Diversity of the Bacterial Community Associated with Hindgut, Malpighian Tubules, and Foam of Nymphs of Two Spittlebug Species (Hemiptera: Aphrophoridae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Foam and Insect Samples
2.2. Microbiological Analyses
2.2.1. Real-Time PCR
2.2.2. Denaturing Gradient Gel Electrophoresis
2.2.3. Sequence Analysis
2.3. Statistical Analysis
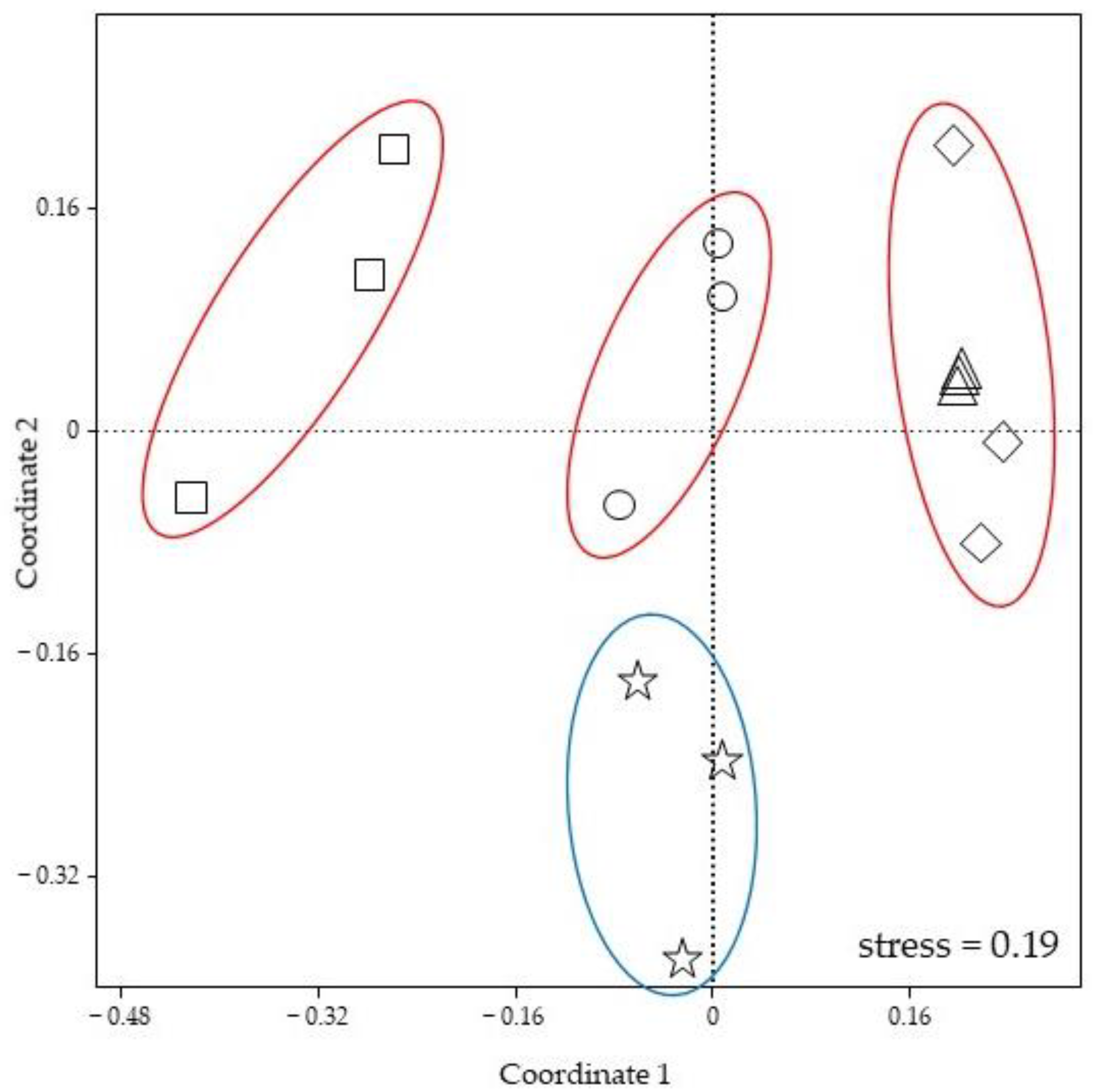
3. Results
3.1. Bacterial Biomass Quantification
3.2. Gut Bacterial Community Composition
3.3. Foam Bacterial Community Composition
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Buchner, P. Endosymbiosis of Animals with Plant Microorganisms; Interscience Publishers: New York, NY, USA, 1965; p. 909. [Google Scholar]
- Moran, N.A.; Baumann, P. Bacterial endosymbionts in animals. Curr. Opin. Microbiol. 2000, 3, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190. [Google Scholar] [CrossRef]
- Baumann, P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189. [Google Scholar] [CrossRef] [PubMed]
- Koga, R.; Bennett, G.M.; Cryan, J.R.; Moran, N.A. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ. Microbiol. 2013, 15, 2073–2081. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A.; Tran, P.; Gerardo, N.M. Symbiosis and insect diversification: An ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microb. 2005, 71, 8802–8810. [Google Scholar] [CrossRef] [PubMed]
- McCutcheon, J.P.; Moran, N.A. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2010, 2, 708–718. [Google Scholar] [CrossRef]
- Koga, R.; Moran, N.A. Swapping symbionts in spittlebugs: Evolutionary replacement of a reduced genome symbiont. Isme J. 2014, 8, 1237–1246. [Google Scholar] [CrossRef] [PubMed]
- Marshall, A.T. Spittle-production and tube-building by cercopid larvae (Homoptera)—IV. Mucopolysaccharide associated with spittle-production. J. Insect Physiol. 1966, 12, 635–644. [Google Scholar] [CrossRef]
- Marshall, A.T. Protein synthesis and secretion by the Malpighian tubules of cercopoid larvae (Homoptera). J. Insect Physiol. 1973, 19, 2317–2326. [Google Scholar] [CrossRef]
- Yurtsever, S. On the polymorphic meadow spittlebug, Philaenus spumarius (L.) (Homoptera: Cercopidae). Turk. J. Zool. 2000, 24, 447–460. [Google Scholar]
- Cornara, D.; Bosco, D.; Fereres, A. Philaenus spumarius: When an old acquaintance becomes a new threat to European agriculture. J. Pest Sci. 2018, 91, 957–972. [Google Scholar] [CrossRef]
- Tonelli, M.; Gomes, G.; Silva, W.D.; Magri, N.T.; Vieira, D.M.; Aguiar, C.L.; Bento, J.M.S. Spittlebugs produce foam as a thermoregulatory adaptation. Sci. Rep. 2018, 8, 4729. [Google Scholar] [CrossRef] [PubMed]
- Balzani, P.; Nencioni, A.; Grillini, M.; Masoni, A.; Zuri, F.; Picchi, M.S.; Frizzi, F.; Sacchetti, P.; Cantini, C.; Santini, G. Spittlebug invisibility cloak: Experimental tests on the antipredatory effect of the froth of Philaenus spumarius. B. Insectol. 2023; 76, in press. [Google Scholar]
- Schöbel, C.; Carvalho, G.S. The “State of Art” of Mahanarva (Hemiptera: Cercopidae) research. An economically important New World spittlebug genus. Appl. Entomol. Zool. 2021, 56, 299–309. [Google Scholar] [CrossRef]
- Thompson, V. Associative nitrogen fixation, C4 photosynthesis, and the evolution of spittlebugs (Hemiptera: Cercopidae) as major pests of neotropical sugarcane and forage grasses. Bull. Entomol. Res. 2004, 94, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liang, A.P. Laboratory rearing of Callitettix versicolor (Hemiptera: Cicadomorpha: Cercopidae), with descriptions of the immature stages. Ann. Entomol. Soc. Am. 2012, 105, 664–670. [Google Scholar] [CrossRef]
- Cornara, D.; Saponari, M.; Zeilinger, A.R.; de Stradis, A.; Boscia, D.; Loconsole, G.; Bosco, D.; Martelli, G.P.; Almeida, R.P.P.; Porcelli, F. Spittlebugs as vectors of Xylella fastidiosa in olive orchards in Italy. J. Pest Sci. 2016, 90, 521–530. [Google Scholar] [CrossRef]
- Loconsole, G.; Boscia, D.; Palmisano, F.; Savino, V.; Potere, O.; Martelli, G.P.; Saponari, M. A Xylella fastidiosa strain with unique biology and phylogeny is associated with a severe disease of olive in Southern Apulia. J. Plant Pathol. 2014, 96, S4. [Google Scholar]
- Kapantaidaki, D.E.; Antonatos, S.; Evangelou, V.; Papachristos, D.P.; Milonas, P. Genetic and endosymbiotic diversity of Greek populations of Philaenus spumarius, Philaenus signatus and Neophilaenus campestris, vectors of Xylella fastidiosa. Sci. Rep. 2021, 11, 3752. [Google Scholar] [CrossRef]
- Lis, A.; Maryańska-Nadachowska, A.; Kajtoch, Ł. Relations of Wolbachia infection with phylogeography of Philaenus spumarius (Hemiptera: Aphrophoridae) populations within and beyond the Carpathian contact zone. Microb. Ecol. 2015, 70, 509–521. [Google Scholar] [CrossRef]
- Wilson, H.A.; Dorsey, C.K. Studies on the composition and microbiology of insect spittle. Ann. Entomol. Soc. Am. 1957, 50, 399–406. [Google Scholar] [CrossRef]
- Tonelli, M.; Cotta, S.R.; Rigotto, A.; Dias, A.C.F.; Andreote, F.D.; Bento, J.M.S. The composition of the bacterial community in the foam produced by Mahanarva fimbriolata is distinct from those at gut and soil. Braz. J. Microbiol. 2020, 51, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.K.; Douglas, A.E. Hype or opportunity? Using microbial symbionts in novel strategies for insect pest control. J. Insect Physiol. 2017, 103, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Dáder, B.; Viñuela, E.; Moreno, A.; Plaza, M.; Garzo, E.; del Estal, P.; Fereres, A. Sulfoxaflor and natural pyrethrin with piperonyl butoxide are effective alternatives to neonicotinoids against juveniles of Philaenus spumarius, the European vector of Xylella fastidiosa. Insects 2019, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Zhang, Y.; Wie, C. Anatomy and fine structure of the alimentary canal of the spittlebug Lepyronia coleopterata (L.) (Hemiptera: Cercopoidea). Arthropod. Struct. Dev. 2013, 42, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Cecil, R. The alimentary canal of Philaenus leucophthalmus L. Ohio J. Sci. 1930, 30, 120–130. [Google Scholar]
- López-Gutiérrez, J.C.; Henry, S.; Hallet, S.; Martin-Laurent, F.; Catroux, G.; Philippot, L. Quantification of a novel group of nitrate-reducing bacteria in the environment by real-time PCR. J. Microbiol. Meth. 2004, 57, 399–407. [Google Scholar] [CrossRef]
- Felske, A.; Engelen, B.; Nübel, U.; Backhaus, H. Direct ribosome isolation from soil to extract bacterial rRNA for community analysis. Appl. Environ. Microb. 1996, 62, 4162–4167. [Google Scholar] [CrossRef]
- Webster, N.S.; Taylor, M.W.; Behnam, F.; Lucker, S.; Rattei, T.; Whalan, S.; Horn, M.; Wagner, M. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ. Microbiol. 2010, 12, 2070–2082. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Palaeontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Ramette, A. Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 2007, 62, 142–160. [Google Scholar] [CrossRef]
- Ishikawa, H. Insect Symbiosis: An introduction. In Insect Symbiosis; Bourtzis, K., Miller, T.A., Eds.; CRC Press: Boca Raton, FL, USA, 2003; Volume 1, pp. 1–21. [Google Scholar]
- Engel, P.; Moran, N.A. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Wei, C. Bacterial communities in digestive and excretory organs of cicadas. Arch. Microbiol. 2020, 202, 539–553. [Google Scholar] [CrossRef]
- Gonella, E.; Negri, I.; Marzorati, M.; Mandrioli, M.; Sacchi, L.; Pajoro, M.; Crotti, E.; Rizzi, A.; Clementi, E.; Tedeschi, R.; et al. Bacterial endosymbiont localization in Hyalesthes obsoletus, the insect vector of bois noir in Vitis vinifera. Appl. Environ. Microbiol. 2011, 77, 1423–1435. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.J.; Hunter, M.S.; Zchori-Fein, E. The emerging diversity of Rickettsia. Proc. R. Soc. B 2006, 273, 2097–2106. [Google Scholar] [CrossRef] [PubMed]
- Lawson, E.T.; Mousseau, T.A.; Klaper, R.; Hunter, M.D.; Werren, J.H. Rickettsia associated with male-killing in a buprestid beetle. Heredity 2001, 86, 497–505. [Google Scholar] [CrossRef]
- Werren, J.H.; Hurst, G.D.D.; Zhang, W.; Breeuwer, J.A.J.; Stouthamer, R.; Majerus, M.E.N. Rickettsial relative associated with male killing in the ladybird beetle (Adalia bipunctata). J. Bacteriol. 1994, 176, 388–394. [Google Scholar] [CrossRef]
- Hagimori, T.; Abe, Y.; Date, S.; Miura, K. The first finding of a Rickettsia bacterium associated with parthenogenesis induction among insects. Curr. Microbiol. 2006, 52, 97–101. [Google Scholar] [CrossRef]
- Gottlieb, Y.; Ghanim, M.; Chiel, E.; Gerling, D.; Portnoy, V.; Steinberg, S.; Tzuri, G.; Horowitz, A.R.; Belausov, E.; Mozes-Daube, N.; et al. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microb. 2006, 72, 3646–3652. [Google Scholar] [CrossRef]
- Sakurai, M.; Koga, R.; Tsuchida, T.; Meng, X.Y.; Fukatsu, T. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: Novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 2005, 71, 4069–4075. [Google Scholar] [CrossRef]
- Davis, M.J.; Ying, Z.T.; Brunner, B.R.; Pantoja, A.; Ferwerda, F.H. Rickettsial relative associated with papaya bunchy top disease. Curr. Microbiol. 1998, 36, 80–84. [Google Scholar] [CrossRef]
- Ishii, Y.; Matsuura, Y.; Kakizawa, S.; Nikoh, N.; Fukatsu, T. Diversity of bacterial endosymbionts associated with Macrosteles leafhoppers vectoring phytopathogenic phytoplasmas. Appl. Environ. Microb. 2013, 79, 5013–5022. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wang, D.; He, H.; Wei, C. Bacterial diversity of bacteriomes and organs of reproductive, digestive and excretory systems in two cicada species (Hemiptera: Cicadidae). PLoS ONE 2017, 12, e0175903. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Huang, Z.; He, H.; Wei, C. Comparative analysis of microbial communities associated with bacteriomes, reproductive organs and eggs of the cicada Subpsaltria yangi. Arch. Microbiol. 2018, 200, 227–235. [Google Scholar] [CrossRef]
- Himler, A.G.; Adachi-Hagimori, T.; Bergen, J.E.; Kozuch, A.; Kelly, S.E.; Tabashnik, B.E.; Chiel, E.; Duckworth, V.E.; Dennehy, T.J.; Zchori-Fein, E.; et al. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 2011, 332, 254–256. [Google Scholar] [CrossRef]
- Brumin, M.; Kontsedalov, S.; Ghanim, M. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 2011, 18, 57–66. [Google Scholar] [CrossRef]
- Lukasik, P.; Guo, H.; Asch, M.; Ferrari, J.; Godfray, H. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 2013, 16, 214–218. [Google Scholar] [CrossRef]
- Brumin, M.; Levy, M.; Ghanim, M. Transovarial transmission of Rickettsia spp. and organ-specific infection of the whitefly Bemisia tabaci. Appl. Environ. Microb. 2012, 78, 5565–5574. [Google Scholar] [CrossRef]
- Charles, H.; Heddi, A.; Rahbe, Y. A putative insect intracellular endosymbiont stem clade, within the Enterobacteriaceae, inferred from phylogenetic analysis based on a heterogeneous model of DNA evolution. C. R. Acad. Sci. III-Vie 2001, 324, 489–494. [Google Scholar] [CrossRef]
- Moran, N.A.; Russell, J.A.; Koga, R.; Fukatsu, T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microb. 2005, 71, 3302–3310. [Google Scholar] [CrossRef]
- Bar-Shmuel, N.; Behar, A.; Segoli, M. What do we know about biological nitrogen fixation in insects? Evidence and implications for the insect and the ecosystem. Insect Sci. 2020, 27, 392–403. [Google Scholar] [CrossRef]
- Tsitko, I. Characterization of Actinobacteria Degrading and Tolerating Organic Pollutants. Ph.D. Thesis, University of Helsinki, Helsinki, Finland, 12 January 2007. [Google Scholar]
- Xu, J.L.; He, J.; Wang, Z.C.; Wang, K.; Li, W.J.; Tang, S.K.; Li, S.P. Rhodococcus qingshengii sp. nov., a carbendazim-degrading bacterium. Int. J. Syst. Evol. Micr. 2007, 57, 2754–2757. [Google Scholar] [CrossRef] [PubMed]
- Brecher, G.; Wigglesworth, V.B. The transmission of Actinomyces rhodnii Erikson in Rhodnius prolixus Stål (Hemiptera) and its influence on the growth of the host. Parasitology 1944, 35, 220–224. [Google Scholar] [CrossRef]
- Pachebat, J.A.; van Keulen, G.; Whitten, M.M.A.; Girdwood, S.; Del Sol, R.; Dyson, P.J.; Facey, P.D. Draft genome sequence of Rhodococcus rhodnii strain LMG5362, a symbiont of Rhodnius prolixus (Hemiptera, Reduviidae, Triatominae), the principle vector of Trypanosoma cruzi. Genome Announc. 2013, 1, e00329-13. [Google Scholar] [CrossRef] [PubMed]
- Kaltenpoth, M. Actinobacteria as mutualists: General healthcare for insects? Trends Microbiol. 2009, 17, 529–535. [Google Scholar] [CrossRef]
- Welch, E.W.; Macias, J.; Bextine, B. Geographic patterns in the bacterial microbiome of the glassy winged sharpshooter, Homalodisca vitripennis (Hemiptera:Cicadellidae). Symbiosis 2015, 66, 1–12. [Google Scholar] [CrossRef]
- Farina, P.; Bedini, S.; Conti, B. Multiple functions of Malpighian tubules in insects: A review. Insects 2022, 13, 1001. [Google Scholar] [CrossRef]
- Li, Q.; Zhong, H.; Zhang, Y.; Wei, C. Comparative morphology of the distal segments of Malpighian tubules in cicadas and spittlebugs, with reference to their functions and evolutionary indications to Cicadomorpha (Hemiptera: Auchenorrhyncha). Zool. Anz. 2015, 258, 54–68. [Google Scholar] [CrossRef]
- Batut, J.; Andersson, S.G.; O’Callaghan, D. The evolution of chronic infection strategies in the α-proteobacteria. Nat. Rev. Microbiol. 2004, 2, 933–945. [Google Scholar] [CrossRef]
- Fritz, I.; Strömpl, C.; Nikitin, D.I.; Lysenko, A.M.; Abraham, W.R. Brevundimonas mediterranea sp. nov., a non-stalked species from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol. 2005, 55, 479–486. [Google Scholar] [CrossRef]
- Zhang, C.; Li, J.; An, H.; Wu, X.; Wu, Y.; Long, Y.; Li, R.; Xing, D. Enhanced elimination of dimethachlon from soils using a novel strain Brevundimonas naejangsanensis J3. J. Environ. Manag. 2020, 255, 109848. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, X.; Wu, Y.; Li, J.; An, H.; Zhang, T. Enhancement of dicarboximide fungicide degradation by two bacterial cocultures of Providencia stuartii JD and Brevundimonas naejangsanensis J3. J. Hazard. Mater. 2021, 403, 123888. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, S.; Kumar, P.S.; Chitra, B.; Rangasamy, G. Biodegradation of textile dye Rhodamine-B by Brevundimonas diminuta and screening of their breakdown metabolites. Chemosphere 2022, 308, 136266. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Zhao, Z.; Liang, Z. Biodegradation of ochratoxin A and ochratoxin B by Brevundimonas naejangsanensis isolated from soil. Food Control 2022, 133, 108611. [Google Scholar] [CrossRef]
- Naqqash, T.; Imran, A.; Hameed, S.; Shahid, M.; Majeed, A.; Iqbal, J.; Hanif, M.K.; Ejaz, S.; Malik, K.A. First report of diazotrophic Brevundimonas spp. as growth enhancer and root colonizer of potato. Sci. Rep. 2020, 10, 12893. [Google Scholar] [CrossRef] [PubMed]
- Rana, A.; Saharan, B.; Joshi, M.; Prasanna, R.; Kumar, K.; Nain, L. Identification of multi-trait PGPR isolates and evaluating their potential as inoculants for wheat. Ann. Microbiol. 2011, 61, 893–900. [Google Scholar] [CrossRef]
- Park, Y.; Je, K.W.; Lee, K.; Jung, S.E.; Choi, T.J. Growth promotion of Chlorella ellipsoidea by co-inoculation with Brevundimonas sp. isolated from the microalga. Hydrobiologia 2008, 598, 219–228. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; McDonald, B.R.; Moran, N.A. Origin of an alternative genetic code in the extremely small and GC–rich genome of a bacterial symbiont. PLoS Genet. 2009, 5, e1000565. [Google Scholar] [CrossRef]
- McCutcheon, J.P.; McDonald, B.R.; Moran, N.A. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. USA 2009, 106, 15394–15399. [Google Scholar] [CrossRef]
- Belouhova, M.; Daskalova, E.; Yotinov, I.; Topalova, Y.; Velkova, L.; Dolashki, A.; Dolashka, P. Microbial diversity of garden snail mucus. MicrobiologyOpen 2022, 11, e1263. [Google Scholar] [CrossRef]

| Site of Collection | GPS Coordinates | Species Nymph or Foam | Host Plant | Number of Collected Nymphs | Overall Amount of Collected Foam (mL) |
|---|---|---|---|---|---|
| Monte Argentario (Grosseto) | 42.37701 N 11.18620 E | P. spumarius | Cistus monspeliensis (Cistaceae) | - | 12 |
| Fabio, Vaiano (Prato) | 43.939682 N 11.142821 E | P. spumarius | Vicia sativa (Fabaceae) | - | 9 |
| Cirsium sp. (Asteraceae) | - | 9 | |||
| Gavigno, Cantagallo (Prato) | 44.040865 N 11.104997 E | P. spumarius | Euphorbia cyparissias (Euphorbiaceae) | - | 10 |
| Ranunculus sp. (Ranunculaceae) | 20 | - | |||
| Montepaldi, San Casciano (Firenze) | 43.667408 N 11.143868 E | L. coleoptrata | Trifolium repens (Fabaceae) | 15 | 30 |
| Spittlebug Species | Host Plant | Bacterial Species | Class, Family | DGGE Band |
|---|---|---|---|---|
| P. spumarius | Cistus monspeliensis | Ciceribacter selenitireducens | α-Proteobacteria, Rhizobiaceae | F-11 |
| Cistus monspeliensis | Ciceribacter selenitireducens | α-Proteobacteria, Rhizobiaceae | F-13 | |
| Cistus monspeliensis | Ciceribacter azotofigens | α-Proteobacteria, Rhizobiaceae | F-16 | |
| Cistus monspeliensis | Ciceribacter azotofigens | α-Proteobacteria, Rhizobiaceae | F-17 | |
| Cistus monspeliensis | Brevundimonas mediterranea | α-Proteobacteria, Caulobacteraceae | F-15 | |
| Cistus monspeliensis | Erwinia rhapontici | γ-Proteobacteria, Enterobacteriaceae | F-14 | |
| Cistus monspeliensis | Stenotrophomonas rhizoplilia | γ-Proteobacteria, Xanthomonadaceae | F-18 | |
| Cirsium sp. | Ciceribacter azotofigens | α-Proteobacteria, Rhizobiaceae | F-31 | |
| Cirsium sp. | Pigmentiphaga humi | β-Proteobacteria, Alcaligenaceae | F-10 | |
| Vicia sativa | Devosia oryziradicis | α-Proteobacteria, Devosiaceae | F-12 | |
| L. coleoptrata | Trifolium repens | Brevundimonas mediterranea | α-Proteobacteria, Caulobacteraceae | F-26 |
| Trifolium repens | Rhizobium skierniewicense | α-Proteobacteria, Rhizobiaceae | F-28, F-29 | |
| Trifolium repens | Sinorhizobium sp. | α-Proteobacteria, Rhizobiaceae | F-7 | |
| Trifolium repens | Erwinia rhapontici | γ-Proteobacteria, Enterobacteriaceae | F-9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nencioni, A.; Pastorelli, R.; Bigiotti, G.; Cucu, M.A.; Sacchetti, P. Diversity of the Bacterial Community Associated with Hindgut, Malpighian Tubules, and Foam of Nymphs of Two Spittlebug Species (Hemiptera: Aphrophoridae). Microorganisms 2023, 11, 466. https://doi.org/10.3390/microorganisms11020466
Nencioni A, Pastorelli R, Bigiotti G, Cucu MA, Sacchetti P. Diversity of the Bacterial Community Associated with Hindgut, Malpighian Tubules, and Foam of Nymphs of Two Spittlebug Species (Hemiptera: Aphrophoridae). Microorganisms. 2023; 11(2):466. https://doi.org/10.3390/microorganisms11020466
Chicago/Turabian StyleNencioni, Anita, Roberta Pastorelli, Gaia Bigiotti, Maria Alexandra Cucu, and Patrizia Sacchetti. 2023. "Diversity of the Bacterial Community Associated with Hindgut, Malpighian Tubules, and Foam of Nymphs of Two Spittlebug Species (Hemiptera: Aphrophoridae)" Microorganisms 11, no. 2: 466. https://doi.org/10.3390/microorganisms11020466
APA StyleNencioni, A., Pastorelli, R., Bigiotti, G., Cucu, M. A., & Sacchetti, P. (2023). Diversity of the Bacterial Community Associated with Hindgut, Malpighian Tubules, and Foam of Nymphs of Two Spittlebug Species (Hemiptera: Aphrophoridae). Microorganisms, 11(2), 466. https://doi.org/10.3390/microorganisms11020466






