Y.; Xiao, R. Enhanced thermostability of a Rhizopus chinensis lipase by in vivo recombination in Pichia pastoris. Microb. Cell Factories 2012, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Yu, H.; Xu, J.; Wang, Z.; Wang, Z.; Kang, T.; Chen, K.; Pu, Z.; Wu, J.; Yang, L. Enhancing thermostability of lipase from Pseudomonas alcaligenes for producing l-menthol by the CREATE strategy. Catal. Sci. Technol. 2022, 12, 2531–2541. [Google Scholar] [CrossRef]
- Ma, D.; Xin, Y.; Guo, Z.; Shi, Y.; Zhang, L.; Li, Y.; Gu, Z.; Ding, Z.; Shi, G. Ancestral sequence reconstruction and spatial structure analysis guided alteration of longer-chain substrate catalysis for Thermomicrobium roseum lipase. Enzym. Microb. Technol. 2022, 156, 109989. [Google Scholar] [CrossRef]
- Tian, M.; Yang, L.; Lv, P.; Wang, Z.; Fu, J.; Miao, C.; Li, Z.; Li, L.; Liu, T.; Du, W. Improvement of methanol tolerance and catalytic activity of Rhizomucor miehei lipase for one-step synthesis of biodiesel by semi-rational design. Bioresour. Technol. 2022, 348, 126769. [Google Scholar] [CrossRef]
- Peng, B.; Chen, F.; Liu, X.; Hu, J.-N.; Zheng, L.-F.; Li, J.; Deng, Z.-Y. Trace water activity could improve the formation of 1, 3-oleic-2-medium chain-rich triacylglycerols by promoting acyl migration in the lipase RM IM catalyzed interesterification. Food Chem. 2020, 313, 126130. [Google Scholar] [CrossRef]
- Zulkeflee, S.A.; Rohman, F.S.; Sata, S.A.; Aziz, N. Temperature and water activity control in a lipase catalyzed esterification process using nonlinear model predictive control. Can. J. Chem. Eng. 2022, 100, 3669–3690. [Google Scholar] [CrossRef]
- Wehtje, E.; Adlercreutz, P. Water activity and substrate concentration effects on lipase activity. Biotechnol. Bioeng. 1997, 55, 798–806. [Google Scholar] [CrossRef]
- Sharma, S.; Kanwar, S.S. Organic solvent tolerant lipases and applications. Sci. World J. 2014, 2014, 625258. [Google Scholar] [CrossRef]
- Yan, J.; Yan, Y.; Madzak, C.; Han, B. Harnessing biodiesel-producing microbes: From genetic engineering of lipase to metabolic engineering of fatty acid biosynthetic pathway. Crit. Rev. Biotechnol. 2017, 37, 26–36. [Google Scholar] [CrossRef]
- Chen, H.; Yu, F.; Shi, N.; Du, P.; Liu, S.; Zhang, X.; Tan, J. Overexpression and Mutation of a Novel Lipase from Serratia marcescens L1 in Escherichia coli. Process Biochem. 2021, 111, 233–240. [Google Scholar] [CrossRef]
- Fitri, R.D.; Illavi, G. Expression of Recombinant Lipase from Serratia marcescens LII61 in Escherichia coli. Jordan J. Biol. Sci. 2022, 15, 199–203. [Google Scholar]
- Duman-Özdamar, Z.E.; Binay, B. Production of industrial enzymes via Pichia pastoris as a cell factory in bioreactor: Current status and future aspects. Protein J. 2021, 40, 367–376. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, S.; Khan, S.A.; Hamayun, M.; Lee, I.-J. The Recent Advances in the Utility of Microbial Lipases: A Review. Microorganisms 2023, 11, 510. https://doi.org/10.3390/microorganisms11020510
Ali S, Khan SA, Hamayun M, Lee I-J. The Recent Advances in the Utility of Microbial Lipases: A Review. Microorganisms. 2023; 11(2):510. https://doi.org/10.3390/microorganisms11020510
Chicago/Turabian StyleAli, Sajid, Sumera Afzal Khan, Muhammad Hamayun, and In-Jung Lee. 2023. "The Recent Advances in the Utility of Microbial Lipases: A Review" Microorganisms 11, no. 2: 510. https://doi.org/10.3390/microorganisms11020510
APA StyleAli, S., Khan, S. A., Hamayun, M., & Lee, I.-J. (2023). The Recent Advances in the Utility of Microbial Lipases: A Review. Microorganisms, 11(2), 510. https://doi.org/10.3390/microorganisms11020510








