Abstract
Rodents and bats are the most diverse mammal group that host Bartonella species. In the Americas, they were described as harboring Bartonella species; however, they were mostly characterized to the genotypic level. We describe here Bartonella isolates obtained from blood samples of one rodent (Peromyscus yucatanicus from San José Pibtuch, Yucatan) and two bat species (Desmodus rotundus from Progreso, and Pteronotus parnellii from Chamela-Cuitzmala) from Mexico. We sequenced and described the genomic features of three Bartonella strains and performed phylogenomic and pangenome analyses to decipher their phylogenetic relationships. The mouse-associated genome was closely related to Bartonella vinsonii. The two bat-associated genomes clustered into a single distinct clade in between lineages 3 and 4, suggesting to be an ancestor of the rodent-associated Bartonella clade (lineage 4). These three genomes showed <95% OrthoANI values compared to any other Bartonella genome, and therefore should be considered as novel species. In addition, our analyses suggest that the B. vinsonii complex should be revised, and all B. vinsonii subspecies need to be renamed and considered as full species. The phylogenomic clustering of the bat-associated Bartonella strains and their virulence factor profile (lack of the Vbh/TraG conjugation system remains of the T4SS) suggest that it should be considered as a new lineage clade (L5) within the Bartonella genus.
1. Introduction
Bartonella species are Gram-negative, facultative intracellular bacteria, transmitted by arthropods, infecting mammalian erythrocytes and endothelial cells [1]. Currently, there are 39 species and three subspecies with a valid taxonomic standing in the genus, at least 20 of which have been associated with human diseases [2,3]. As the greatest diversity of Bartonella species is reported in rodents and bats [4,5,6], rodents and possibly bats are considered as a potential source of emerging zoonotic Bartonella pathogens [7]. Moreover, bats were reported to have a crucial role in the diversification of the Bartonella genus [8].
Several studies described the presence of Bartonella DNA in samples of rodents and bats in the Americas [5,9,10,11,12,13,14,15]. However, the genomic diversity of Bartonella species detected or isolated in mammal hosts, especially in the Americas has not been fully described and has yet to be investigated and elucidated [14,15]. Therefore, this study describes three novel Bartonella genomes acquired from Mexican rodents and bats. We analyzed their phylogenomic relationships and genome divergence among all known Bartonella species.
2. Methods
2.1. Sampling and Isolation
Blood samples were obtained from three different sites in Mexico in previous studies [14,15]. A rodent sample (strain 220B) was collected from San José Pibtuch, Yucatan and two bat samples (strains F02 and A05) were from Chamela-Cuitzmala (F02) and Progreso (A05), respectively. Strain 220B was isolated from the blood of a Yucatan deer mouse (Peromyscus yucatanicus). Strains A05 and F02 were isolated from the blood of a common vampire (Desmodus rotundus) and a Parnell mustached bat (Pteronotus parnellii), respectively. The blood samples from these three strains were cultured on chocolate agar plates using 200 µL blood and M199 growth media with 5% amphotericin B. Suspected Bartonella colonies were re-plated and the DNA was extracted and submitted to conventional PCR to confirm that they belong to the Bartonella genus. Target genes of Bartonella from these samples are available in GenBank (gltA: MN394838, KY629834, KY629884; ftsZ: KY629824, KY629797; rpoB: KY629925, KY629898). After confirmation as Bartonella, the three samples were subjected to WGS.
2.2. Genomic and Library Methods
The genomic DNA extraction was performed following growth on chocolate agar. Isolates were selected and the gDNA was extracted after lysis using Qiagen DNeasy blood and tissue kits [14]. Each sample was used to produce a sequencing library with gDNA that was of sufficient quality (A260/280 and A260/230 > 1.5) with an average insert size of 350 bp according to the KAPA HyperPlus Kit protocol as described previously [16,17,18,19]. All samples were sequenced using HiSeq XTEN Sequencing System (Illumina, San Diego, CA, USA) [20,21].
2.3. Genome Analysis
The whole genome sequences were trimmed, quality filtered, and quality assured using TrimGalore (v.0.5.0) [22]. The trimmed reads were submitted to the comprehensive genome analysis in the PATRIC database using UNICYCLER v.0.4.8 [23]. The resulting assembly was analyzed in PATRIC [24] and MiGa [25] to evaluate the genome features and completeness, contamination, and quality. The genomes were annotated by Bakta (v.1.6.1) [26], Prokka (v.3.2.1) [27], RASTtk (v1.073) [28], as well as PGAP (v.6.4) [29], and the PGAP was chosen as the reference annotation. A whole-genome-based phylogenetic tree was reconstructed using RAxML (v8.2.11) [30] implemented in PATRIC [31]. Whole-genome average nucleotide identity (ANI) was calculated between genomes of members of the genus Bartonella using the software Orthologous Average Nucleotide Identity Tool (OAT v.0.93.1) [18,32]. Delimitation of species using OrthoANI results was based on Goris et al. [33] and Ciufo et al. [34], considering <95% values as novel species [19].
2.4. Virulence Factors Characterization
The Bartonella lineages were classified according to their virulence factors (BaGTA, Flagellum, TrW T4SS, Vbht T4SS, VirB/D4 T4SS) following Wagner and Dehio [35]. The sequences and annotations of all Bartonella genomes were retrieved from the NCBI database and analyzed by Geneious program v.7 (Biomatters, Newark, NJ, USA). All the cassettes of virulence factor genes found in their genomes were evaluated and indicated on a phylogenetic tree.
2.5. Pangenome Analysis
The pangenome of the Bartonella genus was determined by the same genome sequences used in other analyses, including the Brucella abortus as an outgroup. The quality of genome sequences, contamination, completeness, and gene markers of lineage was performed using the CheckM [36]. The Prokka annotation was used as an input for pangenome analysis using Roary v.3.12.0 [37]. Core genes were considered to be those present in >90% of the genomes in the comparison [38]. To evaluate the similarity between the gene sequences and compare shared sequences in the data, the sourmash v.3.2.3 was used [39]. A neighbor-joining phylogeny reconstruction was built using the sourmash matrix output of shared coding gene [40]. The “roary_plots.py” script was used to visualize a matrix with gene presence/absence of core, shell, and accessory genes [41]. Gene presence/absence and the neighbor-joining phylogenetic reconstruction were visualized using Phandango [42].
3. Results
Bartonella isolates were sequenced and their genome features are described in Table 1. Two genomes from bat samples were considered as complete genomes (strains F02 and A05), with one hundred and six essential genes and one nearly complete genome with one hundred and five essential genes, with only the ribosomal protein L34 gene missing (strain 220B) (Table 1). This Whole Genome Shotgun project has been deposited at DDB/ENA/GenBank under accession numbers: JANHOI000000000 (strain 220B); JANHOJ000000000 (strain A05); JANHOK000000000 (strain F02); Bioproject PRJNA8621808; BioSample: SAMN29927589-SAMN29927591.

Table 1.
Genome features of the three strains of Bartonella from one mouse and two bats.
The topology of 137,193 nucleotides from 149 coding genes shows high nodal support (>95 bootstrap, Figure 1) and indicates a diversification of these isolates into two clades. The first is composed of two new strains, Bartonella strains A05 and F02 from bats of the species D. rotundus and P. parnellii, respectively. The topology of these two strains appears as a single branch of lineage 4 located between lineages 3 and 4 suggesting that it is an ancestor clade of the rodent-associated Bartonella (Figure 1). The strain 220B from the mouse Peromyscus yucatanicus is clustered as a sister group with the complex B. vinsonii in lineage 4 clade (Figure 1).
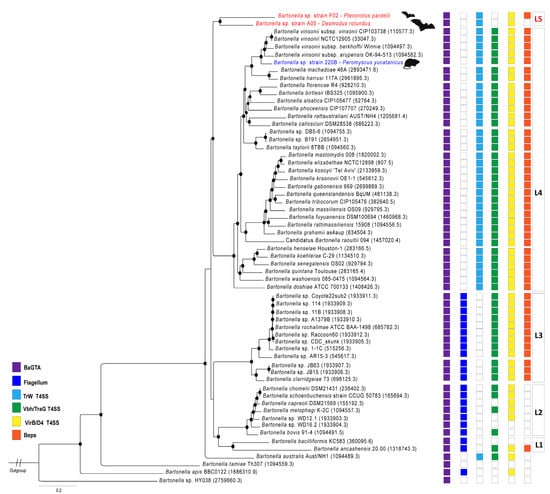
Figure 1.
Whole-genome-based phylogeny. Phylogenetic tree of species of the genus Bartonella based on 149 shared protein genes and 137,193 nucleotide positions. The topology was reconstructed using the RAxML method on PATRIC database. Red branches represent the novel bat-associated Bartonella species. The blue branch represents the novel mouse-associated Bartonella species. Nodal support >95 of bootstrap values is shown as black circles. The genome ID references used in this analysis are described in brackets. The known Bartonella lineages are described on boxes on the right of main clades (L1–L4), the proposal of a new lineage is described in red (L5). The squares represent the virulence factors found in the Bartonella genomes: BaGTA (purple), Flagellum (sky blue), TrW T4SS (light blue), Vbh/TraG T4SS (green), Virb/D4 T4SS (yellow), and Beps (orange). Bar, substitutions per nucleotide position. Brucella abortus strain MC was used as an outgroup. Figure adapted from Wagner and Dehio [35].
The OrthoANI between strains A05 and F02 was 79.8%. The strain A05 had OrthoANI values between 78.6–80.07% with the species B. alsatica, B. vinsonii subsp. vinsonii, B. taylorii, B. henselae, B. quintana, B. doshiae, B. tribocorum, and B. grahamii (Figure 2). The strain F02 had OrthoANI values between 78.2–78.8% with the species B. alsatica, B. vinsonii subsp. vinsonii, B. taylorii, B. henselae, B. quintana, B. doshiae, B. tribocorum, and B. grahamii (Figure 2). The strain 220B had OrthoANI values of 90–91% with the B. vinsonii complex, 85% with B. taylorii, 84% with B. alsatica and B. phoceensis, 83.5% with B. florencae, and 82% with B. rattaustraliani (Figure 2).
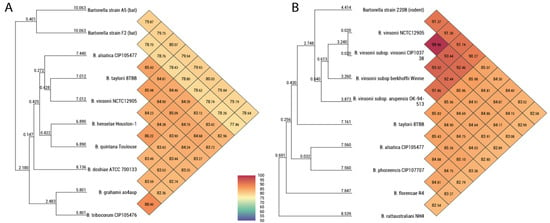
Figure 2.
Heat map generated using OrthoANI values based on whole genome sequences, calculated using OAT software and other closely related species. (A) OrthoANI values of Bartonella strain A05 and strain F02. (B) OrthoANI values of Bartonella strain 220B. The numbers in each diamond at the diagonal lines correspond to the OrthoANI values between the species.
Strain 220B contains the same repertoire of virulence factor genes presented in lineage 4 (Figure 1). The annotation shows the virulence factor genes encoding the BaGTA, TrW T4SS, Vbh/TraG T4SS, Virb/D4 T4SS, and Beps. Two cassettes of complex VirB2-11 were found in this strain. Strain F2 contains genes of the BaGTA, TrW T4SS, Virb/D4 T4SS, and Beps; however, it does not contain genes of toxin Vbh/TraG T4SS. Two cassettes of complex VirB2-11 were detected in this strain. Strain A5 contains the BaGTA, TrW T4SS, and Virb/D4 T4SS virulence factors; however, the cassettes of genes encoding VBh/TraG T4SS and Beps were not identified.
Bartonella pangenome containing 39,431 genes and a core of 27 genes (>99%) and 81 soft core genes (95–99%) was observed for 60 genomes. The known lineages (1–4) were recovered from the 1668 genes (15–95%). The unique genes were the most representative of the genus with 37,655 genes. Strain 220B contains more genes shared with the B. vinsonii complex and with the rodent-associated Bartonella clade. The two bat-associated strains, A05 and F02, do not share the same cassettes of shell genes shared with other lineages. The pangenome analyses show that the Bartonella species are significantly different among each other (Figure S1; Supplementary Data).
4. Discussion
The genus Bartonella includes a great diversity of species harbored by many mammalian hosts worldwide. The genetic diversity of this genus has been described by comparisons of the genetic divergence and multilocus analyses of target genes, which allowed for the description of several species and genotypes [43,44]. In this study, we characterized three strains of Bartonella by whole genome sequencing (WGS) and according to Goris et al. [33], the Bartonella genome analysis and their OrthoANI values (cut-off less than <95%), they should be considered as three novel species of the genus Bartonella. However, to fully describe and name these bacteria as new species according to the International Code of Nomenclature of Prokaryotes (ICNP), the characterization of several phenotypic, biological, and biochemical aspects of the bacteria must be carried out in addition to the isolation and WGS [45].
Fischedick et al. [15] described Bartonella genotypes from the Yucatan dear mouse (P. yucatanicus) based on target genes and named strain 220B as Bartonella vinsonii subsp. yucatanensis; however, according to our analysis, its OrthoANI values of 90–91% indicate that it is a distinct species within the B. vinsonii complex. Our results support recent findings described by Amaral et al. [46] suggesting that the B. vinsonii complex needs to be revised, as the subspecies already described and named in this complex have OrthoANI values less than 95% between them (see Figure 2). This is particularly true for B. vinsonii subsp. berkhoffii that has mainly a carnivore reservoir rather than a rodent reservoir as described for the other subspecies [47]. Considering the virulence factors, the mouse-associated strain 220B has the cassette genes of BaGTA, TrW T4SS, Vbh/TraG T4SS, VirB/D4, and Beps (Figure 1). This is the same repertoire of virulence factors found in lineage 4 of the Bartonella genus. This lineage is commonly found in rodents [7,48]. The host of Bartonella strain 220B belongs to the genus Peromyscus. This rodent genus is distributed across North and Central America and is considered a natural reservoir of zoonotic viral and bacterial pathogens, such as Hantavirus and Borrelia burgdorferi [49,50]. Three species of Peromyscus have already been found to host Bartonella DNA in northwestern, central, and eastern Mexico including P. leucopus, P. maniculatus, and P. yucatanicus [14,51].
Two bat-associated complete genomes were annotated herein and indicated that they are new species with high divergence across the Bartonella species. The first ever bat-associated Bartonella genome to be sequenced was named Bartonella sp. HY038. This genome was sequenced from bat faeces in China, and is the most ancient ancestor of all Bartonella species (see Figure 1). The two described bat-associated genomes herein were the first genomes sequenced from isolates of bat blood samples. In the comparison between Bartonella sp. HY038 and strains A5 and F2, OrthoANI values of 68% and 69%, respectively, were retrieved, indicating that the great diversity of bat-associated Bartonella genomes is still underestimated. The bat-associated Bartonella genomes clustered into a single clade, distinct from the L3 and L4 lineages (Figure 1). The L4 lineage is the most diverse and mostly represented by rodent-associated Bartonella species described to date, as described in the mouse-associated 220B strain in this study, while the L3 has only two species, B. clarridgeiae and B. rochalimae with a valid status, mainly associated with domestic and wild carnivores [52]. The two novel species described in this study reveal a new lineage of Bartonella. The bat-associated genomes shared the TrW T4SS, the genetic cassette acquired following the loss of the flagellum [35]; however, both strains do not have the genes encoding Vbh/traG conjugation systems (Figure 1). According to Harms and Dehio [53], these genes have an important role in the pathogenesis of Bartonella species, and the complete cassette of Vbh conjugation systems are found in the Bartonella chromosomes or plasmids [35]. Most of the Bartonella species contain VbhA toxin and TraG genes in their genomes, two homologous regions known as part of the VbhT T4SS system; however, these genes were not identified in any bat-associated genome to date. The bat-associated strains have the VirB-like genes (Figure 1). This type IV secretion system is responsible for the translocation of Bartonella effector proteins (Beps) in different niches of mammalian host cells [53]. The Beps were detected in strain F02, but not in strain A05. Due to this high divergence between the virulence factors in these new strains and lineage 4, and their phylogenomic clustering into a single clade, distinct from lineages 3 and 4, herein we propose these strains as representative of a new lineage, lineage 5, within the Bartonella genus (Figure 1). Bats form a diverse group as rodents, and act in the dispersion of Bartonella to other mammalian hosts, which consequently contribute to the adaptation and diversification of these bacteria [8]. P. parnelli is distributed throughout North America and northern South America. The strain F02 isolated in this insectivorous bat was described by Stuckey et al. [14], with eight out of thirty (26.7%) specimens reported to be positive for Bartonella DNA. Previously, only flies and ticks collected from this species have been described to contain Bartonella DNA, both in Mexico and French Guiana [54,55]. The vampire bat D. rotundus is widely distributed from southern North America to southern South America and is known to be one of the main natural hosts of the rabies virus [56]. Due to its exclusive diet of mammalian blood, the ecological interaction between this species and other domestic and food producing animals is one of the biggest concerns regarding the spillover of pathogens in Latin America [57]. In addition, this species is known to roost with other bat species and share vectors, and therefore potentially cause a spill over of pathogens to other bat species [58]. From these ecological and dietary aspects, D. rotundus is an important host of Bartonella spp., with a high prevalence and great diversity of genotypes reported from Central and South America [5,9,14,58,59,60]. Previous studies have reported that Bartonella genotypes found in vampire bats were mainly associated with lineage 2 (L2), the ruminant-associated Bartonella spp. [9,60]. However, herein we describe a new Bartonella strain isolated from D. rotundus which was not associated with L2, and belongs probably to a new lineage, distant from those described in previous studies.
According to the pangenome analysis that was carried out in this study, the Bartonella species showed high variability of unique genes compared with shared and core genes. The pangenome analysis indicated that the strains described herein should be considered as new species. Additionally, the genetic distance cut-off (OrthoANI values) and core genes similarity (%) should be established for Bartonella, as previously conducted for Hungatella hathewayi, a necessary step toward evaluating the intrageneric diversity [18].
5. Conclusions
A deeper understanding of the evolutionary diversification observed in this genus is made possible by the characterization and description of additional genomes in rodents and bats. Our study using genomic approach shows that in Mexico, there are at least three novel Bartonella species described herein that need further characterization in order to be fully named according to the ICNP. Based on phylogenomic clustering and virulence factors, the bat-associated Bartonella species, presented in this study, suggest the possibility of a new fifth lineage in the genus. Additionally, the analysis of OrthoANI values carried out herein suggests that there is a need to revise the B. vinsonii complex.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11020340/s1, Figure S1: Pan-genome analysis of 60 Bartonella genomes: The neighbor-joining phylogenomic tree to the left, colored according to the described lineages: (black: ancestors; brown: lineage 1; green: lineage 2; orange: lineage 3; blue, lineage 4; red: lineage 5). The matrix on the right is the calculated pan-genome for Bartonella genomes.
Author Contributions
Conceptualization, J.G.-O., R.G., B.B.C. and S.H.; methodology, J.G.-O., R.G., C.L.S., B.C.W., B.C.H., B.B.C. and S.H.; formal analysis, J.G.-O., R.G., C.L.S. and B.C.W.; investigation, J.G.-O., R.G., Y.N.-B., D.A.J., A.A.-S., H.-J.B., B.B.C. and S.H.; resources, D.A.J., B.C.W., B.B.C. and S.H.; data curation, J.G.-O., R.G. and C.L.S.; writing—original draft preparation, J.G.-O., R.G., Y.N.-B., B.B.C. and S.H.; writing—review and editing, J.G.-O., R.G., D.A.J., C.L.S., A.A.-S., H.-J.B., Y.N.-B., B.C.H., B.C.W., B.B.C. and S.H.; visualization, J.G.-O., R.G., Y.N.-B., C.L.S. and B.C.W.; supervision, B.B.C. and S.H.; project administration, B.B.C. and S.H.; funding acquisition, B.B.C. and S.H. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the “UC Davis—Israel Collaborations in Research” grant for Shimon Harrus and Bruno Chomel.
Data Availability Statement
The data is published on NCBI by the Bioproject link: https://www.ncbi.nlm.nih.gov/bioproject/861808/.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chomel, B.B.; Boulouis, H.-J.; Breitschwerdt, E.B.; Kasten, R.W.; Vayssier-Taussat, M.; Birtles, R.J.; Koehler, J.E.; Dehio, C. Ecological fitness and strategies of adaptation of Bartonella species to their hosts and vectors. Vet. Res. 2009, 40, 22–29. [Google Scholar] [CrossRef]
- Okaro, U.; Addisu, A.; Casanas, B.; Anderson, B. Bartonella Species, an Emerging Cause of Blood-Culture-Negative Endocarditis. Clin. Microbiol. Rev. 2017, 30, 709–746. [Google Scholar] [CrossRef]
- Cheslock, M.A.; Embers, M.E. Human Bartonellosis: An Underappreciated Public Health Problem? Trop. Med. Infect. Dis. 2019, 4, 69. [Google Scholar] [CrossRef]
- Gutiérrez, R.; Krasnov, B.; Morick, D.; Gottlieb, Y.; Khokhlova, I.S.; Harrus, S. Bartonella Infection in Rodents and Their Flea Ectoparasites: An Overview. Vector-Borne Zoonotic Dis. 2015, 15, 27–39. [Google Scholar] [CrossRef]
- Stuckey, M.J.; Chomel, B.B.; de Fleurieu, E.C.; Aguilar-Setién, A.; Boulouis, H.-J.; Chang, C.-C. Bartonella, bats and bugs: A review. Comp. Immunol. Microbiol. Infect. Dis. 2017, 55, 20–29. [Google Scholar] [CrossRef]
- Han, H.; Li, Z.; Li, X.; Liu, J.; Peng, Q.; Wang, R.; Gu, X.; Jiang, Y.; Zhou, C.; Li, D.; et al. Bats and their ectoparasites (Nycteribiidae and Spinturnicidae) carry diverse novel Bartonella genotypes, China. Transbound. Emerg. Dis. 2021, 69, e845–e858. [Google Scholar] [CrossRef]
- Krügel, M.; Król, N.; Kempf, V.A.J.; Pfeffer, M.; Obiegala, A. Emerging rodent-associated Bartonella: A threat for human health? Parasites Vectors 2022, 15, 113. [Google Scholar] [CrossRef]
- McKee, C.D.; Bai, Y.; Webb, C.T.; Kosoy, M.Y. Bats are key hosts in the radiation of mammal-associated Bartonella bacteria. Infect. Genet. Evol. 2021, 89, 104719. [Google Scholar] [CrossRef]
- André, M.R.; Gutiérrez, R.; Ikeda, P.; Amaral, R.B.D.; de Sousa, K.C.M.; Nachum-Biala, Y.; Lima, L.; Teixeira, M.M.G.; Machado, R.Z.; Harrus, S. Genetic diversity of Bartonella spp. in vampire bats from Brazil. Transbound. Emerg. Dis. 2019, 66, 2329–2341. [Google Scholar] [CrossRef]
- Müller, A.; Gutiérrez, R.; Seguel, M.; Monti, G.; Otth, C.; Bittencourt, P.; Sepúlveda, P.; Alabí, A.; Nachum-Biala, Y.; Harrus, S. Molecular survey of Bartonella spp. in rodents and fleas from Chile. Acta Trop. 2020, 212, 105672. [Google Scholar] [CrossRef]
- Ikeda, P.; Seki, M.C.; Carrasco, A.O.T.; Rudiak, L.V.; Miranda, J.M.D.; Gonçalves, S.M.M.; Hoppe, E.G.L.; Albuquerque, A.C.A.; Teixeira, M.M.G.; Passos, C.E.; et al. Evidence and molecular characterization of Bartonella spp. and hemoplasmas in neotropical bats in Brazil. Epidemiol. Infect. 2017, 145, 2038–2052. [Google Scholar] [CrossRef]
- de Oliveira, J.G.; Rozental, T.; Guterres, A.; Teixeira, B.R.; Andrade-Silva, B.E.; da Costa-Neto, S.F.; Furtado, M.C.; Moratelli, R.; D’Andrea, P.S.; Lemos, E.R.S. Investigation of Bartonella spp. in brazilian mammals with emphasis on rodents and bats from the Atlantic Forest. Int. J. Parasitol. Parasites Wildl. 2020, 13, 80–89. [Google Scholar] [CrossRef]
- Gonçalves, L.R.; Favacho, A.R.D.M.; Roque, A.L.R.; Mendes, N.S.; Fidelis, O.L., Jr.; Benevenute, J.L.; Herrera, H.M.; D’Andrea, P.S.; de Lemos, E.R.S.; Machado, R.Z.; et al. Association of Bartonella Species with Wild and Synanthropic Rodents in Different Brazilian Biomes. Appl. Environ. Microbiol. 2016, 82, 7154–7164. [Google Scholar] [CrossRef]
- Stuckey, M.J.; Chomel, B.B.; Obregón-Morales, C.; Leyva, J.I.O.; Arechiga-Ceballos, N.; Moreno-Sandoval, H.; Aguilar-Setién, A.; Salas-Rojas, M.; Galvez-Romero, G. Bartonella Infection in Hematophagous, Insectivorous, and Phytophagous Bat Populations of Central Mexico and the Yucatan Peninsula. Am. J. Trop. Med. Hyg. 2017, 97, 413–422. [Google Scholar] [CrossRef]
- Fischedick, F.B.S.; Stuckey, M.J.; Aguilar-Setién, A.; Moreno-Sandoval, H.; Galvez-Romero, G.; Salas-Rojas, M.; Arechiga-Ceballos, N.; Overgaauw, P.A.M.; Kasten, R.W.; Chomel, B.B. Identification of Bartonella Species Isolated from Rodents from Yucatan, Mexico, and Isolation of Bartonella vinsonii subsp. yucatanensis subsp. nov. Vector-Borne Zoonotic Dis. 2016, 16, 636–642. [Google Scholar] [CrossRef]
- Weis, A.M.; Storey, D.B.; Taff, C.C.; Townsend, A.K.; Huang, B.C.; Kong, N.T.; Clothier, K.A.; Spinner, A.; Byrne, B.A.; Weimer, B.C. Genomic Comparison of Campylobacter spp. and Their Potential for Zoonotic Transmission between Birds, Primates, and Livestock. Appl. Environ. Microbiol. 2016, 82, 7165–7175. [Google Scholar] [CrossRef]
- Weimer, B.C. 100K Pathogen Genome Project. Genome Announc. 2017, 5, e00594-17. [Google Scholar] [CrossRef]
- Hernández-Juárez, L.E.; Camorlinga, M.; Méndez-Tenorio, A.; Calderón, J.F.; Huang, B.C.; Bandoy, D.D.R.; Weimer, B.C.; Torres, J. Analyses of publicly available Hungatella hathewayi genomes revealed genetic distances indicating they belong to more than one species. Virulence 2021, 12, 1950–1964. [Google Scholar] [CrossRef]
- Flores-Valdez, M.; Ares, M.A.; Rosales-Reyes, R.; Torres, J.; Girón, J.A.; Weimer, B.C.; Mendez-Tenorio, A.; De la Cruz, M.A. Whole Genome Sequencing of Pediatric Klebsiella pneumoniae Strains Reveals Important Insights Into Their Virulence-Associated Traits. Front. Microbiol. 2021, 12, 711577. [Google Scholar] [CrossRef]
- Chen, P.; Reiter, T.; Huang, B.; Kong, N.; Weimer, B.C. Prebiotic Oligosaccharides Potentiate Host Protective Responses against L. Monocytogenes Infection. Pathogens 2017, 6, 68. [Google Scholar] [CrossRef]
- Aguilar-Zamora, E.; Weimer, B.C.; Torres, R.C.; Gómez-Delgado, A.; Ortiz-Olvera, N.; Aparicio-Ozores, G.; Barbero-Becerra, V.J.; Torres, J.; Camorlinga-Ponce, M. Molecular Epidemiology and Antimicrobial Resistance of Clostridioides difficile in Hospitalized Patients From Mexico. Front. Microbiol. 2022, 12, 787451. [Google Scholar] [CrossRef]
- Trim Galore. Available online: https://github.com/FelixKrueger/TrimGalore (accessed on 18 March 2022).
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- PATRIC. Pathosystems Resource Integration Center. Available online: https://www.patricbrc.org/ (accessed on 18 May 2022).
- MiGa. Microbial Genome Atlas Online. Available online: http://microbial-genomes.org/ (accessed on 7 October 2022).
- Schwengers, O.; Jelonek, L.; Dieckmann, M.A.; Beyvers, S.; Blom, J.; Goesmann, A. Bakta: Rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb. Genom. 2021, 7, 000685. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Dehio, C. Role of distinct type-IV-secretion systems and secreted effector sets in host adaptation by pathogenic Bartonella species. Cell. Microbiol. 2019, 21, e13004. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Available online: https://github.com/sanger-pathogens/Roary (accessed on 12 November 2022).
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Irber, L. sourmash: A library for MinHash sketching of DNA. J. Open Source Softw. 2016, 1, 27. [Google Scholar] [CrossRef]
- Howe, K.; Bateman, A.; Durbin, R. QuickTree: Building huge Neighbour-Joining trees of protein sequences. Bioinformatics 2002, 18, 1546–1547. [Google Scholar] [CrossRef]
- Roary_plots: Gene Presence and Absence, Frequency Plot and Piechart of Pangenome. Available online: https://github.com/sanger-pathogens/Roary/tree/master/contrib/roary_plots (accessed on 12 November 2022).
- Hadfield, J.; Croucher, N.J.; Goater, R.J.; AbuDahab, K.; Aanensen, D.M.; Harris, S.R. Phandango: An interactive viewer for bacterial population genomics. Bioinformatics 2018, 34, 292–293. [Google Scholar] [CrossRef]
- La Scola, B.; Zeaiter, Z.; Khamis, A.; Raoult, D. Gene-sequence-based criteria for species definition in bacteriology: The Bartonella paradigm. Trends Microbiol. 2003, 11, 318–321. [Google Scholar] [CrossRef]
- Kosoy, M.; Mckee, C.; Albayrak, L.; Fofanov, Y. Genotyping of Bartonella bacteria and their animal hosts: Current status and perspectives. Parasitology 2018, 145, 543–562. [Google Scholar] [CrossRef]
- Rainey, F.A. How to Describe New Species of Prokaryotes. Methods Microbiol. 2011, 38, 7–14. [Google Scholar] [CrossRef]
- Amaral, R.B.D.; Cardozo, M.V.; Varani, A.D.M.; Furquim, M.E.C.; Dias, C.M.; de Assis, W.O.; da Silva, A.R.; Herrera, H.M.; Machado, R.Z.; André, M.R. First Report of Bartonella spp. in Marsupials from Brazil, with a Description of Bartonella harrusi sp. nov. and a New Proposal for the Taxonomic Reclassification of Species of the Genus Bartonella. Microorganisms 2022, 10, 1609. [Google Scholar] [CrossRef]
- Kordick, D.L.; Swaminathan, B.; Greene, C.E.; Wilson, K.H.; Whitney, A.M.; O’Connor, S.; Hollis, D.G.; Matar, G.M.; Steigerwalt, A.G.; Malcolm, G.B.; et al. Bartonella vinsonii subsp. berkhoffii subsp. nov., Isolated from Dogs; Bartonella vinsonii subsp. vinsonii; and Emended Description of Bartonella vinsonii. Int. J. Syst. Evol. Microbiol. 1996, 46, 704–709. [Google Scholar] [CrossRef]
- Buffet, J.-P.; Kosoy, M.; Vayssier-Taussat, M. Natural history of Bartonella-infecting rodents in light of new knowledge on genomics, diversity and evolution. Futur. Microbiol. 2013, 8, 1117–1128. [Google Scholar] [CrossRef]
- Milholland, M.T.; Castro-Arellano, I.; Suzán, G.; Garcia-Peña, G.E.; Lee, T.E.; Rohde, R.E.; Aguirre, A.A.; Mills, J.N. Global Diversity and Distribution of Hantaviruses and Their Hosts. EcoHealth 2018, 15, 163–208. [Google Scholar] [CrossRef]
- Ostfeld, R.S.; Brisson, D.; Oggenfuss, K.; Devine, J.; Levy, M.Z.; Keesing, F. Effects of a zoonotic pathogen, Borrelia burgdorferi, on the behavior of a key reservoir host. Ecol. Evol. 2018, 8, 4074–4083. [Google Scholar] [CrossRef]
- Rubio, A.V.; Ávila-Flores, R.; Osikowicz, L.M.; Bai, Y.; Suzán, G.; Kosoy, M.Y. Prevalence and Genetic Diversity of Bartonella Strains in Rodents from Northwestern Mexico. Vector-Borne Zoonotic Dis. 2014, 14, 838–845. [Google Scholar] [CrossRef]
- Kosoy, M.; Goodrich, I. Comparative Ecology of Bartonella and Brucella Infections in Wild Carnivores. Front. Vet. Sci. 2019, 5, 322. [Google Scholar] [CrossRef]
- Harms, A.; Dehio, C. Intruders below the Radar: Molecular Pathogenesis of Bartonella spp. Clin. Microbiol. Rev. 2012, 25, 42–78. [Google Scholar] [CrossRef]
- Davoust, B.; Marié, J.-L.; Dahmani, M.; Berenger, J.-M.; Bompar, J.-M.; Blanchet, D.; Cheuret, M.; Raoult, D.; Mediannikov, O. Evidence of Bartonella spp. in Blood and Ticks (Ornithodoros hasei) of Bats, in French Guiana. Vector-Borne Zoonotic Dis. 2016, 16, 516–519. [Google Scholar] [CrossRef]
- Moskaluk, A.E.; Stuckey, M.J.; Jaffe, D.A.; Kasten, R.W.; Aguilar-Setién, A.; Olave-Leyva, J.I.; Galvez-Romero, G.; Obregón-Morales, C.; Salas-Rojas, M.; García-Flores, M.M.; et al. Molecular Detection ofBartonellaSpecies in Blood-Feeding Bat Flies from Mexico. Vector-Borne Zoonotic Dis. 2018, 18, 258–265. [Google Scholar] [CrossRef]
- Johnson, N.; Aréchiga-Ceballos, N.; Aguilar-Setién, A. Vampire Bat Rabies: Ecology, Epidemiology and Control. Viruses 2014, 6, 1911–1928. [Google Scholar] [CrossRef] [PubMed]
- Benavides, J.A.; Valderrama, W.; Recuenco, S.; Uieda, W.; Suzán, G.; Avila-Flores, R.; Velasco-Villa, A.; Almeida, M.; De Andrade, F.A.; Molina-Flores, B.; et al. Defining New Pathways to Manage the Ongoing Emergence of Bat Rabies in Latin America. Viruses 2020, 12, 1002. [Google Scholar] [CrossRef]
- Wray, A.K.; Olival, K.J.; Morán, D.; Lopez, M.R.; Alvarez, D.; Navarrete-Macias, I.; Liang, E.; Simmons, N.B.; Lipkin, W.I.; Daszak, P.; et al. Viral Diversity, Prey Preference, and Bartonella Prevalence in Desmodus rotundus in Guatemala. EcoHealth 2016, 13, 761–774. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Kosoy, M.; Recuenco, S.; Alvarez, D.; Moran, D.; Turmelle, A.; Ellison, J.; Garcia, D.L.; Estevez, A.; Lindblade, K.; et al. Bartonella spp. in Bats, Guatemala. Emerg. Infect. Dis. 2011, 17, 1269–1272. [Google Scholar] [CrossRef] [PubMed]
- Becker, D.J.; Bergner, L.; Bentz, A.; Orton, R.J.; Altizer, S.; Streicker, D.G. Genetic diversity, infection prevalence, and possible transmission routes of Bartonella spp. in vampire bats. PLoS Neglected Trop. Dis. 2018, 12, e0006786. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).