Colonization and Healthcare-Associated Infection of Carbapenem-Resistant Enterobacteriaceae, Data from Polish Hospital with High Incidence of Carbapenem-Resistant Enterobacteriaceae, Does Active Target Screening Matter?
Abstract
1. Introduction
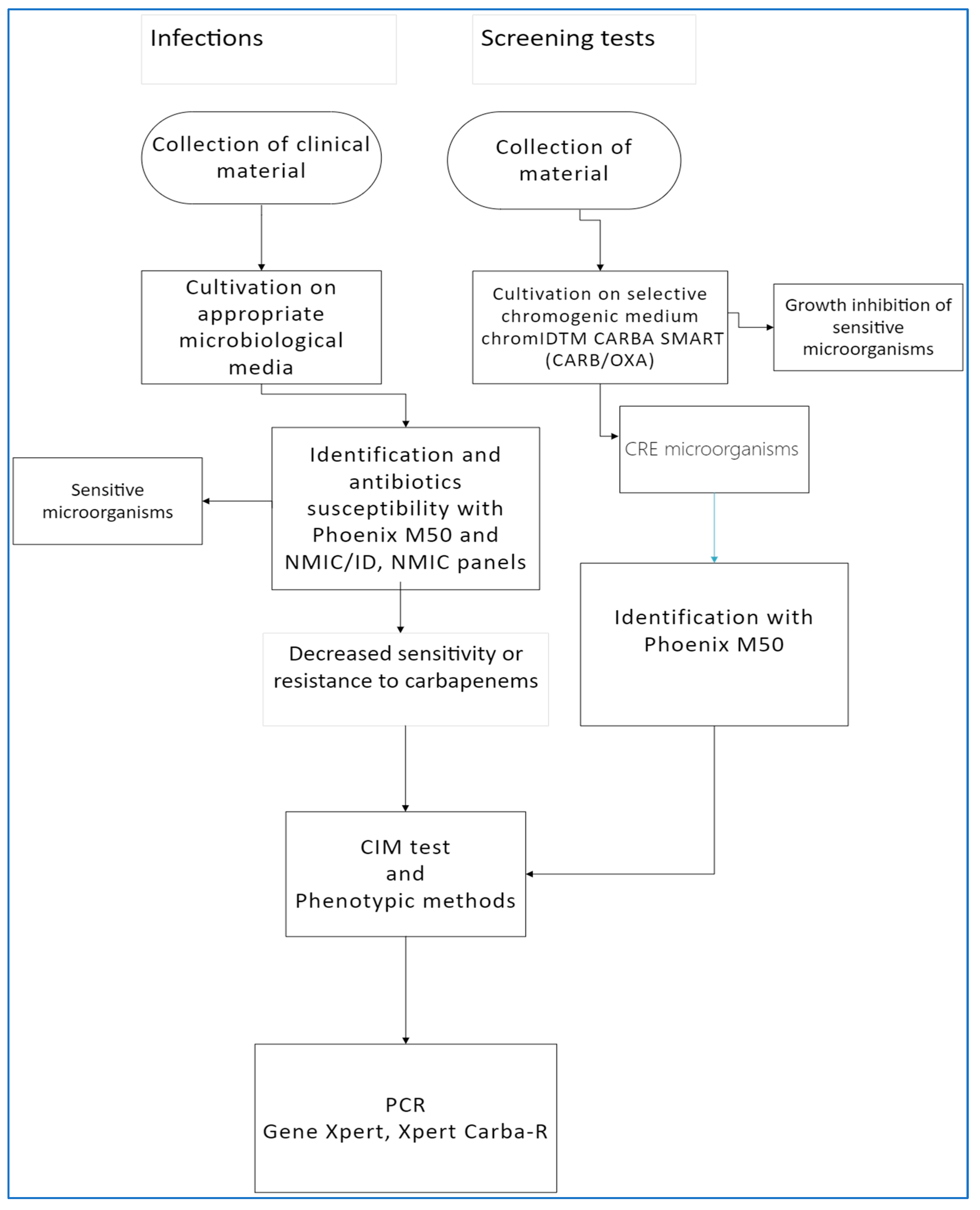
2. Materials and Methods
- At admission to hospital, in case of suspected CRE colonization, e.g., antibiotic therapy, previous hospitalization, stay in the long-term-care facilities.
- Exposition or with close contact to patients with confirmed CRE infection during hospitalization.
- In an outbreak investigation, in the analysed period, there were 8 epidemics caused by CR Klebsiella pneumoniae, including 3 in Intensive Care Unit (ICU); most often (5 events, 62.5%) the cause was the KPC+ strain, followed by OXA-48.
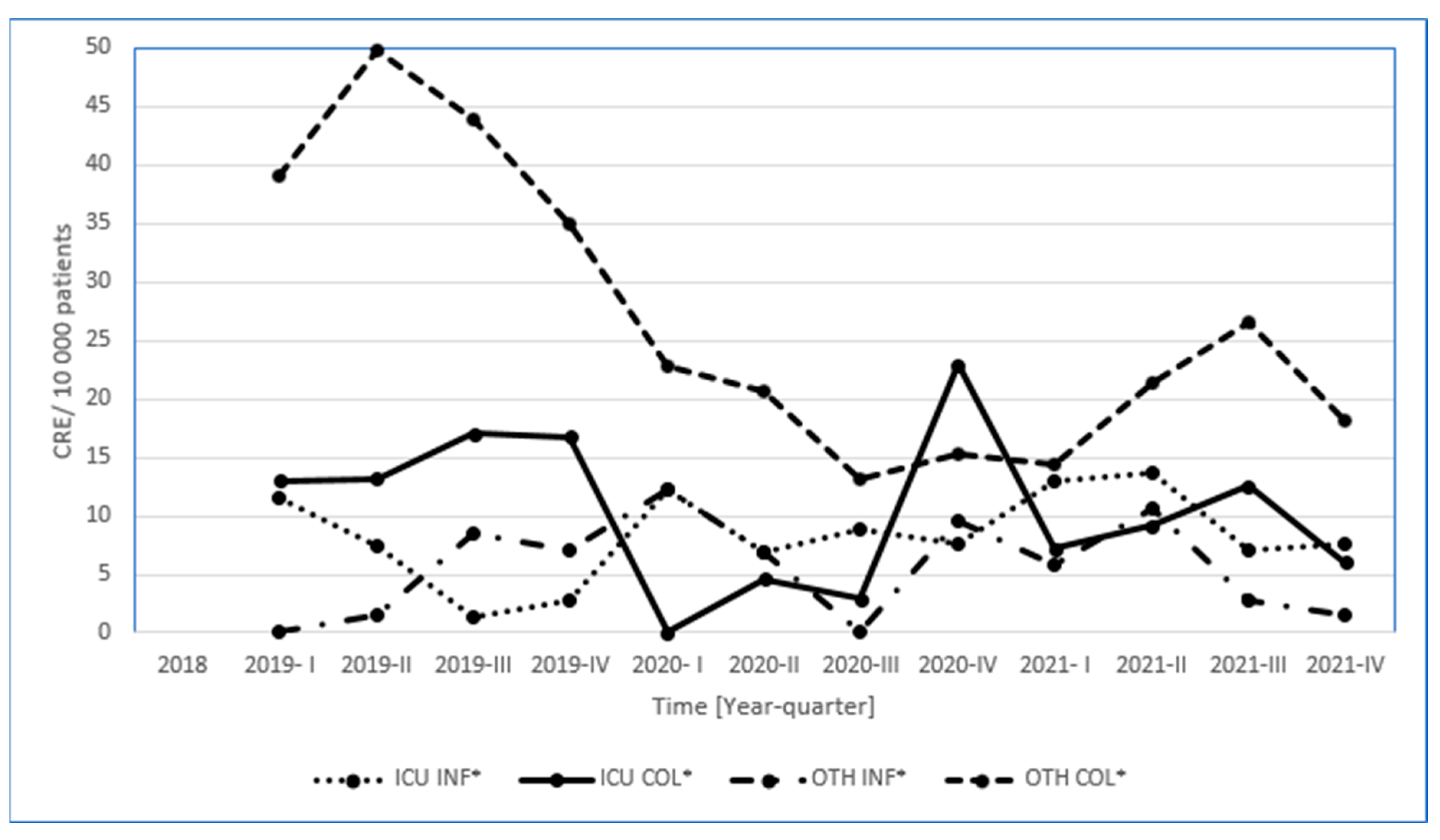
3. Results
4. Discussion
5. Strengths and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- El-Gamal, M.I.; Brahim, I.; Hisham, N.; Aladdin, R.; Mohammed, H.; Bahaaeldin, A. Recent updates of carbapenem antibiotics. Eur. J. Med. Chem. 2017, 5, 185–195. [Google Scholar] [CrossRef]
- Papp-Wallace, K.M.; Endimiani, A.; Taracila, M.A.; Bonomo, R.A. Carbapenems: Past, Present, and Future. Antimicrob. Agents Chemother. 2011, 55, 4943–4960. [Google Scholar] [CrossRef]
- Bianco, G.; Boattini, M.; Comini, S.; Casale, R.; Iannaccone, M.; Cavallo, R.; Costa, C. Occurrence of multi-carbapenemases producers among carbapenemase-producing Enterobacterales and in vitro activity of combinations including cefiderocol, ceftazidime-avibactam, meropenem-vaborbactam, and aztreonam in the COVID-19 era. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Farfour, E.; Lecuru, M.; Dortet, L.; Le Guen, M.; Cerf, C.; Karnycheff, F.; Bonnin, R.A.; Vasse, M.; Lesprit, P. Carbapenemase-producing Enterobacterales outbreak: Another dark side of COVID-19. Am. J. Infect. Control. 2020, 48, 1533–1536. [Google Scholar] [PubMed]
- Ghosh, S.; Bornman, C.; Zafer, M.M. Antimicrobial Resistance Threats in the emerging COVID-19 pandemic: Where do we stand? J. Infect. Public Health 2021, 14, 555–560. [Google Scholar] [PubMed]
- Khan, S.; Hasan, S.S.; Bond, S.E.; Conway, B.R.; Aldeyab, M.A. Antimicrobial consumption in patients with COVID-19: A systematic review and meta-analysis. Expert Rev. Anti-Infect. Ther. 2022, 20, 749–772. [Google Scholar] [CrossRef]
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed. Available online: https://www.who.int/es/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 1 December 2022).
- European Centre for Disease Prevention and Control. Rapid Risk Assessment: Carbapenem-Resistant Enterobacteriaceae—8 April 2016; ECDC: Stockholm, Sweden, 2016. [CrossRef]
- Logan, L.K.; Weinstein, R.A. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017, 215, S28–S36. [Google Scholar] [CrossRef] [PubMed]
- Karampatakis, T.; Antachopoulos, C.; Iosifidis, E.; Tsakris, A.; Roilides, E. Molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in Greece. Future Microbiol. 2016, 11, 809–823. [Google Scholar] [CrossRef]
- EUCAST. Clinical Breakpoints and Dosing of Antibiotics; EUCAST: Seongnam-si, Republic of Korea, 2023. [Google Scholar]
- van der Zwaluw, K.; de Haan, A.; Pluister, G.N. The Carbapenem Inactivation Method (CIM), a Simple and Low-Cost Alternative for the Carba NP Test to Assess Phenotypic Carbapenemase Activity in Gram-Negative Rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef]
- Jernigan, J.A.; Hatfield, K.M.; Wolford, H.; Nelson, R.E.; Olubajo, B.; Reddy, S.C.; McCarthy, N.; Paul, P.; McDonald, L.C.; Kallen, A.; et al. Multidrug-resistant bacterial infections in US hospitalized patients, 2012–2017. N. Engl. J. Med. 2020, 382, 1309–1319. [Google Scholar] [CrossRef]
- Otter, J.A.; Mookerjee, S.; Davies, F.; Bolt, F.; Dyakova, E.; Shersing, Y.; Boonyasiri, A.; Weiße, A.Y.; Gilchrist, M.; Galletly, T.J.; et al. Detecting carbapenemase-producing Enterobacterales (CPE): An evaluation of an enhanced CPE infection control and screening programme in acute care. J. Antimicrob. Chemother. 2020, 75, 2670–2676. [Google Scholar] [CrossRef] [PubMed]
- Ura, L.; Deja-Makara, B.; Pajdziński, M.; Gottwald, L.M. The occurrence and pathogenicity of B-class Carbapenemase—producing Enterobacteriaceae—Klebsiella pneumoniae strains (MBL/NDM) in patients hospitalized and treated in Mazowiecki Memorial Hospital of Radom between 2016–2018. Term Care Nurs. 2020, 5, 239–249. [Google Scholar] [CrossRef]
- Mathers, A.J.; Poulter, M.; Dirks, D.; Carroll, J.; Sifri, C.D.; Hazen, K.C. Clinical microbiology costs for methods of active surveillance for Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Infect. Control. Hosp. Epidemiol. 2014, 35, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Kardaś-Słoma, L.; Fournier, S.; Dupont, J.-C.; Rochaix, L.; Birgand, G.; Zahar, J.-R.; Lescure, F.-X.; Kernéis, S.; Durand-Zaleski, I.; Lucet, J.-C. Cost-effectiveness of strategies to control the spread of carbapenemase-producing Enterobacterales in hospitals: A modelling study. Antimicrob. Resist. Infect. Control. 2022, 19, 117. [Google Scholar] [CrossRef]
- Antimicrobal Resistance Surveillance ECDC Report. Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2022-2020-data (accessed on 1 December 2022).
- Potter, R.F.; D’Souza, A.W.; Dantas, G. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist. 2016, 29, 30–46. [Google Scholar] [CrossRef]
- Han, R.; Shi, Q.; Wu, S.; Yin, D.; Peng, M.; Dong, D.; Zheng, Y.; Guo, Y.; Zhang, R.; Hu, F.; et al. China Antimicrobial Surveillance Network (CHINET) Study Group. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated From Adult and Children Patients in China. Front. Cell. Infect. Microbiol. 2020, 10, 314. [Google Scholar] [CrossRef]
- Hosseinzadeh, Z.; Ebrahim-Saraie, H.S.; Sarvari, J.; Mardaneh, J.; Dehghani, B.; Rokni-Hosseini, S.M.H.; Motamedifar, M. Emerge of blaNDM-1 and blaOXA-48-like harboring carbapenem-resistant Klebsiella pneumoniae isolates from hospitalized patients in southwestern Iran. J. Chin. Med. Assoc. 2018, 81, 536–540. [Google Scholar] [CrossRef]
- Nordmann, P.; Poirel, L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin. Microbiol. Infect. 2014, 20, 821–830. [Google Scholar]
- Karampatakis, T.; Tsergouli, K.; Iosifidis, E.; Antachopoulos, C.; Karapanagiotou, A.; Karyoti, A.; Gritsi-Gerogianni, N.; Tsakris, A.; Roilides, E. Impact of active surveillance and infection control measures on carbapenem-resistant Gram-negative bacterial colonization and infections in intensive care. J. Hosp. Infect. 2018, 99, 396–404. [Google Scholar] [CrossRef]
- Connor, M.O.; Mc Namara, C.; Doody, O. Healthcare workers’ experiences of caring for patients colonized with carbapenemase-producing Enterobacterales (CRE) in an acute hospital setting—A scoping review. J. Hosp. Infect. 2022, 131, 181–189. [Google Scholar] [CrossRef]
- Wałaszek, M.; Kołpa, M.; Różańska, A.; Wolak, Z.; Bulanda, M.; Wójkowska-Mach, J. Practice of hand hygiene and use of protective gloves: Differences in the perception between patients and medical staff. Am. J. Infect. Control. 2018, 46, 1074–1076. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Kim, G.R.; Jeong, J.; Kim, S.; Shin, J.H. Prevalence and Characteristics of Carbapenemase-Producing Enterobacteriaceae in Three Tertiary-Care Korean University Hospitals between 2017 and 2018. Jpn. J. Infect. Dis. 2020, 24, 431–436. [Google Scholar] [CrossRef]
- Oteo, J.; Miró, E.; Pérez-Vázquez, M.; Navarro, F. Evolution of carbapenemase-producing Enterobacteriaceae at the global and national level: What should be expected in the future? Enferm. Infect. Microbiol. Clin. 2014, 32, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef] [PubMed]
- Fritzenwanker, M.; Imirzalioglu, C.; Herold, S.; Wagenlehner, F.M.; Zimmer, K.P.; Chakraborty, T. Treatment options for carbapenem- resistant gram-negative infections. Dtsch. Arztebl. Int. 2018, 115, 345–352. [Google Scholar] [CrossRef]
- van Duin, D.; Kaye, K.S.; Neuner, E.A.; Bonomo, R.A. Carbapenem-resistant Enterobacteriaceae: A review of treatment and outcomes. Diagn. Microbiol. Infect. Dis. 2013, 75, 115–120. [Google Scholar] [CrossRef]
- Sheu, C.-C.; Chang, Y.-T.; Lin, S.-Y.; Chen, Y.-H.; Hsueh, P.-R. Infections Caused by Carbapenem-Resistant Enterobacteriaceae: An Update on Therapeutic Options. Front. Microbiol. 2019, 30, 80. [Google Scholar]
| Specimen | No. CRE | Incidence Rate Per 10,000 Hospitalizations | |||
|---|---|---|---|---|---|
| 2019 | 2020 | 2021 | Total (%) | ||
| Respiratory specimens | 0 | 12 | 17 | 29 (7) | 3.7 |
| Urine | 16 | 11 | 15 | 42 (11) | 5.4 |
| Skin and soft tissue infections | 4 | 5 | 3 | 12 (3) | 1.5 |
| Blood | 8 | 3 | 7 | 18 (5) | 2.3 |
| Subtotal | 28 | 33 | 43 | 104 (26) | 13.3 |
| Screening swabs | 159 | 57 | 79 | 295 (74) | |
| Total | 187 | 88 | 121 | 399 (100) | |
| Quarter | Year | No. Admission | No. Screening Tests | Screening Tests (%) | Results CRE+ | Results CRE+ (%) | CRE-HAI | Incidence CRE-HAI * | CRE+/CRE-HAI ** |
|---|---|---|---|---|---|---|---|---|---|
| I | 2019 | 6914 | 1877 | 27.1 | 36 | 1.9 | 8 | 11.6 | 4.5 |
| II | 2019 | 6817 | 1951 | 28.6 | 43 | 2.2 | 6 | 8.8 | 7.2 |
| III | 2019 | 7050 | 1889 | 26.8 | 43 | 2.3 | 7 | 9.9 | 6.1 |
| IV | 2019 | 7151 | 1615 | 22.6 | 37 | 2.3 | 7 | 9.8 | 5.3 |
| I | 2020 | 6551 | 1433 | 21.9 | 15 | 1.0 | 10 | 15.3 | 1.5 |
| II | 2020 | 4366 | 712 | 16.3 | 11 | 1.5 | 6 | 13.7 | 1.8 |
| III | 2020 | 6804 | 1143 | 16.8 | 11 | 1.0 | 6 | 8.8 | 1.8 |
| IV | 2020 | 5237 | 423 | 8.1 | 20 | 4.7 | 9 | 17.2 | 2.2 |
| I | 2021 | 6914 | 660 | 9.5 | 15 | 2.3 | 13 | 18.8 | 1.2 |
| II | 2021 | 6571 | 906 | 13.8 | 20 | 2.2 | 16 | 24.3 | 1.3 |
| III | 2021 | 7137 | 1165 | 16.3 | 28 | 2.4 | 7 | 9.8 | 4.0 |
| IV | 2021 | 6628 | 940 | 14.2 | 16 | 1.7 | 6 | 9.1 | 2.7 |
| Species | No. CRE | Incidence Rate Per 10,000 Hospitalizations | |||
|---|---|---|---|---|---|
| 2019 | 2020 | 2021 | Total (%) | ||
| Klebsiella pneumoniae | 25 | 32 | 40 | 97 (93) | 12.4 |
| Klebsiella oxytoca | 0 | 0 | 1 | 1 (1) | 0.1 |
| Klebsiella aerogenes | 0 | 0 | 1 | 1 (1) | 0.1 |
| Escherichia coli | 3 | 0 | 0 | 3 (3) | 0.4 |
| Serratia marcesces | 0 | 1 | 0 | 1 (1) | 0.1 |
| Proteus mirabilis | 0 | 0 | 1 | 1 (1) | 0.1 |
| Total | 28 | 33 | 42 | 104 (100) | 13.3 |
| Isolated Strains N | ||||
|---|---|---|---|---|
| Screening | HAI | Total | Prevalence (%) | |
| Klebsiella spp. n = 393 | ||||
| OXA-48 | 215 | 64 | 279 | 76.6 |
| NDM | 4 | 6 | 10 | 3.3 |
| KPC | 75 | 27 | 102 | 28.8 |
| OXA-48, KPC | 0 | 2 | 2 | 1.0 |
| Escherichia coli n = 4 | ||||
| OXA-48 | 1 | 3 | 4 | 100.0 |
| NDM | 0 | 0 | 0 | 0.0 |
| KPC | 0 | 0 | 0 | 0.0 |
| OXA-48, KPC | 0 | 0 | 0 | 0.0 |
| Proteus mirabilis n = 1 | ||||
| OXA-48 | 0 | 0 | 0 | 0.0 |
| NDM | 0 | 0 | 0 | 0.0 |
| KPC | 0 | 0 | 0 | 0.0 |
| OXA-48, KPC | 0 | 0 | 0 | 0.0 |
| VIM | 0 | 1 | 1 | 1.0 |
| Serratia marcescens n = 1 | ||||
| OXA-48 | 0 | 1 | 1 | 100.0 |
| NDM | 0 | 0 | 0 | 0 |
| KPC | 0 | 0 | 0 | 0 |
| OXA-48, KPC | 0 | 0 | 0 | 0 |
| VIM | 0 | 0 | 0 | 0 |
| Antibiotics | Klebsiella Pneumoniae | Other * | Total | ||
|---|---|---|---|---|---|
| 2019 | 2020 | 2021 | 2019–2021 | 2019–2021 | |
| (n = 25) (%) | (n = 30) (%) | (n = 39) (%) | (n = 7) (%) | (n = 104) (%) | |
| β-Lactam Antibacterials, Penicillins | |||||
| ampicillin | 100 | 100 | 100 | 100 | 100 |
| amoxicillin + clavulanic acid | 100 | 100 | 100 | 100 | 100 |
| piperacillin + tazobactam | 100 | 100 | 100 | 100 | 100 |
| Cephalosporins | |||||
| cefuroxime | 100 | 100 | 100 | 100 | 100 |
| cefotaxime | 100 | 100 | 100 | 100 | 100 |
| ceftazidime | 100 | 100 | 100 | 100 | 100 |
| ceftriaxone | 100 | 100 | 100 | 100 | 100 |
| cefepime | 100 | 100 | 100 | 100 | 100 |
| Cephalosporin+ Non-β-Lactam β-Lactamase Inhibitor | |||||
| ceftazidime-avibactam | 2 | 8 | 6 | 0 | 0.7 |
| Carbapenems | |||||
| IPM | 65 | 91 | 89 | 85 | 84.5 |
| MEM | 63 | 75 | 67 | 68 | 68.2 |
| ETP | 100 | 100 | 100 | 100 | 100 |
| Aminoglycosides | |||||
| amikacin | 8 | 8 | 11 | 0 | 2 |
| gentamicin | 88 | 82 | 40 | 50 | 53.3 |
| tobramycin | 98 | 98 | 93 | 52 | 59.3 |
| netilmicin | 87 | NT | NT | 100 *** | 96.5 |
| Fluoroquinolones | |||||
| ciprofloxacin | 100 | 100 | 95 | 72 | 76.3 |
| levofloxacin | 100 | 100 | 95 | 72 | 76.3 |
| Other Antibacterials | |||||
| aztreonam | 100 | 95 | 95 | 44.3 ** | 57.8 |
| colistin | 75 | 92 | 73 | 60 | 61.2 |
| fosfomycin | 62 | 63 | 33 | NT | 55 |
| nitrofurantoin | NT | NT | NT | 25 *** | 25 |
| trimethoprim-sulfamethoxazole | 98 | 92 | 96 | 40 | 49.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pawłowska, I.; Ziółkowski, G.; Jachowicz-Matczak, E.; Stasiowski, M.; Gajda, M.; Wójkowska-Mach, J. Colonization and Healthcare-Associated Infection of Carbapenem-Resistant Enterobacteriaceae, Data from Polish Hospital with High Incidence of Carbapenem-Resistant Enterobacteriaceae, Does Active Target Screening Matter? Microorganisms 2023, 11, 437. https://doi.org/10.3390/microorganisms11020437
Pawłowska I, Ziółkowski G, Jachowicz-Matczak E, Stasiowski M, Gajda M, Wójkowska-Mach J. Colonization and Healthcare-Associated Infection of Carbapenem-Resistant Enterobacteriaceae, Data from Polish Hospital with High Incidence of Carbapenem-Resistant Enterobacteriaceae, Does Active Target Screening Matter? Microorganisms. 2023; 11(2):437. https://doi.org/10.3390/microorganisms11020437
Chicago/Turabian StylePawłowska, Iwona, Grzegorz Ziółkowski, Estera Jachowicz-Matczak, Michał Stasiowski, Mateusz Gajda, and Jadwiga Wójkowska-Mach. 2023. "Colonization and Healthcare-Associated Infection of Carbapenem-Resistant Enterobacteriaceae, Data from Polish Hospital with High Incidence of Carbapenem-Resistant Enterobacteriaceae, Does Active Target Screening Matter?" Microorganisms 11, no. 2: 437. https://doi.org/10.3390/microorganisms11020437
APA StylePawłowska, I., Ziółkowski, G., Jachowicz-Matczak, E., Stasiowski, M., Gajda, M., & Wójkowska-Mach, J. (2023). Colonization and Healthcare-Associated Infection of Carbapenem-Resistant Enterobacteriaceae, Data from Polish Hospital with High Incidence of Carbapenem-Resistant Enterobacteriaceae, Does Active Target Screening Matter? Microorganisms, 11(2), 437. https://doi.org/10.3390/microorganisms11020437






