The Pneumococcal Protein SufC Binds to Host Plasminogen and Promotes Its Conversion into Plasmin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Methods
2.2. Construction of rGdhA and rRny
2.3. Construction of rSufC
2.4. SDS-PAGE and Western Blotting
2.5. ELISA
2.6. Binding Analysis Assay Using Surface Plasmon Resonance
2.7. Plasminogen Activation Analysis
2.8. rSufC Interaction with Plasmin
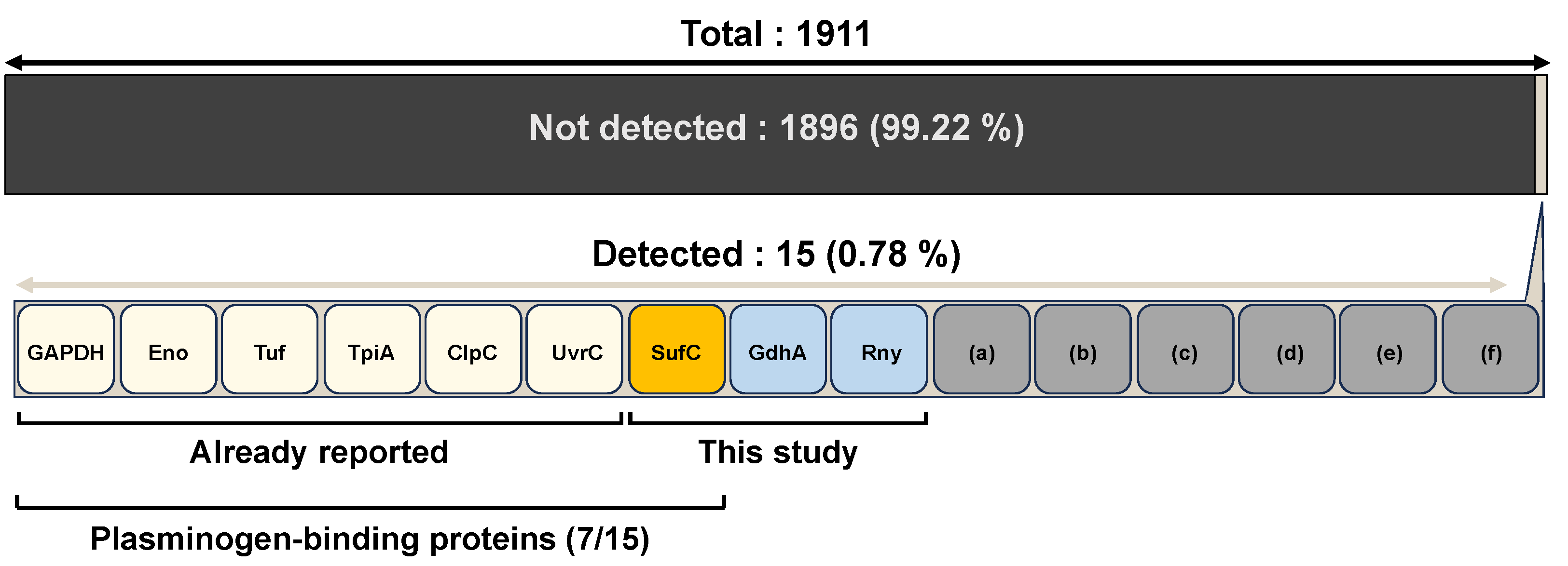
2.9. Analysis of Bacterial Surface Proteins
2.10. Real-Time PCR
2.11. Statistical Analysis
3. Results
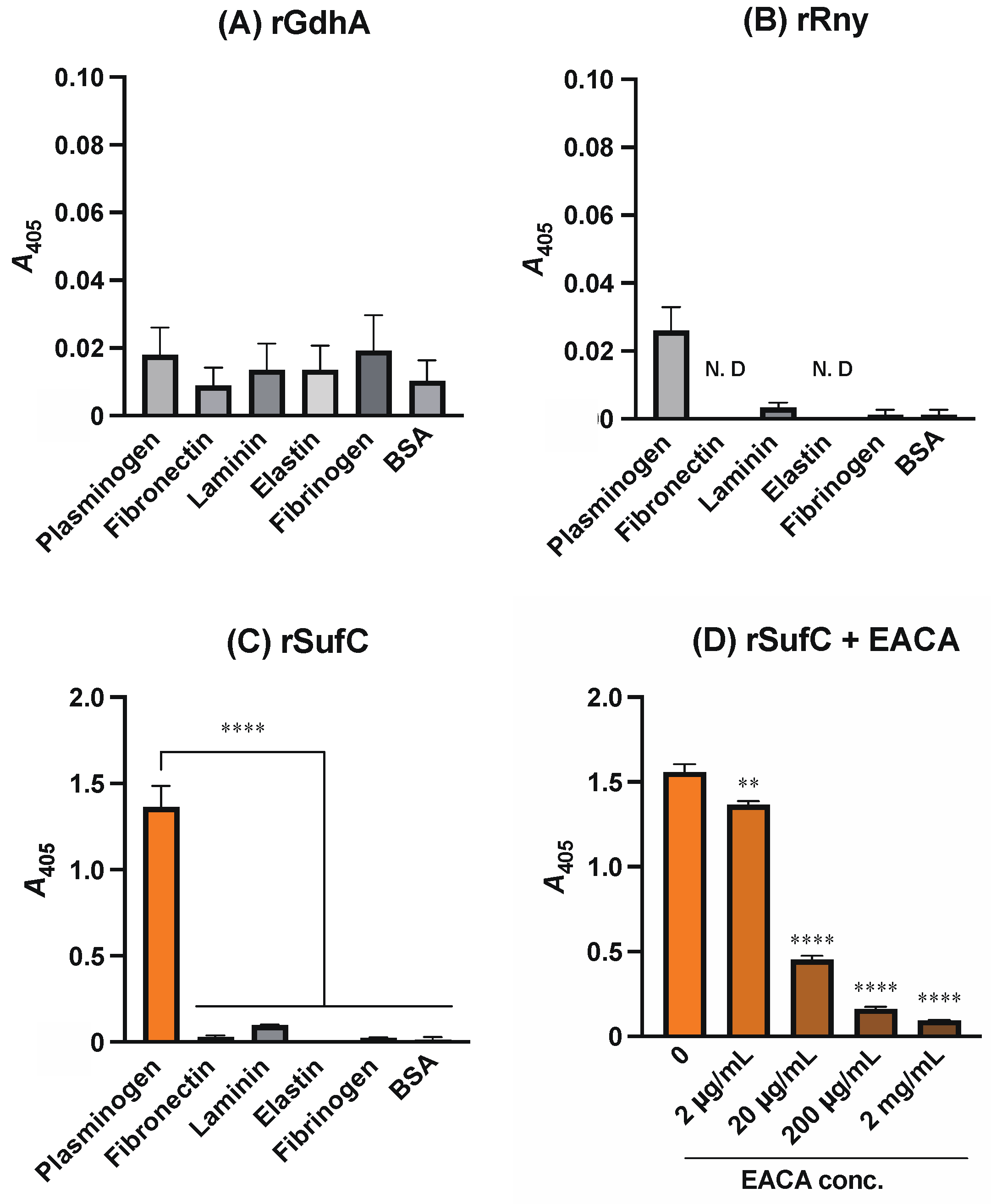
3.1. rSufC Binds to Plasminogen, Which Is Inhibited by Lysine Analogs
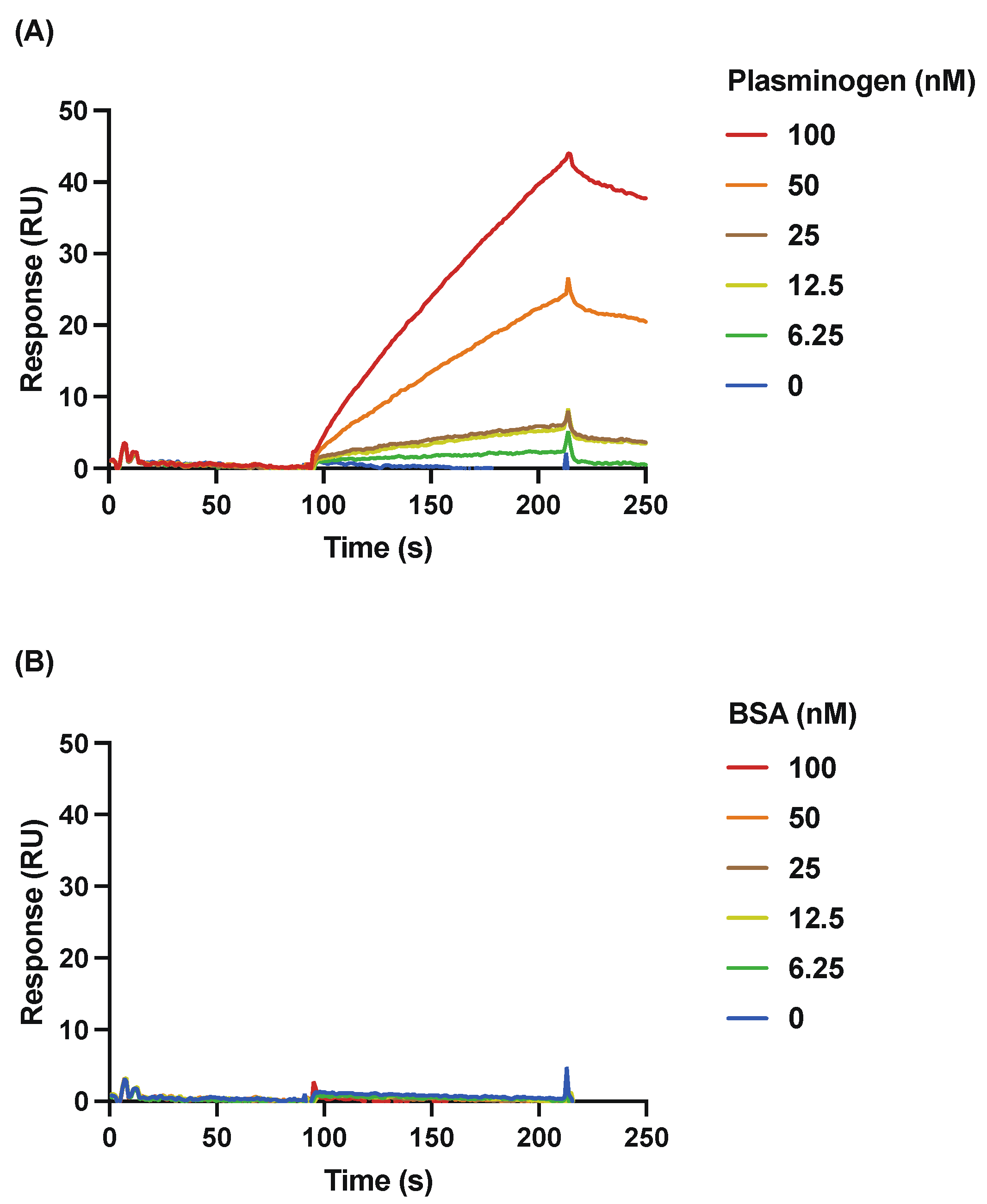
3.2. Detailed Analysis of the Binding of rSufC to Plasminogen Using SPR
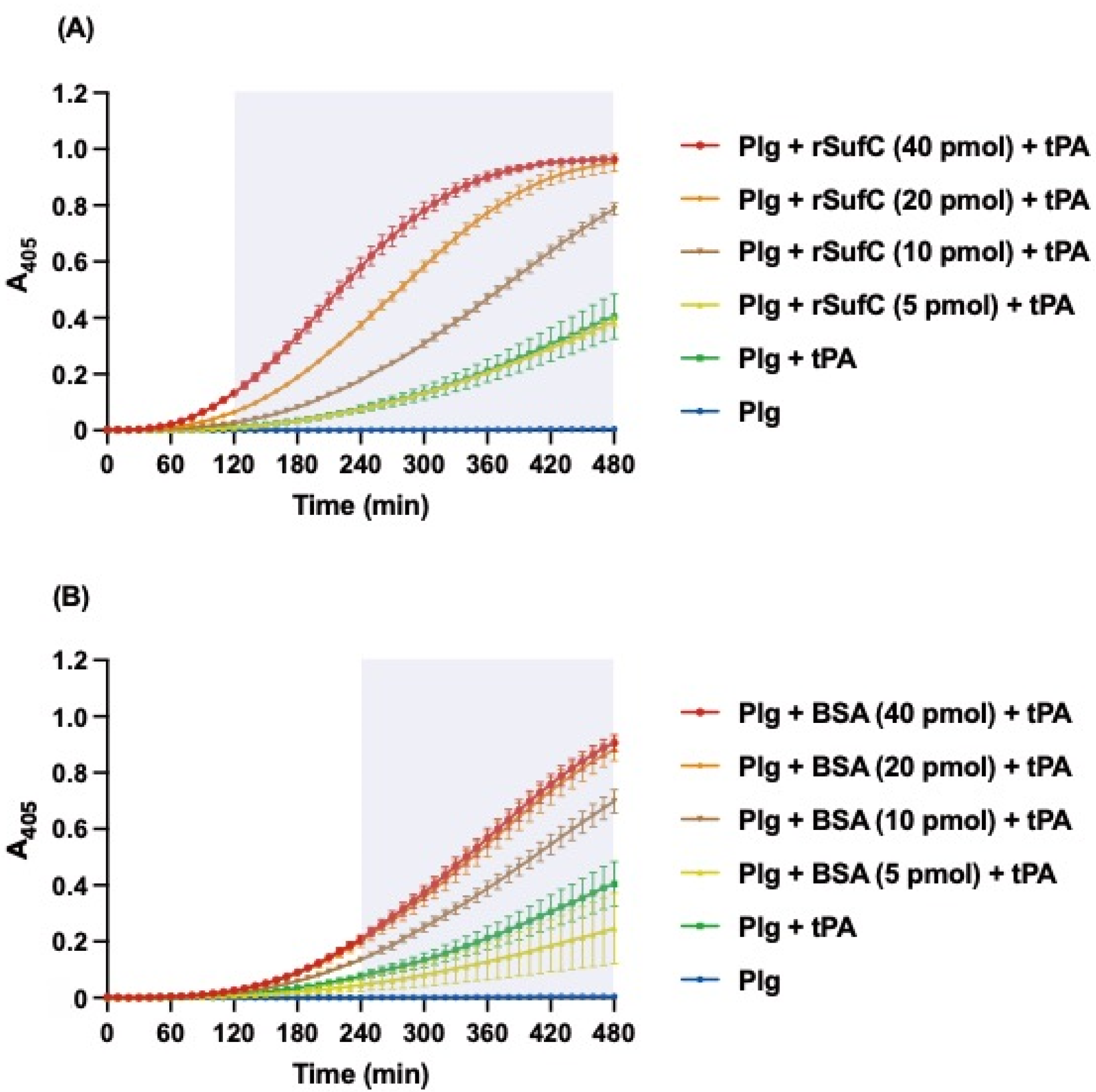
3.3. rSufC Promotes Plasminogen Activation
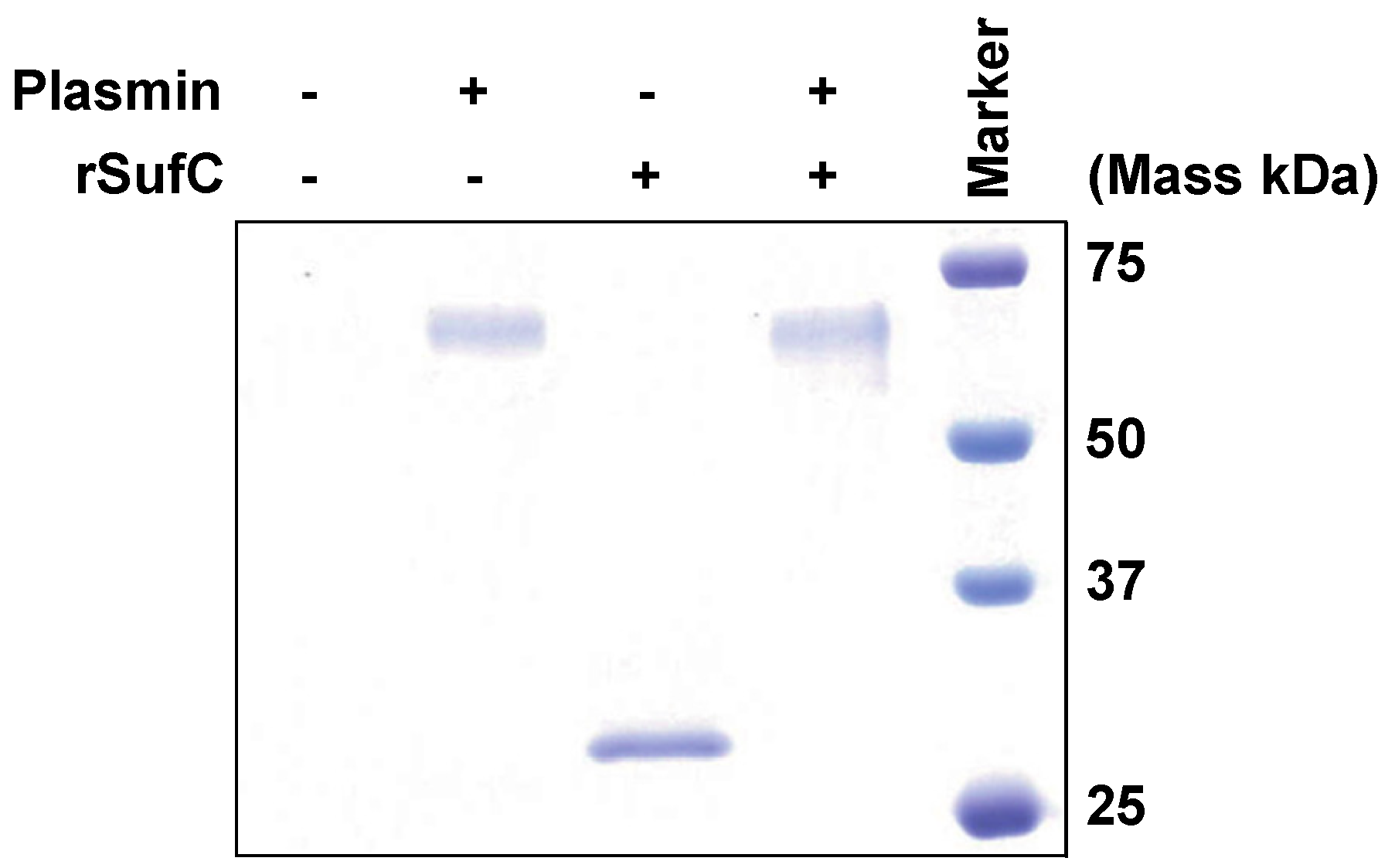
3.4. rSufC Is Degraded by Plasmin
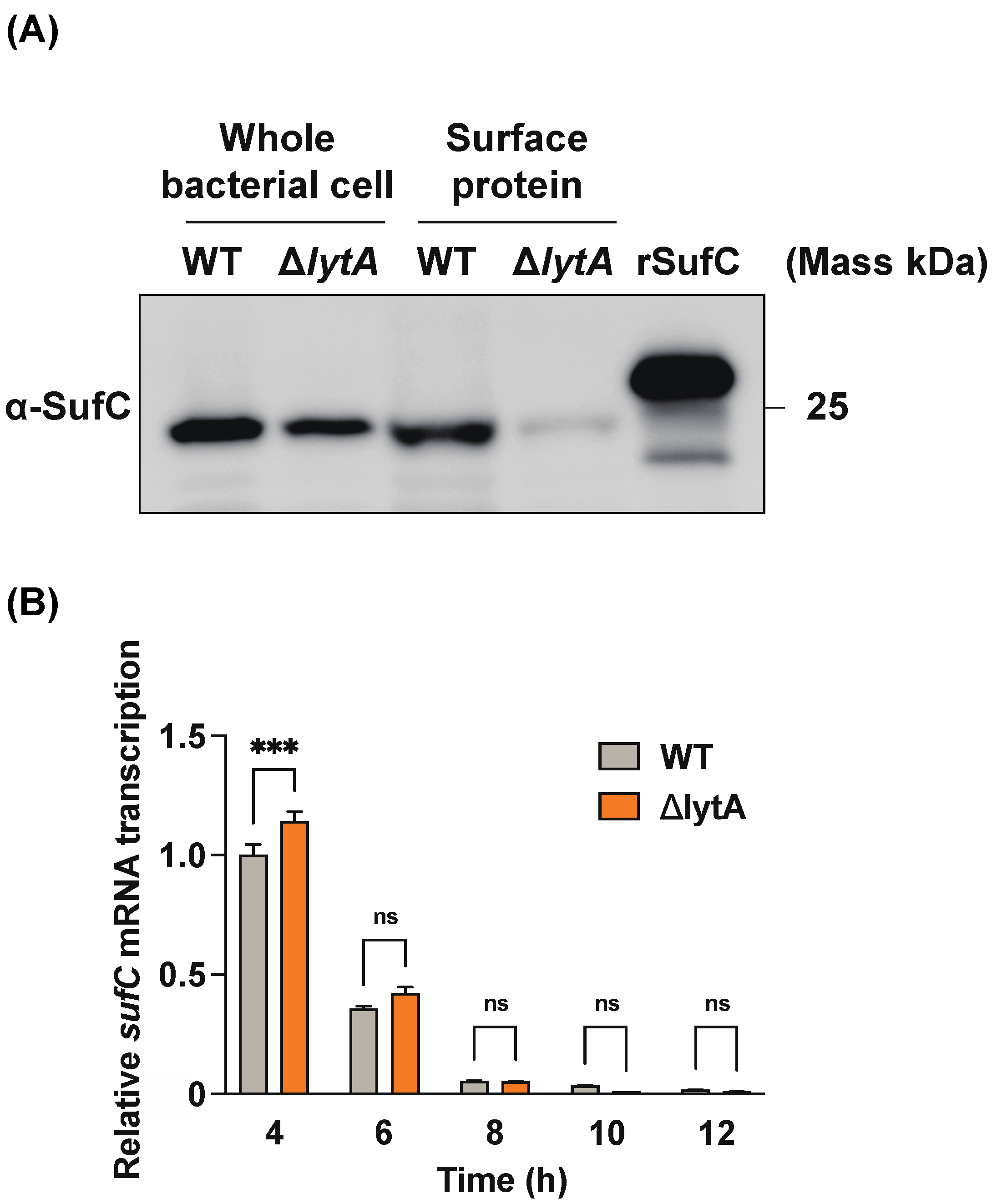
3.5. LytA-Mediated Autolysis Has a Profound Effect on the Extracellular Release of SufC in S. pneumoniae
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duke, J.A.; Avci, F.Y. Emerging vaccine strategies against the incessant pneumococcal disease. NPJ Vaccines 2023, 8, 122. [Google Scholar] [CrossRef]
- Henriques-Normark, B.; Tuomanen, E.I. The pneumococcus: Epidemiology, microbiology, and pathogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a010215. [Google Scholar] [CrossRef]
- See, K.C. Pneumococcal Vaccination in Adults: A Narrative Review of Considerations for Individualized Decision-Making. Vaccines 2023, 11, 908. [Google Scholar] [CrossRef]
- Kaur, R.; Pham, M.; Yu, K.O.; Pichichero, M.E. Rising Pneumococcal Antibiotic Resistance in the Post–13-Valent Pneumococcal Conjugate Vaccine Era in Pediatric Isolates From a Primary Care Setting. Clin. Infect. Dis. 2021, 72, 797–805. [Google Scholar] [CrossRef]
- Nakano, S.; Fujisawa, T.; Ito, Y.; Chang, B.; Matsumura, Y.; Yamamoto, M.; Suga, S.; Ohnishi, M.; Nagao, M. Whole-Genome Sequencing Analysis of Multidrug-Resistant Serotype 15A Streptococcus pneumoniae in Japan and the Emergence of a Highly Resistant Serotype 15A-ST9084 Clone. Antimicrob. Agents Chemother. 2019, 63, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.R.; Stephens, D.S. Macrolide Resistance in Streptococcus pneumoniae. Front. Cell Infect. Microbiol. 2016, 6, 98. [Google Scholar] [CrossRef] [PubMed]
- Lähteenmäki, K.; Kuusela, P.; Korhonen, T.K. Bacterial plasminogen activators and receptors. FEMS Microbiol. Rev. 2001, 25, 531–552. [Google Scholar] [CrossRef] [PubMed]
- Pancholi, V.; Fontan, P.; Jin, H. Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells. Microb. Pathog. 2003, 35, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Sumitomo, T.; Nakata, M.; Higashino, M.; Yamaguchi, M.; Kawabata, S. Erratum: Group A Streptococcus exploits human plasminogen for bacterial translocation across epithelial barrier via tricellular tight junctions. Sci. Rep. 2017, 7, 20069. [Google Scholar] [CrossRef] [PubMed]
- Lentini, G.; Midiri, A.; Firon, A.; Galbo, R.; Mancuso, G.; Biondo, C.; Mazzon, E.; Passantino, A.; Romeo, L.; Trieu-Cuot, P.; et al. The plasminogen binding protein PbsP is required for brain invasion by hypervirulent CC17 Group B streptococci. Sci. Rep. 2018, 8, 14322. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, V.; Andrade, E.B.; Alves, J.; Ribeiro, A.; Kim, K.S.; Lima, M.; Trieu-Cuot, P.; Ferreira, P. Group B Streptococcus hijacks the host plasminogen system to promote brain endothelial cell invasion. PLoS ONE 2013, 8, e63244. [Google Scholar] [CrossRef] [PubMed]
- Lähteenmäki, K.; Edelman, S.; Korhonen, T.K. Bacterial metastasis: The host plasminogen system in bacterial invasion. Trends Microbiol. 2005, 13, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Satala, D.; Bednarek, A.; Kozik, A.; Rapala-Kozik, M.; Karkowska-Kuleta, J. The Recruitment and Activation of Plasminogen by Bacteria—The Involvement in Chronic Infection Development. Int. J. Mol. Sci. 2023, 24, 10436. [Google Scholar] [CrossRef] [PubMed]
- Furuya, H.; Ikeda, R. Interaction of triosephosphate isomerase from Staphylococcus aureus with plasminogen. Microbiol. Immunol. 2011, 55, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, S.; Domon, H.; Hiyoshi, T.; Isono, T.; Tamura, H.; Sasagawa, K.; Takizawa, F.; Terao, Y. Triosephosphate isomerase of Streptococcus pneumoniae is released extracellularly by autolysis and binds to host plasminogen to promote its activation. FEBS Open Bio. 2022, 12, 1206–1219. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Hirayama, S.; Yasui, Y.; Sasagawa, K.; Domon, H.; Terao, Y. Pneumococcal proteins ClpC and UvrC as novel host plasminogen binding factors. Microbiol. Immunol. 2022, 67, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Gazioglu, O.; Kareem, B.O.; Afzal, M.; Shafeeq, S.; Kuipers, O.P.; Ulijasz, A.T.; Andrew, P.W.; Yesilkaya, H. Glutamate Dehydrogenase (GdhA) of Streptococcus pneumoniae Is Required for High Temperature Adaptation. Infect. Immun. 2021, 89, e0040021. [Google Scholar] [CrossRef]
- Sinha, D.; Frick, J.P.; Clemons, K.; Winkler, M.E.; De Lay, N.R. Pivotal Roles for Ribonucleases in Streptococcus pneumoniae Pathogenesis. mBio 2021, 12, e0238521. [Google Scholar] [CrossRef]
- Nachin, L.; Loiseau, L.; Expert, D.; Barras, F. SufC: An unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 2003, 22, 427–437. [Google Scholar] [CrossRef]
- Domon, H.; Isono, T.; Hiyoshi, T.; Tamura, H.; Sasagawa, K.; Maekawa, T.; Hirayama, S.; Yanagihara, K.; Terao, Y. Clarithromycin Inhibits Pneumolysin Production via Downregulation of ply Gene Transcription despite Autolysis Activation. Microbiol. Spectr. 2021, 9, e0031821. [Google Scholar] [CrossRef]
- Hirayama, S.; Nakao, R. Glycine significantly enhances bacterial membrane vesicle production: A powerful approach for isolation of LPS-reduced membrane vesicles of probiotic Escherichia coli. Microb. Biotechnol. 2020, 13, 1162–1178. [Google Scholar] [CrossRef]
- Nagai, K.; Domon, H.; Maekawa, T.; Oda, M.; Hiyoshi, T.; Tamura, H.; Yonezawa, D.; Arai, Y.; Yokoji, M.; Tabeta, K.; et al. Pneumococcal DNA-binding proteins released through autolysis induce the production of proinflammatory cytokines via toll-like receptor 4. Cell Immunol. 2018, 325, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Terao, Y.; Mori, Y.; Yamaguchi, M.; Shimizu, Y.; Ooe, K.; Hamada, S.; Kawabata, S. Group A streptococcal cysteine protease degrades C3 (C3b) and contributes to evasion of innate immunity. J. Biol. Chem. 2008, 283, 6253–6260. [Google Scholar] [CrossRef] [PubMed]
- Murakami, J.; Terao, Y.; Morisaki, I.; Hamada, S.; Kawabata, S. Group A streptococcus adheres to pharyngeal epithelial cells with salivary proline-rich proteins via GrpE chaperone protein. J. Biol. Chem. 2012, 287, 22266–22275. [Google Scholar] [CrossRef] [PubMed]
- Terao, Y.; Kawabata, S.; Kunitomo, E.; Nakagawa, I.; Hamada, S. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect. Immun. 2002, 70, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Terao, Y.; Kawabata, S.; Nakata, M.; Nakagawa, I.; Hamada, S. Molecular characterization of a novel fibronectin-binding protein of Streptococcus pyogenes strains isolated from toxic shock-like syndrome patients. J. Biol. Chem. 2002, 277, 47428–47435. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Rohde, M.; Chhatwal, G.S.; Hammerschmidt, S. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 2001, 40, 1273–1287. [Google Scholar] [CrossRef]
- Whiting, G.; Evans, J.; Patel, S.; Gillespie, S. Purification of native alpha-enolase from Streptococcus pneumoniae that binds plasminogen and is immunogenic. J. Med. Microbiol. 2002, 51, 837–843. [Google Scholar] [CrossRef]
- Bergmann, S.; Wild, D.; Diekmann, O.; Frank, R.; Bracht, D.; Chhatwal, G.S.; Hammerschmidt, S. Identification of a novel plasmin(ogen)-binding motif in surface displayed alpha-enolase of Streptococcus pneumoniae. Mol. Microbiol. 2003, 49, 411–423. [Google Scholar] [CrossRef]
- Bergmann, S.; Rohde, M.; Chhatwal, G.S.; Hammerschmidt, S. Characterization of plasmin(ogen) binding to Streptococcus pneumoniae. Indian J. Med. Res. 2004, 119, 29–32. [Google Scholar]
- Ehinger, S.; Schubert, W.-D.; Bergmann, S.; Hammerschmidt, S.; Heinz, D.W. Plasmin(ogen)-binding alpha-enolase from Streptococcus pneumoniae: Crystal structure and evaluation of plasmin(ogen)-binding sites. J. Mol. Biol. 2004, 343, 997–1005. [Google Scholar] [CrossRef]
- Kolberg, J.; Aase, A.; Bergmann, S.; Herstad, T.K.; Rødal, G.; Frank, R.; Rohde, M.; Hammerschmidt, S. Streptococcus pneumoniae enolase is important for plasminogen binding despite low abundance of enolase protein on the bacterial cell surface. Microbiology 2006, 152, 1307–1317. [Google Scholar] [CrossRef]
- Mori, Y.; Yamaguchi, M.; Terao, Y.; Hamada, S.; Ooshima, T.; Kawabata, S. alpha-Enolase of Streptococcus pneumoniae induces formation of neutrophil extracellular traps. J. Biol. Chem. 2012, 287, 10472–10481. [Google Scholar] [CrossRef]
- Bergmann, S.; Rohde, M.; Hammerschmidt, S. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect. Immun. 2004, 72, 2416–2419. [Google Scholar] [CrossRef]
- Mohan, S.; Hertweck, C.; Dudda, A.; Hammerschmidt, S.; Skerka, C.; Hallström, T.; Zipfel, P.F. Tuf of Streptococcus pneumoniae is a surface displayed human complement regulator binding protein. Mol. Immunol. 2014, 62, 249–264. [Google Scholar] [CrossRef]
- Kitaoka, S.; Wada, K.; Hasegawa, Y.; Minami, Y.; Fukuyama, K.; Takahashi, Y. Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery. FEBS Lett. 2005, 580, 137–143. [Google Scholar] [CrossRef]
- Domon, H.; Oda, M.; Maekawa, T.; Nagai, K.; Takeda, W.; Terao, Y. Streptococcus pneumoniae disrupts pulmonary immune defence via elastase release following pneumolysin-dependent neutrophil lysis. Sci. Rep. 2016, 6, 38013. [Google Scholar] [CrossRef]
- Rouault, T.A. The indispensable role of mammalian iron sulfur proteins in function and regulation of multiple diverse metabolic pathways. BioMetals 2019, 32, 343–353. [Google Scholar] [CrossRef]
- Blahut, M.; Sanchez, E.; Fisher, C.E.; Outten, F.W. Fe-S cluster biogenesis by the bacterial Suf pathway. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118829. [Google Scholar] [CrossRef]
- Przybyla-Toscano, J.; Roland, M.; Gaymard, F.; Couturier, J.; Rouhier, N. Roles and maturation of iron–sulfur proteins in plastids. JBIC J. Biol. Inorg. Chem. 2018, 23, 545–566. [Google Scholar] [CrossRef]
- Tokumoto, U.; Kitamura, S.; Fukuyama, K.; Takahashi, Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 2004, 136, 199–209. [Google Scholar] [CrossRef]
- Takahashi, Y.; Tokumoto, U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 2002, 277, 28380–28383. [Google Scholar] [CrossRef]
- Wang, M.; Qi, L.; Xiao, Y.; Qin, C.; Zhang, H.; Sheng, Y.; Du, H. SufC may promote the survival of Salmonella enterica serovar Typhi in macrophages. Microb. Pathog. 2015, 85, 40–43. [Google Scholar] [CrossRef]
- Attali, C.; Frolet, C.; Durmort, C.; Offant, J.; Vernet, T.; Di Guilmi, A.M. Streptococcus pneumoniae choline-binding protein E interaction with plasminogen/plasmin stimulates migration across the extracellular matrix. Infect. Immun. 2008, 76, 466–476. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Terao, Y.; Mori, Y.; Hamada, S.; Kawabata, S. PfbA, a novel plasmin- and fibronectin-binding protein of Streptococcus pneumoniae, contributes to fibronectin-dependent adhesion and antiphagocytosis. J. Biol. Chem. 2008, 283, 36272–36279. [Google Scholar] [CrossRef]
- Papasergi, S.; Garibaldi, M.; Tuscano, G.; Signorino, G.; Ricci, S.; Peppoloni, S.; Pernice, I.; Passo, C.L.; Teti, G.; Felici, F.; et al. Plasminogen- and fibronectin-binding protein B is involved in the adherence of streptococcus pneumoniae to human epithelial cells. J. Biol. Chem. 2010, 285, 7517–7524. [Google Scholar] [CrossRef]
- Agarwal, V.; Kuchipudi, A.; Fulde, M.; Riesbeck, K.; Bergmann, S.; Blom, A.M. Streptococcus pneumoniae endopeptidase O (PepO) is a multifunctional plasminogen- and fibronectin-binding protein, facilitating evasion of innate immunity and invasion of host cells. J. Biol. Chem. 2013, 288, 6849–6863. [Google Scholar] [CrossRef]
- Fulde, M.; Bernardo-García, N.; Rohde, M.; Nachtigall, N.; Frank, R.; Preissner, K.T.; Klett, J.; Morreale, A.; Chhatwal, G.S.; Hermoso, J.A.; et al. Pneumococcal phosphoglycerate kinase interacts with plasminogen and its tissue activator. Thromb. Haemost. 2014, 112, 401–416. [Google Scholar] [CrossRef]
- Meinel, C.; Spartà, G.; Dahse, H.-M.; Hörhold, F.; König, R.; Westermann, M.; Coldewey, S.M.; Cseresnyés, Z.; Figge, M.T.; Hammerschmidt, S.; et al. Streptococcus pneumoniae From Patients With Hemolytic Uremic Syndrome Binds Human Plasminogen via the Surface Protein PspC and Uses Plasmin to Damage Human Endothelial Cells. J. Infect. Dis. 2017, 217, 358–370. [Google Scholar] [CrossRef]
- Hirayama, S.; Hiyoshi, T.; Yasui, Y.; Domon, H.; Terao, Y. C-Terminal Lysine Residue of Pneumococcal Triosephosphate Isomerase Contributes to Its Binding to Host Plasminogen. Microorganisms 2023, 11, 1198. [Google Scholar] [CrossRef]
- Cesarman-Maus, G.; Hajjar, K.A. Molecular mechanisms of fibrinolysis. Br. J. Haematol. 2005, 129, 307–321. [Google Scholar] [CrossRef]
- Keragala, C.B.; Medcalf, R.L. Plasminogen: An enigmatic zymogen. Blood 2021, 137, 2881–2889. [Google Scholar] [CrossRef]
- Lahteenmaki, K.; Virkola, R.; Sarén, A.; Emody, L.; Korhonen, T.K. Expression of plasminogen activator pla of Yersinia pestis enhances bacterial attachment to the mammalian extracellular matrix. Infect. Immun. 1998, 66, 5755–5762. [Google Scholar] [CrossRef]
- Attali, C.; Durmort, C.; Vernet, T.; Di Guilmi, A.M. The interaction of Streptococcus pneumoniae with plasmin mediates transmigration across endothelial and epithelial monolayers by intercellular junction cleavage. Infect. Immun. 2008, 76, 5350–5356. [Google Scholar] [CrossRef]
- Rooijakkers, S.; van Wamel, W.; Ruyken, M.; van Kessel, K.; van Strijp, J. Anti-opsonic properties of staphylokinase. Microbes Infect. 2005, 7, 476–484. [Google Scholar] [CrossRef]
- Nitzsche, R.; Köhler, J.; Kreikemeyer, B.; Oehmcke-Hecht, S. Streptococcus pyogenes Escapes Killing from Extracellular Histones through Plasminogen Binding and Activation by Streptokinase. J. Innate Immun. 2016, 8, 589–600. [Google Scholar] [CrossRef]
| Name | Sequence (5′-3′) | Reference |
|---|---|---|
| gdhA_BamHI_F | CTACGGATCCACATCTGCTAAAGAATATATCCAAAGC | This study |
| gdhA_KpnI_R | CTCGGGTACCTTAAACAATACCTTGTGCAATCATAG | This study |
| rny_BamHI_F | TCTAGGATCCGAAATCATGTCGCTTGCG | This study |
| rny_KpnI_R | TCTAGGTACCTTATTTAGCATAATCTACTGCACGAAG | This study |
| pQE30-ins-F | CGCGATATCAGGATTCGG | [17] |
| pQE30-ins-R | CTGAACAAATCCAGATGGAG | [17] |
| pBIC_sufC_F w/o ATG | TCAGTATTAGAGATCAAAGATCTTCACG | This study |
| pBIC_sufC_R | GATACGAGGGAATTACAATTCTTCC | This study |
| pBIC-ins-F | CGCGATATCAGGATTCGG | [15] |
| pBIC-ins-R | CAATGTAATTGTTCCCTACCTGC | [15] |
| sufC_qPCR_F | TCATCACTCACTACCAACGTCTTTT | This study |
| sufC_qPCR_R | GTTCTTCAGCTAATTTTGCGTATCCT | This study |
| sufC_qPCR_probe | (FAM) GGTCGTGTTGTCCTTTCTGGT (TAMRA) | This study |
| 16S_qPCR_F | TCCTACGGGAGGCAGCAGT | [17] |
| 16S_qPCR_R | GGACTACCAGGGTATCTAATCCTGTT | [17] |
| 16S_qPCR_probe | (FAM) CGTATTACCGCGGCTGCTGGCAC (TAMRA) | [17] |
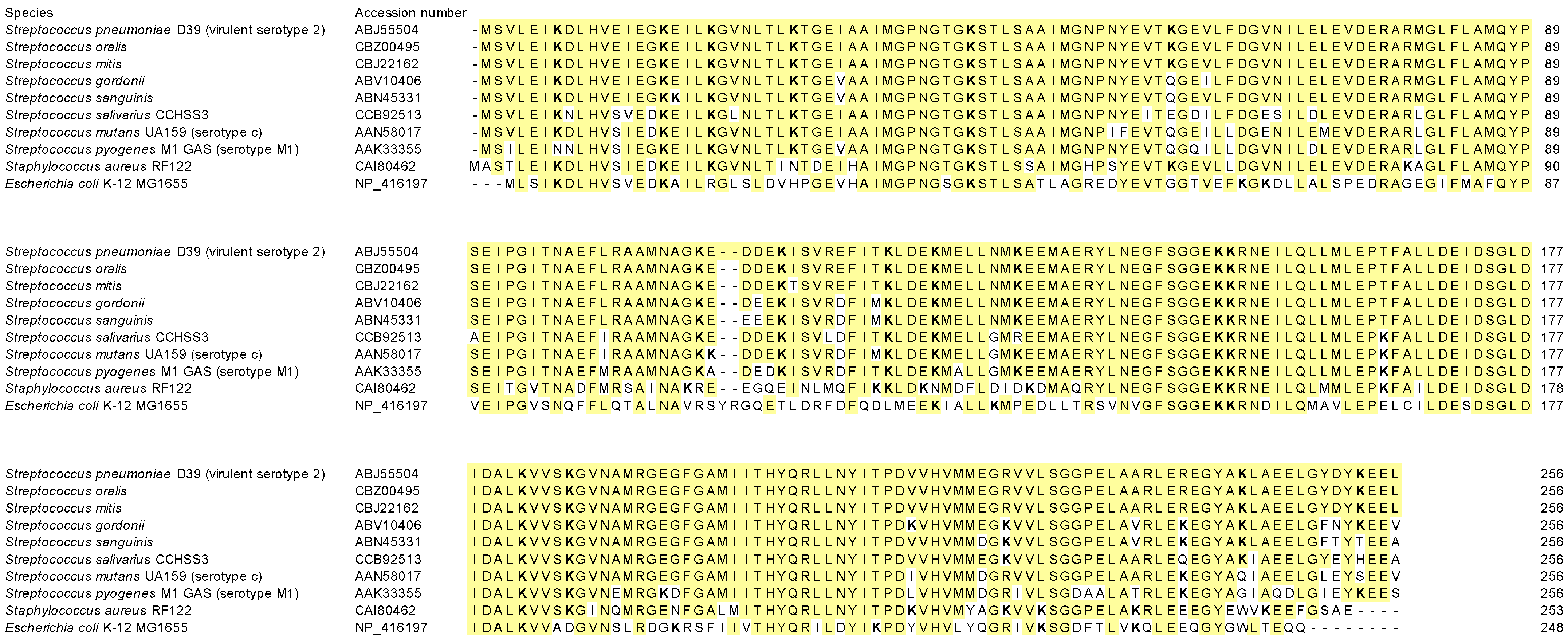
| Species | Accession Number | Identity (%) | Positives (%) | Gap (%) |
|---|---|---|---|---|
| Streptococcus oralis | CBZ00495 | 100 | 100.0 | 0.0 |
| Streptococcus mitis | CBJ22162 | 99.6 | 99.6 | 0.0 |
| Streptococcus gordonii | ABV10406 | 94.9 | 98.8 | 0.0 |
| Streptococcus sanguinis | ABN45331 | 94.5 | 98.4 | 0.0 |
| Streptococcus salivarius CCHSS3 | CCB92513 | 90.2 | 97.3 | 0.0 |
| Streptococcus mutans UA159 (serotype c) | AAN58017 | 90.2 | 96.1 | 0.0 |
| Streptococcus pyogenes M1 GAS (serotype M1) | AAK33355 | 85.9 | 94.1 | 0.0 |
| Staphylococcus aureus RF122 | CAI80462 | 75.8 | 88.3 | 0.0 |
| Escherichia coli K-12 MG1655 | NP_416197 | 52.2 | 70.9 | 0.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yasui, Y.; Hirayama, S.; Hiyoshi, T.; Isono, T.; Domon, H.; Maekawa, T.; Tabeta, K.; Terao, Y. The Pneumococcal Protein SufC Binds to Host Plasminogen and Promotes Its Conversion into Plasmin. Microorganisms 2023, 11, 2969. https://doi.org/10.3390/microorganisms11122969
Yasui Y, Hirayama S, Hiyoshi T, Isono T, Domon H, Maekawa T, Tabeta K, Terao Y. The Pneumococcal Protein SufC Binds to Host Plasminogen and Promotes Its Conversion into Plasmin. Microorganisms. 2023; 11(12):2969. https://doi.org/10.3390/microorganisms11122969
Chicago/Turabian StyleYasui, Yoshihito, Satoru Hirayama, Takumi Hiyoshi, Toshihito Isono, Hisanori Domon, Tomoki Maekawa, Koichi Tabeta, and Yutaka Terao. 2023. "The Pneumococcal Protein SufC Binds to Host Plasminogen and Promotes Its Conversion into Plasmin" Microorganisms 11, no. 12: 2969. https://doi.org/10.3390/microorganisms11122969
APA StyleYasui, Y., Hirayama, S., Hiyoshi, T., Isono, T., Domon, H., Maekawa, T., Tabeta, K., & Terao, Y. (2023). The Pneumococcal Protein SufC Binds to Host Plasminogen and Promotes Its Conversion into Plasmin. Microorganisms, 11(12), 2969. https://doi.org/10.3390/microorganisms11122969






