Mucin and Agitation Shape Predation of Escherichia coli by Lytic Coliphage
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria and Bacteriophage
2.2. Mucin
2.3. Plate Motility Assays E. coli
2.4. In Tube Assays
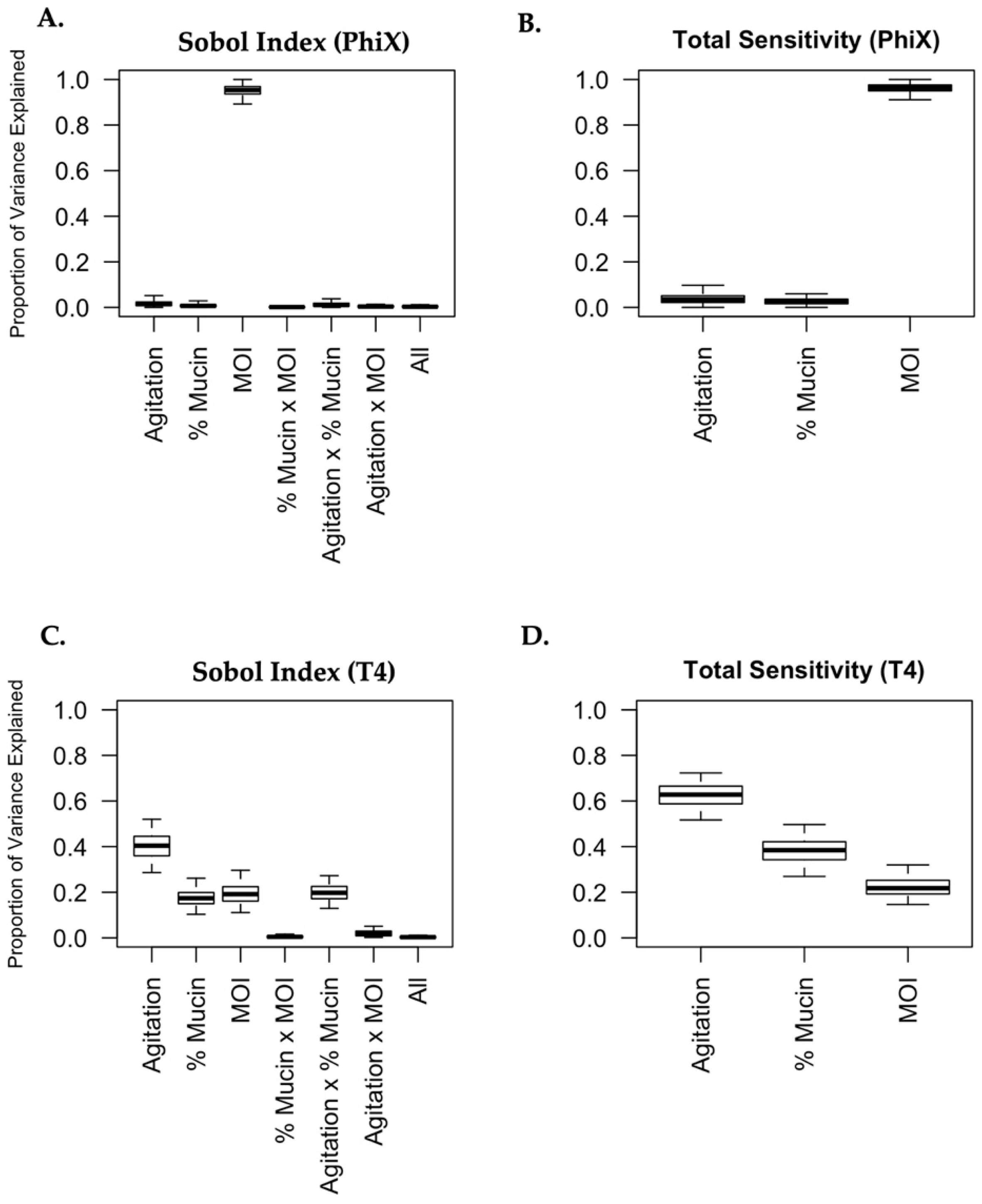
2.5. Generation of Sobol Plots
3. Results
3.1. Effect of Mucin Concentration on Bacterial Motility
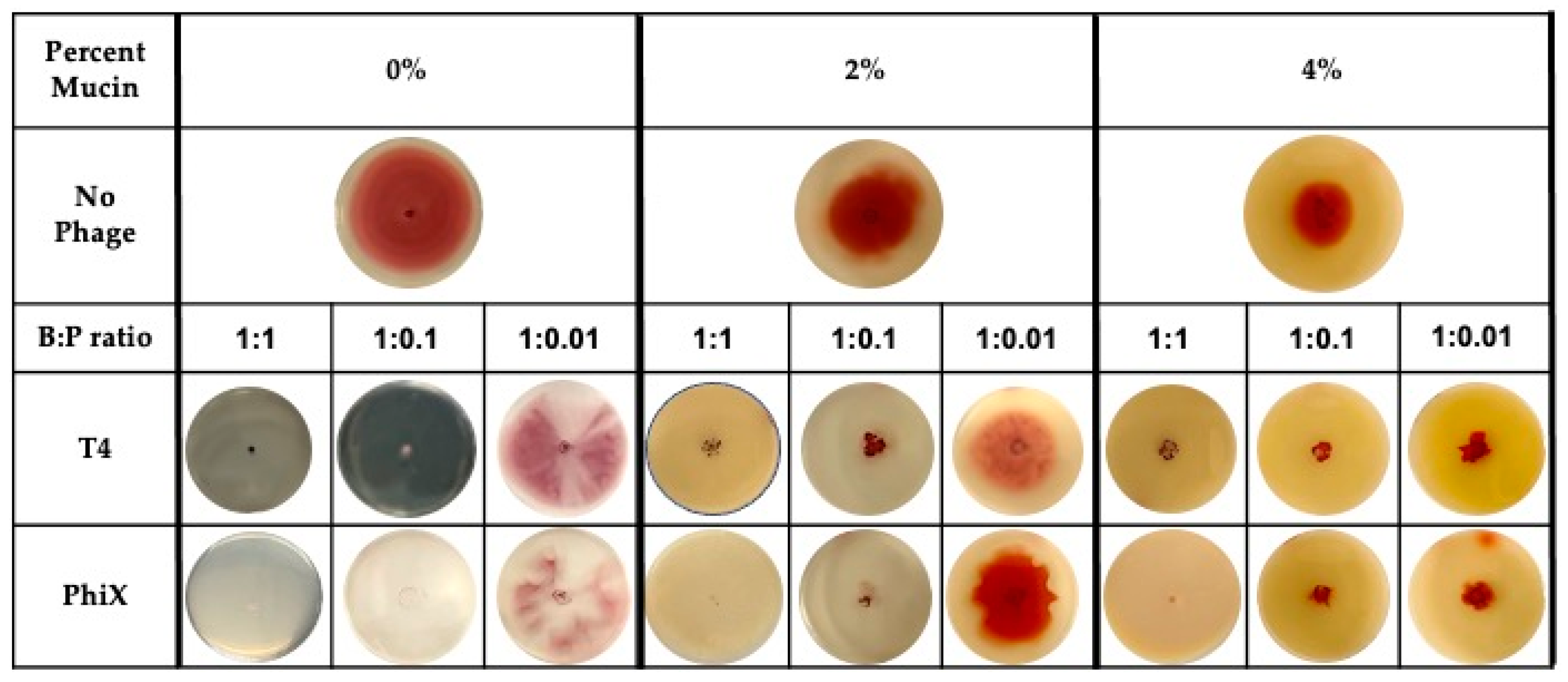
3.2. Effect of Phages on Ability of Bacteria to Spread
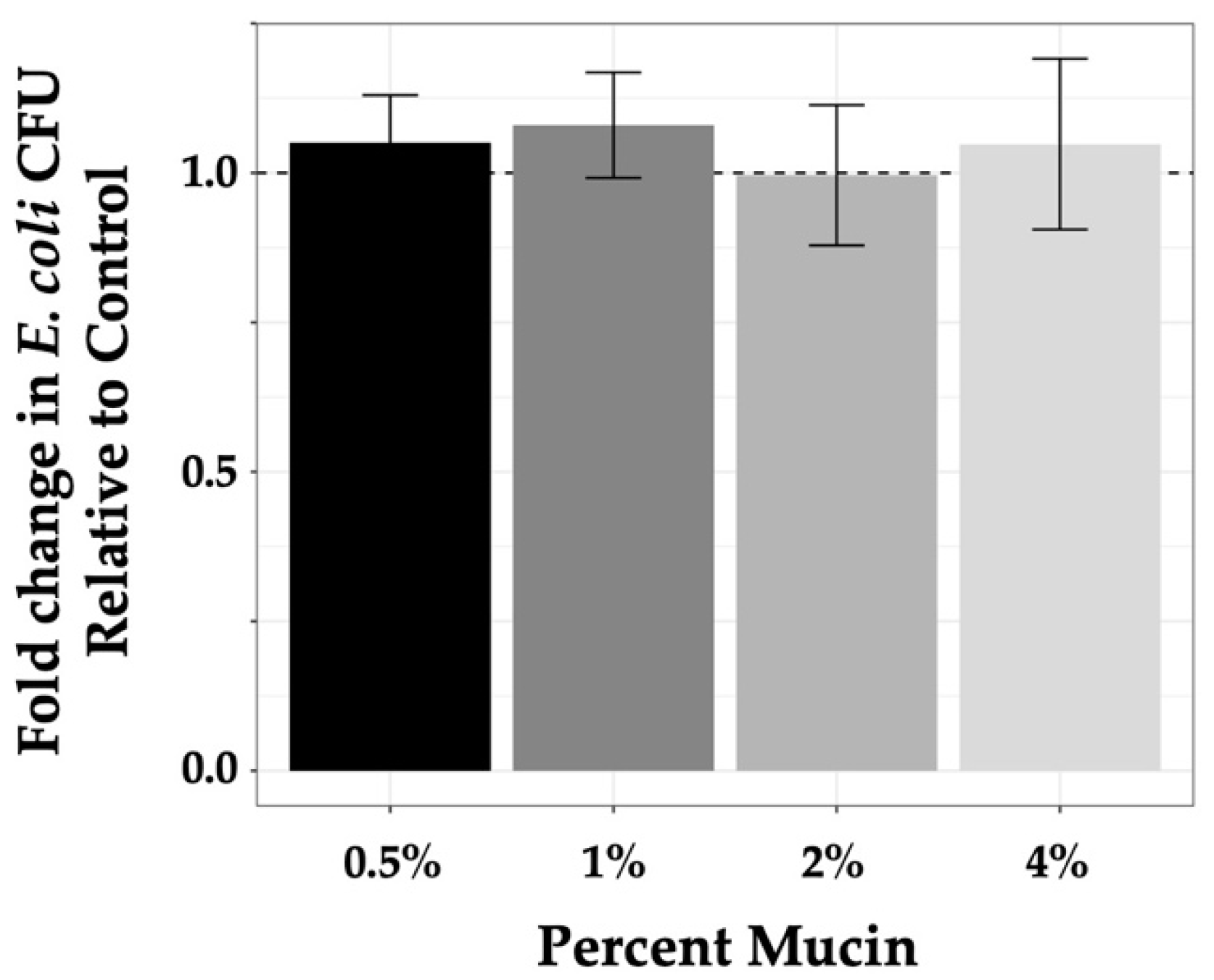
3.3. Effect of Mucin Concentration on Bacterial Growth
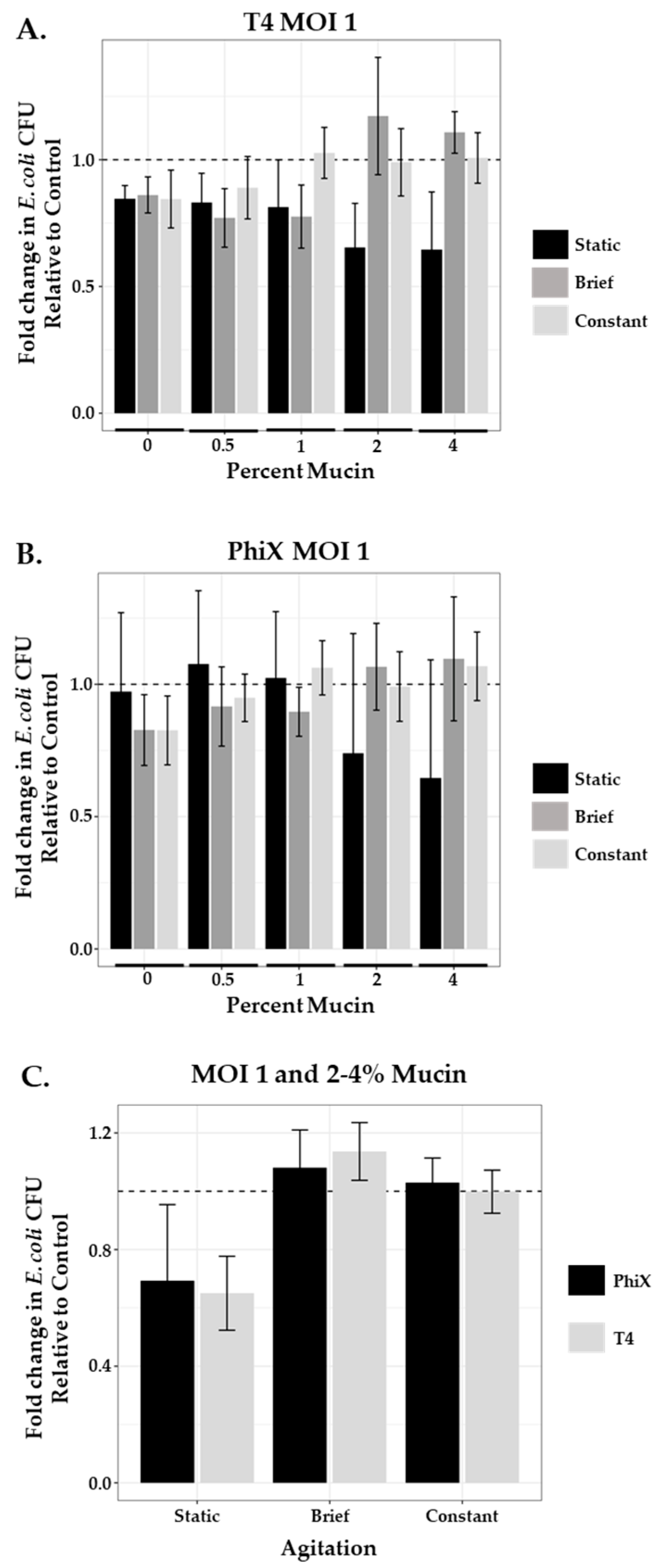
3.4. Phage Predation at MOI 1 in a Static System
3.5. Effect of Agitation on Phage Predation at MOI 1
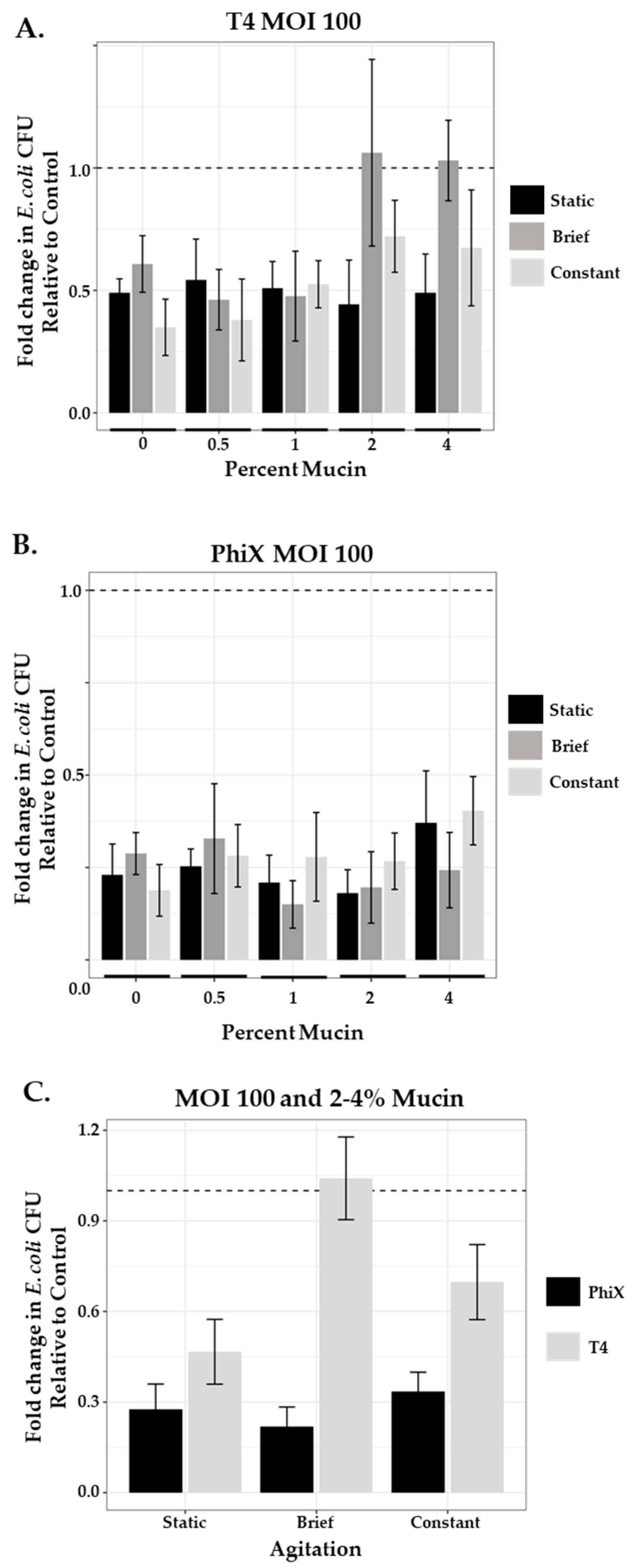
3.6. Phage Predation and Agitation in a Phage Therapy Setting
3.7. Mucin and Agitation Influence Phage Predation in a Phage- and MOI-Specific Manner
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Díaz-Muñoz, S.L.; Koskella, B. Bacteria–Phage Interactions in Natural Environments. Adv. Appl. Microbiol. 2014, 89, 135–183. [Google Scholar] [CrossRef] [PubMed]
- Olszak, T.; Latka, A.; Roszniowski, B.; Valvano, M.A.; Drulis-Kawa, Z. Phage Life Cycles Behind Bacterial Biodiversity. Curr. Med. Chem. 2017, 24, 3987–4001. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Kumar, P.; Sahu, R.; Singh, A.K.; Kumar, A. Bacteriophages concept and applications: A review on phage therapy. Curr. Pharm. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Zagaliotis, P.; Michalik-Provasek, J.; Gill, J.; Walsh, T. Therapeutic Bacteriophages for Gram-Negative Bacterial Infections in Animals and Humans. Pathog. Immun. 2022, 7, 1–45. [Google Scholar] [CrossRef] [PubMed]
- Danis-Wlodarczyk, K.; Dąbrowska, K.; Abedon, S.T. Phage Therapy: The Pharmacology of Antibacterial Viruses. Curr. Issues Mol. Biol. 2021, 40, 81–164. [Google Scholar] [CrossRef]
- Lin, D.M.; Koskella, B.; Lin, H.C. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 162–173. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef]
- Simpson, D.; Sacher, J.; Szymanski, C.M. Exploring the interactions between bacteriophage-encoded glycan binding proteins and carbohydrates. Curr. Opin. Struct. Biol. 2015, 34, 69–77. [Google Scholar] [CrossRef]
- Cazares, D.; Cazares, A.; Figueroa, W.; Guarneros, G.; Edwards, R.A.; Vinuesa, P. A Novel Group of Promiscuous Podophages Infecting Diverse Gammaproteobacteria from River Communities Exhibits Dynamic Intergenus Host Adaptation. Msystems 2021, 6, e00773-20. [Google Scholar] [CrossRef]
- Andrews, B.; Fields, S. Balance between promiscuity and specificity in phage lambda host range. ISME J. 2021, 15, 2195–2205. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Campanacci, V.; Blangy, S.; Moineau, S.; Tegoni, M.; Cambillau, C. Modular Structure of the Receptor Binding Proteins of Lactococcus lactis Phages. J. Biol. Chem. 2006, 281, 14256–14262. [Google Scholar] [CrossRef]
- Ly-Chatain, M.H. The factors affecting effectiveness of treatment in phages therapy. Front. Microbiol. 2014, 5, 51. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages—Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Iebba, V.; Totino, V.; Gagliardi, A.; Santangelo, F.; Cacciotti, F.; Trancassini, M.; Mancini, C.; Cicerone, C.; Corazziari, E.; Pantanella, F.; et al. Eubiosis and dysbiosis: The two sides of the microbiota. New Microbiol. 2016, 39, 1–12. [Google Scholar]
- Chibani-Chennoufi, S.; Sidoti, J.; Bruttin, A.; Kutter, E.; Sarker, S.; Brüssow, H. In Vitro and In Vivo Bacteriolytic Activities of Escherichia coli Phages: Implications for Phage Therapy. Antimicrob. Agents Chemother. 2004, 48, 2558–2569. [Google Scholar] [CrossRef]
- Carroll-Portillo, A.; Lin, H. Exploring Mucin as Adjunct to Phage Therapy. Microorganisms 2021, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Ermund, A.; Schütte, A.; Johansson, M.E.V.; Gustafsson, J.K.; Hansson, G.C. Studies of mucus in mouse stomach, small intestine, and colon. I. Gastrointestinal mucus layers have different properties depending on location as well as over the Peyer’s patches. Am. J. Physiol. Gastrointest. Liver Physiol. 2013, 305, G341–G347. [Google Scholar] [CrossRef]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in Intestinal Mucosal Defense and Inflammation: Learning From Clinical and Experimental Studies. Front. Immunol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Johansson, M.E.; Hansson, G.C. Keeping Bacteria at a Distance. Science 2011, 334, 182–183. [Google Scholar] [CrossRef]
- Corfield, A.P.; Carroll, D.; Myerscough, N.; Probert, C.S. Mucins in the gastrointestinal tract in health and disease. Front. Biosci. 2001, 6, D1321–D1357. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.V.; Sjövall, H.; Hansson, G.C. The gastrointestinal mucus system in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Pelaseyed, T.; Bergström, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.H.; Schütte, A.; van der Post, S.; Svensson, F.; Rodríguez-Piñeiro, A.M.; Nyström, E.E.L.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol. Rev. 2014, 260, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Sicard, J.-F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef] [PubMed]
- Lepage, P.; Colombet, J.; Marteau, P.; Sime-Ngando, T.; Doré, J.; Leclerc, M. Dysbiosis in inflammatory bowel disease: A role for bacteriophages? Gut 2008, 57, 424–425. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J.; Auro, R.; Furlan, M.; Whiteson, K.L.; Erb, M.L.; Pogliano, J.; Stotland, A.; Wolkowicz, R.; Cutting, A.S.; Doran, K.S.; et al. Bacteriophage Adhering to Mucus Provide a Non-Host-Derived Immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 10771–10776. [Google Scholar] [CrossRef]
- Almeida, G.M.F.; Laanto, E.; Ashrafi, R.; Sundberg, L.-R. Bacteriophage Adherence to Mucus Mediates Preventive Protection against Pathogenic Bacteria. Mbio 2019, 10, e01984-19. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, L.-R.; Rantanen, N.; Almeida, G.M.D.F. Mucosal Environment Induces Phage Susceptibility in Streptococcus mutans. Phage 2022, 3, 128–135. [Google Scholar] [CrossRef]
- Rothschild-Rodriguez, D.; Hedges, M.; Kaplan, M.; Karav, S.; Nobrega, F.L. Phage-encoded carbohydrate-interacting proteins in the human gut. Front. Microbiol. 2023, 13, 1083208. [Google Scholar] [CrossRef]
- Allen, A.; Flemstrom, G.; Garner, A.; Kivilaakso, E. Gastroduodenal mucosal protection. Physiol. Rev. 1993, 73, 823–857. [Google Scholar] [CrossRef]
- Kang, Y.; Park, H.; Choe, B.-H.; Kang, B. The Role and Function of Mucins and Its Relationship to Inflammatory Bowel Disease. Front. Med. 2022, 9, 848344. [Google Scholar] [CrossRef]
- Henen, S.; Denton, C.; Teckman, J.; Borowitz, D.; Patel, D. Review of Gastrointestinal Motility in Cystic Fibrosis. J. Cyst. Fibros. 2021, 20, 578–585. [Google Scholar] [CrossRef]
- Sjogren, R.W. Gastrointestinal motility disorders in scleroderma. Arthritis Rheum. 1994, 37, 1265–1282. [Google Scholar] [CrossRef] [PubMed]
- Belizário, J.E.; Faintuch, J. Microbiome and Gut Dysbiosis. Exp. Suppl. 2018, 109, 459–476. [Google Scholar] [PubMed]
- Singh, R.; Zogg, H.; Wei, L.; Bartlett, A.; Ghoshal, U.C.; Rajender, S.; Ro, S. Gut Microbial Dysbiosis in the Pathogenesis of Gastrointestinal Dysmotility and Metabolic Disorders. J. Neurogastroenterol. Motil. 2021, 27, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 2020, 28, 724–740.e8. [Google Scholar] [CrossRef] [PubMed]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 285–299.e8. [Google Scholar] [CrossRef]
- De Paepe, M.; Leclerc, M.; Tinsley, C.R.; Petit, M.A. Bacteriophages: An underestimated role in human and animal health? Front. Cell Infect. Microbiol. 2014, 4, 39. [Google Scholar] [CrossRef]
- Manrique, P.; Dills, M.; Young, M.J. The Human Gut Phage Community and Its Implications for Health and Disease. Viruses 2017, 9, 141. [Google Scholar] [CrossRef]
- Bachmann, B.J. Derivations and genotypes of some mutant derivatives of Escherichia coli. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Curtis, R., Ingraham, J.L., Lin, J.C., Low, K.B., Magasanik, B., Reznikoff, W.S., Riley, M., Schaechter, M., Umbarger, H.E., Eds.; ASM Press: Washington, DC, USA, 1996; Volume 2. [Google Scholar]
- Ye, J.; Ma, N.; Madden, T.L.; Ostell, J.M. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013, 41, W34–W40. [Google Scholar] [CrossRef]
- Rickert, C.A.; Lutz, T.M.; Marczynski, M.; Lieleg, O. Several Sterilization Strategies Maintain the Functionality of Mucin Glycoproteins. Macromol. Biosci. 2020, 20, e2000090. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kong, F. Simulating human gastrointestinal motility in dynamic in vitro models. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3804–3833. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Francom, D.; Sanso, B.; Kupresanin, A.; Johannesson, G. Sensitivity Analysis and Emulation for Functional Data using Bayesian Adaptive Splines. Stat. Sin. 2018, 28, 791–816. [Google Scholar] [CrossRef]
- Sobol′, I.M. Global sensitivity indices for nonlinear mathematical models and their Monte Carlo estimates. Math. Comput. Simul. 2001, 55, 271–280. [Google Scholar] [CrossRef]
- Saltelli, A.; Ratto, M.; Andres, T.; Campolongo, F.; Cariboni, J.; Gatelli, D.; Saisana, M.; Tarantola, S. Global Sensitivity Analysis. The Primer; John Wiley & Sons, Ltd: London, UK, 2007. [Google Scholar]
- Francom, D. BASS: Bayesian Adaptive Spline Surfaces; R Package(Version 1.2.1); R Core Team: Vienna, Austria, 2020. [Google Scholar]
- Francom, D.; Sansó, B. BASS: An R Package for Fitting and Performing Sensitivity Analysis of Bayesian Adaptive Spline Surfaces. J. Stat. Softw. 2020, 94, 1–36. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Joiner, K.L.; Baljon, A.; Barr, J.; Rohwer, F.; Luque, A. Impact of bacteria motility in the encounter rates with bacteriophage in mucus. Sci. Rep. 2019, 9, 16427. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.J.; Auro, R.; Sam-Soon, N.; Kassegne, S.; Peters, G.; Bonilla, N.; Hatay, M.; Mourtada, S.; Bailey, B.; Youle, M.; et al. Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters. Proc. Natl. Acad. Sci. USA 2015, 112, 13675. [Google Scholar] [CrossRef]
- Guenther, S.; Huwyler, D.; Richard, S.; Loessner, M.J. Virulent Bacteriophage for Efficient Biocontrol of Listeria monocytogenes in Ready-To-Eat Foods. Appl. Environ. Microbiol. 2009, 75, 93–100. [Google Scholar] [CrossRef]
- Marcó, M.B.; Reinheimer, J.; Quiberoni, A. Phage adsorption to Lactobacillus plantarum: Influence of physiological and environmental factors. Int. J. Food Microbiol. 2010, 138, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, S.; Musmeci, E.; Candeliere, F.; Amaretti, A.; Rossi, M. Identification of mucin degraders of the human gut microbiota. Sci. Rep. 2021, 11, 11094. [Google Scholar] [CrossRef] [PubMed]
- Conway, T.; Cohen, P.S. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiol. Spectr. 2015, 3, 343–362. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.-E.; Smalley, D.J.; Tucker, D.L.; Leatham, M.P.; Norris, W.E.; Stevenson, S.J.; Anderson, A.B.; Grissom, J.E.; Laux, D.C.; Cohen, P.S.; et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 2004, 101, 7427–7432. [Google Scholar] [CrossRef]
- Kim, S.K. SMALL INTESTINE TRANSIT TIME IN THE NORMAL SMALL BOWEL STUDY. Am. J. Roentgenol. 1968, 104, 522–524. [Google Scholar] [CrossRef]
- Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Jończyk-Matysiak, E.; Wojciechowska, R.; Górski, A. Bacteriophages in the gastrointestinal tract and their implications. Gut Pathog. 2017, 9, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Park, E.-J.; Roh, S.W.; Bae, J.-W. Diversity and Abundance of Single-Stranded DNA Viruses in Human Feces. Appl. Environ. Microbiol. 2011, 77, 8062–8070. [Google Scholar] [CrossRef]
- Deloose, E.; Janssen, P.; Depoortere, I.; Tack, J. The migrating motor complex: Control mechanisms and its role in health and disease. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 271–285. [Google Scholar] [CrossRef] [PubMed]
- Cremer, J.; Segota, I.; Yang, C.-Y.; Arnoldini, M.; Sauls, J.T.; Zhang, Z.; Gutierrez, E.; Groisman, A.; Hwa, T. Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel. Proc. Natl. Acad. Sci. USA 2016, 113, 11414–11419. [Google Scholar] [CrossRef]
- Barr, J.J.; Youle, M.; Rohwer, F. Innate and acquired bacteriophage-mediated immunity. Bacteriophage 2013, 3, e25857. [Google Scholar] [CrossRef]
- Zhang, X.; Trame, M.N.; Lesko, L.J.; Schmidt, S. Sobol Sensitivity Analysis: A Tool to Guide the Development and Evaluation of Systems Pharmacology Models. CPT Pharmacomet. Syst. Pharmacol. 2015, 4, 69–79. [Google Scholar] [CrossRef]
- Arnoldini, M.; Cremer, J.; Hwa, T. Bacterial growth, flow, and mixing shape human gut microbiota density and composition. Gut Microbes 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Cremer, J.; Arnoldini, M.; Hwa, T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc. Natl. Acad. Sci. USA 2017, 114, 6438–6443. [Google Scholar] [CrossRef]
- Su, C.; Padra, M.; Constantino, M.A.; Sharba, S.; Thorell, A.; Lindén, S.K.; Bansil, R. Influence of the viscosity of healthy and diseased human mucins on the motility of Helicobacter pylori. Sci. Rep. 2018, 8, 9710. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.; Pelaseyed, T.; Svensson, F.; Johansson, M.E.V. Study of mucin turnover in the small intestine by in vivo labeling. Sci. Rep. 2018, 8, 5760. [Google Scholar] [CrossRef] [PubMed]
- Kočevar-Nared, J.; Kristl, J.; Šmid-Korbar, J. Comparative rheological investigation of crude gastric mucin and natural gastric mucus. Biomaterials 1997, 18, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Soudah, H.C.; Hasler, W.L.; Owyang, C. Effect of Octreotide on Intestinal Motility and Bacterial Overgrowth in Scleroderma. N. Engl. J. Med. 1991, 325, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.; Buxbaum, J. Gastrointestinal Manifestations of Cystic Fibrosis. Dig. Dis. Sci. 2015, 60, 1903–1913. [Google Scholar] [CrossRef]
- Etienne-Mesmin, L.; Chassaing, B.; Desvaux, M.; De Paepe, K.; Gresse, R.; Sauvaitre, T.; Forano, E.; Van De Wiele, T.; Schüller, S.; Juge, N.; et al. Experimental models to study intestinal microbes–mucus interactions in health and disease. FEMS Microbiol. Rev. 2019, 43, 457–489. [Google Scholar] [CrossRef]
- Paone, P.; Cani, P. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
| % Mucin | 0% | 0.5% | 1% | 2% | 4% |
|---|---|---|---|---|---|
| Bacterial motility (SEM) 1 | 23.87 (0.82) | 18.62 (1.34) | 17.54 (0.74) | 11.99 (0.87) | 8.66 (1.3) |
| Dunnett’s p-value 2 | NA | 0.046 | <0.0001 | <0.0001 | <0.0001 |
| % Mucin | 0.5% | 1% | 2% | 4% |
|---|---|---|---|---|
| Fold Change in growth (SEM) | 1.05 (0.04) | 1.08 (0.04) | 0.996 (0.05) | 1.05 (0.06) |
| % Mucin | 0% | 0.5% | 1% | 2% | 4% |
|---|---|---|---|---|---|
| T4 mean (SEM) | 0.846 (0.02) | 0.832 (0.05) | 0.813 (0.08) | 0.654 (0.08) | 0.646 (0.1) |
| PhiX mean (SEM) | 0.973 (0.12) | 1.077 (0.11) | 1.024 (0.1) | 0.740 (0.18) | 0.646 (0.17) |
| Static Mean (SEM) | Brief Mean (SEM) | Constant Mean (SEM) | Tukey’s p-Value 1 (Brief/Constant) | |
|---|---|---|---|---|
| T4 2% mucin | 0.654 (0.08) | 1.172 (0.1) | 0.990 (0.06) | 0.003/<0.0001 |
| T4 4% mucin | 0.646 (0.1) | 1.108 (0.04) | 1.007 (0.04) | 0.012/0.002 |
| PhiX 2% mucin | 0.740 (0.18) | 1.066 (0.07) | 0.991 (0.06) | 0.02/0.08 |
| PhiX 4% mucin | 0.646 (0.17) | 1.096 (0.1) | 1.068 (0.06) | 0.02/0.04 |
| % Mucin | 0% | 0.5% | 1% | 2% | 4% |
|---|---|---|---|---|---|
| T4 mean (SEM) | 0.490 (0.03) | 0.543 (0.07) | 0.509 (0.05) | 0.443 (0.08) | 0.490 (0.07) |
| PhiX mean (SEM) | 0.230 (0.04) | 0.253 (0.02) | 0.209 (0.04) | 0.181 (0.03) | 0.371 (0.06) |
| Static Mean (SEM) | Brief Mean (SEM) | Constant Mean (SEM) | Tukey’s p-Value 1 (Brief/Constant) | |
|---|---|---|---|---|
| T4 2% mucin | 0.443 (0.08) | 1.062 (0.14) | 0.721 (0.06) | 0.015/0.004 |
| T4 4% mucin | 0.490 (0.07) | 1.030 (0.07) | 0.674 (0.1) | 0.001/0.016 |
| PhiX 2% mucin | 0.181 (0.03) | 0.196 (0.04) | 0.267 (0.03) | 0.951/0.234 |
| PhiX 4% mucin | 0.371 (0.06) | 0.243 (0.05) | 0.404 (0.04) | 0.174/0.89 |
| Agitation Level | % Mucin | MOI | ||||
|---|---|---|---|---|---|---|
| Estimate | 95% CI 1 | Estimate | 95% CI 1 | Estimate | 95% CI 1 | |
| PhiX | 0.038 | (0.00, 0.10) | 0.027 | (0.00, 0.06) | 0.961 | (0.91, 1.00) |
| T4 | 0.625 | (0.52, 0.72) | 0.383 | (0.27, 0.50) | 0.223 | (0.15, 0.32) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carroll-Portillo, A.; Rumsey, K.N.; Braun, C.A.; Lin, D.M.; Coffman, C.N.; Alcock, J.A.; Singh, S.B.; Lin, H.C. Mucin and Agitation Shape Predation of Escherichia coli by Lytic Coliphage. Microorganisms 2023, 11, 508. https://doi.org/10.3390/microorganisms11020508
Carroll-Portillo A, Rumsey KN, Braun CA, Lin DM, Coffman CN, Alcock JA, Singh SB, Lin HC. Mucin and Agitation Shape Predation of Escherichia coli by Lytic Coliphage. Microorganisms. 2023; 11(2):508. https://doi.org/10.3390/microorganisms11020508
Chicago/Turabian StyleCarroll-Portillo, Amanda, Kellin N. Rumsey, Cody A. Braun, Derek M. Lin, Cristina N. Coffman, Joe A. Alcock, Sudha B. Singh, and Henry C. Lin. 2023. "Mucin and Agitation Shape Predation of Escherichia coli by Lytic Coliphage" Microorganisms 11, no. 2: 508. https://doi.org/10.3390/microorganisms11020508
APA StyleCarroll-Portillo, A., Rumsey, K. N., Braun, C. A., Lin, D. M., Coffman, C. N., Alcock, J. A., Singh, S. B., & Lin, H. C. (2023). Mucin and Agitation Shape Predation of Escherichia coli by Lytic Coliphage. Microorganisms, 11(2), 508. https://doi.org/10.3390/microorganisms11020508







