Antibiotic Susceptibility and Molecular Typing of Invasive Haemophilus influenzae Isolates, with Emergence of Ciprofloxacin Resistance, 2017–2021, Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain Collection and PCR Capsular Genotyping
2.2. Antibiotic Susceptibility Testing and Characterization of Resistance Genes to β-Lactams
2.3. MLST, WGS and Phylogenetic Analysis
3. Results
3.1. Serotyping
3.2. Antibiotic Susceptibility Testing
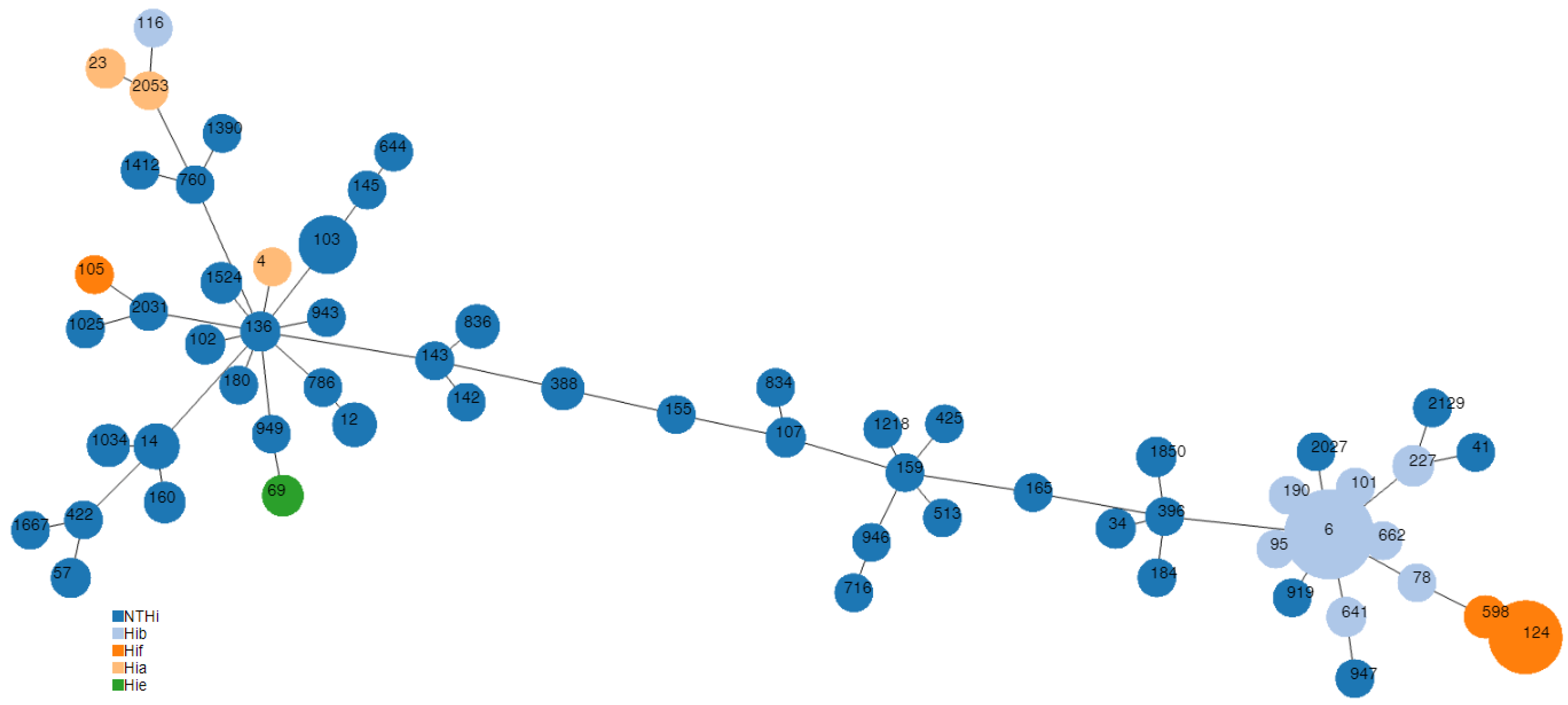
3.3. Molecular Typing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogaert, D.; Keijser, B.; Huse, S.; Rossen, J.; Veenhoven, R.; van Gils, E.; Bruin, J.; Montijn, R.; Bonten, M.; Sanders, E. Variability and diversity of nasopharyngeal microbiota in children: A metagenomics analysis. PLoS ONE 2011, 6, e17035. [Google Scholar] [CrossRef] [PubMed]
- Lemon, K.P.; Klepac-Ceraj, V.; Schiffer, H.K.; Brodie, E.L.; Lynch, S.V.; Kolter, R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. Mbio 2010, 1, e00129-10. [Google Scholar] [CrossRef] [PubMed]
- Turk, D.C. The pathogenicity of Haemophilus influenzae. J. Med. Microbiol. 1984, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Peltola, H. Worldwide Haemophilus influenzae type b disease at the beginning of the 21st century: Global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 2000, 13, 302–317. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control (ECDC). Haemophilus influenzae Annual Epidemiological Report for 2018; European Centre for Disease Prevention and Control (ECDC): Stockholm, Sweden, 2020. [Google Scholar]
- Slack, M.P.E.; Cripps, A.W.; Grimwood, K.; Mackenzie, G.A.; Ulanova, M. Invasive Haemophilus influenzae infections after 3 decades of Hib protein conjugate vaccine use. Clin. Microbiol. Rev. 2021, 34, e0002821. [Google Scholar] [CrossRef]
- Thornsberry, C.; Kirven, L.A. Ampicillin resistance in Haemophilus influenzae as determined by a rapid test for b-lactamase production. Antimicrob. Agents Chemother. 1974, 6, 653–654. [Google Scholar] [CrossRef]
- Tristram, S.; Jacobs, M.R.; Appelbaum, P.C. Antimicrobial resistance in Haemophilus influenzae. Clin. Microbiol. Rev. 2007, 20, 368–389. [Google Scholar] [CrossRef]
- Takeuchi, N.; Ohkusu, M.; Hoshino, T.; Yamamoto, S.; Segawa, S.; Murata, S.; Ishiwada, N. Emergence of Haemophilus influenzae with low susceptibility to quinolones isolated from pediatric patients in Japan. J. Infect. Chemother. 2021, 27, 1020–1026. [Google Scholar] [CrossRef]
- Bakaletz, L.O.; Novotny, L.A. Nontypeable Haemophilus influenzae (NTHi). Trends Microbiol. 2018, 26, 727–728. [Google Scholar] [CrossRef]
- European Commission. Commission Implementing Decision 2012/506/EU Amending Decision 2002/253/EC Laying Down Case Definitions for Reporting Communicable Diseases to the Community Network under Decision No 2119/98/EC of the European Parliament and of the Council. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32012D0506&qid= (accessed on 29 September 2022).
- Falle, T.J.; Crook, D.W.; Brophy, L.N.; Maskell, D.; Kroll, J.S.; Moxon, E.R. PCR for capsular typing of Haemophilus influenzae. J. Clin. Microbiol. 1994, 32, 2382–2386. [Google Scholar] [CrossRef]
- Hobson, R.P.; Williams, A.; Rawal, K.; Pennington, T.H.; Forbes, K.J. Incidence and spread of Haemophilus influenzae on an Antarctic base determined using the polymerase chain reaction. Epidemiol. Infect. 1995, 114, 93–103. [Google Scholar] [CrossRef]
- EUCAST Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 12.0. 2022. Available online: http://www.eucast.org (accessed on 29 September 2022).
- Cerquetti, M.; Cardines, R.; Giufrè, M.; Mastrantonio, P.; Hi Study Group. Antimicrobial susceptibility of Haemophilus influenzae strains isolated from invasive disease in Italy. J. Antimicrob. Chemother. 2004, 54, 1139–1143. [Google Scholar] [CrossRef]
- Cerquetti, M.; Giufrè, M.; Cardines, R.; Mastrantonio, P. First characterization of heterogeneous resistance to imipenem in invasive nontypeable Haemophilus influenzae isolates. Antimicrob. Agents Chemother. 2007, 51, 3155–3161. [Google Scholar] [CrossRef]
- Deghmane, A.E.; Hong, E.; Chehboub, S.; Terrade, A.; Falguières, M.; Sort, M.; Harrison, O.; Jolley, K.A.; Taha, M.K. High diversity of invasive Haemophilus influenzae isolates in France and the emergence of resistance to third generation cephalosporins by alteration of ftsI gene. J. Infect. 2019, 79, 7–14. [Google Scholar] [CrossRef]
- Ubukata, K.; Shibasaki, Y.; Yamamoto, K.; Chiba, N.; Hasegawa, K.; Takeuchi, Y.; Sunakawa, K.; Inoue, M.; Konno, M. Association of amino acid substitutions in penicillin-binding protein 3 with b-lactam resistance in b-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 2001, 45, 1693–1699. [Google Scholar] [CrossRef]
- Dabernat, H.; Delmas, C.; Seguy, M.; Pelissier, R.; Faucon, G.; Bennamani, S.; Pasquier, C. Diversity of b-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 2002, 46, 2208–2218. [Google Scholar] [CrossRef]
- Nürnberg, S.; Claus, H.; Krone, M.; Vogel, U.; Lâm, T.T. Cefotaxime resistance in invasive Haemophilus influenzae isolates in Germany 2016–2019: Prevalence, epidemiology and relevance of PBP3 substitutions. J. Antimicrob. Chemother. 2021, 76, 920–929. [Google Scholar] [CrossRef]
- Pérez-Vázquez, M.; Román, F.; Aracil, B.; Cantón, R.; Campos, J. Laboratory detection of Haemophilus influenzae with decreased susceptibility to nalidixic acid, ciprofloxacin, levofloxacin, and moxifloxacin due to GyrA and ParC mutations. J. Clin. Microbiol. 2004, 42, 1185–1191. [Google Scholar] [CrossRef]
- Ribeiro-Gonçalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carriço, J.A. PHYLOViZ Online: Web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees. Nucleic Acids Res 2016, 44, W246–W251. [Google Scholar] [CrossRef]
- Giufrè, M.; Fabiani, M.; Cardines, R.; Riccardo, F.; Caporali, M.G.; D’Ancona, F.; Pezzotti, P.; Cerquetti, M. Increasing trend in invasive non-typeable Haemophilus influenzae disease and molecular characterization of the isolates, Italy, 2012–2016. Vaccine 2018, 36, 6615–6622. [Google Scholar] [CrossRef]
- Heliodoro, C.I.M.; Bettencourt, C.R.; Bajanca-Lavado, M.P.; Portuguese Group for the Study of Haemophilus influenzae invasive infection. Molecular epidemiology of invasive Haemophilus influenzae disease in Portugal: An update of the post-vaccine period, 2011–2018. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Carrera-Salinas, A.; González-Díaz, A.; Calatayud, L.; Mercado-Maza, J.; Puig, C.; Berbel, D.; Càmara, J.; Tubau, F.; Grau, I.; Domínguez, M.Á.; et al. Epidemiology and population structure of Haemophilus influenzae causing invasive disease. Microb. Genom. 2021, 7, 000723. [Google Scholar] [CrossRef] [PubMed]
- Farrell, D.J.; Morrissey, I.; Bakker, S.; Buckridge, S.; Felmingham, D. Global distribution of TEM-1 and ROB-1 beta-lactamases in Haemophilus influenzae. J. Antimicrob. Chemother. 2005, 56, 773–776. [Google Scholar] [CrossRef] [PubMed]
- San Millan, A.; Garcia-Cobos, S.; Escudero, J.A.; Hidalgo, L.; Gutierrez, B.; Carrilero, L.; Campos, J.; Gonzalez-Zorn, B. Haemophilus influenzae clinical isolates with plasmid pB1000 bearing blaROB-1: Fitness cost and interspecies dissemination. Antimicrob. Agents Chemother. 2010, 54, 1506–1511. [Google Scholar] [CrossRef] [PubMed]
- Potts, C.C.; Rodriguez-Rivera, L.D.; Retchless, A.C.; Buono, S.A.; Chen, A.T.; Marjuki, H.; Blain, A.E.; Wang, X. Antimicrobial Susceptibility Survey of Invasive Haemophilus influenzae in the United States in 2016. Microbiol. Spectr. 2022, 10, e0257921. [Google Scholar] [CrossRef]
- McElligott, M.; Meyler, K.; Bennett, D.; Mulhall, R.; Drew, R.J.; Cunney, R. Epidemiology of Haemophilus influenzae in the Republic of Ireland, 2010–2018. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2335–2344. [Google Scholar] [CrossRef]
- Hegstad, K.; Mylvaganam, H.; Janice, J.; Josefsen, E.; Sivertsen, A.; Skaare, D. Role of horizontal gene transfer in the development of multidrug resistance in Haemophilus influenzae. mSphere 2020, 5, e00969-19. [Google Scholar] [CrossRef]
- Tanaka, T.; Wajima, Y.; Hirai, Y.; Nakaminami, H.; Noguchi, N. Dissemination of quinolone low-susceptible Haemophilus influenzae ST422 in Tokyo, Japan. J. Infect. Chemother. 2021, 27, 962–966. [Google Scholar] [CrossRef]
- Tønnessen, R.; García, I.; Debech, N.; Lindstrøm, J.C.; Wester, A.L.; Skaare, D. Molecular epidemiology and antibiotic resistance profiles of invasive Haemophilus influenzae from Norway 2017–2021. Front. Microbiol. 2022, 13, 973257. [Google Scholar] [CrossRef]
- Andersson, M.; Resman, F.; Eitrem, R.; Drobni, P.; Riesbeck, K.; Kahlmeter, G.; Sundqvist, M. Outbreak of a beta-lactam resistant non-typeable Haemophilus influenzae sequence type 14 associated with severe clinical outcomes. BMC Infect. Dis. 2015, 15, 581. [Google Scholar] [CrossRef]
- Tsang, R.S.W.; Shuel, M.; Whyte, K.; Hoang, L.; Tyrrell, G.; Horsman, G.; Wylie, J.; Jamieson, F.; Lefebvre, B.; Haldane, D.; et al. Antibiotic susceptibility and molecular analysis of invasive Haemophilus influenzae in Canada, 2007 to 2014. J. Antimicrob. Chemother. 2017, 72, 1314–1319. [Google Scholar] [CrossRef]
| MIC (mg/L) | Susceptibility Category | Serotype | ||||||
|---|---|---|---|---|---|---|---|---|
| Antibiotic Agent | MIC50 | MIC90 | Range | Susceptible | Resistant | Capsulated | ||
| No.; % | No.; % | NT-Hi (No. = 305) | Hib (No. = 49) | Non-Hib (No. = 38) | ||||
| Resistant Isolates | ||||||||
| Ampicillin | 0.38 | 16 | 0.064–≥256 | 307; 78.3% | 85; 21.7% | 81; 26.6% | 3; 6.1% | 1; 2.6% |
| Amoxicillin/ clavulanate | 0.50 | 2 | 0.008–12 | 374; 95.4% | 17; 4.3% | 17; 5.6% | 0 | 0 |
| Cefotaxime | 0.023 | 0.064 | 0.006–1.5 | 388; 99.0% | 4; 1.0% | 4; 1.3% | 0 | 0 |
| Ciprofloxacin | 0.008 | 0.016 | 0.002–≥32 | 386; 98.5% | 6; 1.5% | 5; 1.6% | 0 | 1; 2.6% |
| Meropenem | 0.064 | 0.19 | 0.012–0.38 | 392; 100% | 0; 0.0% | 0 | 0 | 0 |
| Isolate | AM | AMC | ST | CC | ftsI Allele | PBP3 Group | E347K | I348V | D350N | S357N | M377I | S385T | L389F | A437S | I449V | G490E | A502V | R517H | N526K | A530S | T532S | V547I | N569S |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BLNAR | |||||||||||||||||||||||
| Hi685 | 2 | 0.5 | 14 | 3 | 209 | I | * | * | * | * | * | * | * | ||||||||||
| Hi687 | 1.5 | 0.75 | 2027 | 137 | m | * | * | * | |||||||||||||||
| Hi728 | 1.5 | 2 | 14 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi729 | 2 | 1.5 | 1034 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi733 | 2 | 8 | 12 | 12 | 2 | IIb | * | * | * | * | * | * | |||||||||||
| Hi736 | 1.5 | 3 | 136 | 3 | 2 | IIb | * | * | * | * | * | * | |||||||||||
| Hi754 | 1.5 | 2 | 1034 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi756 | 1.5 | 1 | 1025 | 1021 | 21 | IIc | T | * | * | * | |||||||||||||
| Hi778 | 1.5 | 2 | 1412 | 37 | IIa | * | |||||||||||||||||
| Hi792 | 2 | 2 | 142 | 142 | 26 | III-like+ | I | * | * | * | * | * | * | * | * | ||||||||
| Hi811 | 3 | 4 | 12 | 12 | 2 | IIb | * | * | * | * | * | * | |||||||||||
| Hi825 | 1.5 | 4 | 12 | 12 | 227 | m | * | * | * | * | * | ||||||||||||
| Hi837 | 2 | 6 | 14 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi879 | 1.5 | 2 | 107 | 107 | 43 | m | * | * | * | * | |||||||||||||
| Hi883 | 1.5 | 1.5 | 1034 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi885 | 1.5 | 2 | 1218 | 107 | 97 | I | * | * | |||||||||||||||
| Hi928 | 1.5 | 2 | 1390 | 2 | IIb | * | * | * | * | * | * | ||||||||||||
| Hi934 | 2 | 3 | 102 | 3 | 2 | IIb | * | * | * | * | * | * | |||||||||||
| Hi937 | 1.5 | 2 | 14 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi940 | 3 | 2 | 41 | 41 | 156 | IIb | * | * | * | ||||||||||||||
| Hi961 | 2 | 3 | 102 | 3 | 32 | III-like | * | * | * | * | * | * | * | * | |||||||||
| Hi974 | 2 | 3 | 145 | 11 | 24 | IIc | * | T | * | * | * | ||||||||||||
| Hi984 | 2 | 1.5 | 14 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi985 | 1.5 | 2 | 396 | 396 | 5 | IId | * | * | * | * | |||||||||||||
| Hi995 | 1.5 | 2 | 425 | 425 | 38 | IIc | * | * | T | * | * | * | |||||||||||
| Hi1016 | 2 | 1.5 | 12 | 12 | 33 | III-like | * | * | * | * | * | * | * | * | |||||||||
| Hi1029 | 2 | 4 | 947 | 24 | IIc | * | T | * | * | * | |||||||||||||
| Hi1030 | 1.5 | 1.5 | 34 | 34 | 71 | m | * | ||||||||||||||||
| Hi1032 | 2 | 4 | 12 | 12 | 42 | IIb | * | * | * | * | * | ||||||||||||
| Hi1035 | 1.5 | 2 | 159 | 107 | 13 | IIc | * | T | * | * | * | ||||||||||||
| Hi1047 | 2 | 1.5 | 1034 | 3 | 1 | IIb | * | * | * | * | * | * | |||||||||||
| Hi1058 | 1.5 | 1.5 | 107 | 107 | 43 | m | * | * | * | * | |||||||||||||
| Hi1061 | 2 | 1.5 | 834 | 390 | 23 | IIb | * | * | * | * | * | * | * | ||||||||||
| Hi1074 | 3 | 3 | 14 | 3 | 1 | IIb | * | * | * | * | |||||||||||||
| Hi1086 | 1.5 | 4 | 949 | 17 | IIb | * | * | * | * | ||||||||||||||
| Hi1108 | 2 | 4 | 919 | 48 | IIb | * | * | ||||||||||||||||
| Hi1109 | 1.5 | 2 | 57 | 57 | 113 | IIc | T | * | |||||||||||||||
| BLPACR | |||||||||||||||||||||||
| Hi854 | 256 | 8 | 836 | 836 | 33 | III-like | * | * | * | * | * | * | |||||||||||
| Hi1031 | 256 | 4 | 422 | 422 | 43 | m | * | * | * | ||||||||||||||
| AMCR | |||||||||||||||||||||||
| Hi691 | 1 | 12 | 2031 | 3 | 32 | III-like | * | * | * | * | * | * | * | * | |||||||||
| Hi744 | 1 | 3 | 136 | 3 | 2 | IIb | * | * | * | * | * | * | |||||||||||
| Isolate | Year | Source | Serotype | ST | CC | MIC (mg/mL) | QRDR Substitutions | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CIP | AM | AMC | CTX | MER | β-lac | GyrA | ParC | ||||||
| Hi805 | 2018 | Blood | NTHi | 1524 | 3 | 32 | 0.25 | 0.3 | 0.016 | 0.023 | − | S84L−D88G | S84I |
| Hi900 | 2018 | Blood | NTHi | 143 | 3 | 32 | 0.25 | 0.2 | 0.016 | 0.094 | − | S84L−D88Y | S84I |
| Hi914 | 2019 | Blood | Hif | 124 | 124 | 0.125 | 0.38 | 0.5 | 0.023 | 0.094 | − | − D88N | − |
| Hi1010 | 2019 | Blood | NTHi | 1524 | 3 | 32 | 0.19 | 0.2 | 0.008 | 0.047 | − | S84L−D88G | S84I |
| Hi1031 | 2020 | CSF | NTHi | 422 | 422 | 0.5 | 256 | 4 | 0.047 | 0.25 | + | S84L− | S84R |
| Hi1062 | 2020 | Blood | NTHi | 1524 | 3 | 32 | 0.19 | 0.3 | 0.016 | 0.047 | − | S84L−D88G | S84I |
| β-Lactamase Producers | BLNAR | ||
|---|---|---|---|
| ST | n | ST | n |
| 6 | 3 | 12 | 5 |
| 34 | 1 | 14 | 6 |
| 57 | 1 | 34 | 1 |
| 69 | 1 | 41 | 1 |
| 103 | 15 | 57 | 1 |
| 155 | 1 | 102 | 2 |
| 160 | 3 | 107 | 2 |
| 165 | 1 | 136 | 1 |
| 180 | 1 | 142 | 1 |
| 184 | 1 | 145 | 1 |
| 388 | 4 | 159 | 1 |
| 422 | 1 | 396 | 1 |
| 513 | 1 | 425 | 1 |
| 644 | 1 | 834 | 1 |
| 716 | 1 | 919 | 1 |
| 760 | 1 | 947 | 1 |
| 786 | 1 | 949 | 1 |
| 836 | 4 | 1025 | 1 |
| 943 | 1 | 1034 | 4 |
| 946 | 1 | 1218 | 1 |
| 1667 | 1 | 1390 | 1 |
| 1850 | 2 | 1412 | 1 |
| 2129 | 1 | 2027 | 1 |
| Total | 48 | 37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giufrè, M.; Cardines, R.; Marra, M.; Carollo, M.; Cerquetti, M.; Stefanelli, P. Antibiotic Susceptibility and Molecular Typing of Invasive Haemophilus influenzae Isolates, with Emergence of Ciprofloxacin Resistance, 2017–2021, Italy. Microorganisms 2023, 11, 315. https://doi.org/10.3390/microorganisms11020315
Giufrè M, Cardines R, Marra M, Carollo M, Cerquetti M, Stefanelli P. Antibiotic Susceptibility and Molecular Typing of Invasive Haemophilus influenzae Isolates, with Emergence of Ciprofloxacin Resistance, 2017–2021, Italy. Microorganisms. 2023; 11(2):315. https://doi.org/10.3390/microorganisms11020315
Chicago/Turabian StyleGiufrè, Maria, Rita Cardines, Manuela Marra, Maria Carollo, Marina Cerquetti, and Paola Stefanelli. 2023. "Antibiotic Susceptibility and Molecular Typing of Invasive Haemophilus influenzae Isolates, with Emergence of Ciprofloxacin Resistance, 2017–2021, Italy" Microorganisms 11, no. 2: 315. https://doi.org/10.3390/microorganisms11020315
APA StyleGiufrè, M., Cardines, R., Marra, M., Carollo, M., Cerquetti, M., & Stefanelli, P. (2023). Antibiotic Susceptibility and Molecular Typing of Invasive Haemophilus influenzae Isolates, with Emergence of Ciprofloxacin Resistance, 2017–2021, Italy. Microorganisms, 11(2), 315. https://doi.org/10.3390/microorganisms11020315







